Significance
Super enhancers (SEs) are enhancer-dense regions found near genes that play key roles in determining cellular identity. Using global nuclear run-on sequencing (GRO-Seq), we find extensive regulation of enhancer RNAs (eRNAs) within SEs in response to lipopolysaccharide (LPS) treatment in macrophages. Both activation and repression of gene expression are associated with SEs and eRNA transcription dynamics. Furthermore, we find that each SE acts as a single regulatory unit within which eRNA and genic transcripts are coordinately regulated. We also find that transcription factor (TF) composition within an SE determines regulatory properties of each SE and associated eRNAs. We propose that signal-dependent SEs and their eRNAs function as molecular rheostats integrating the binding profiles of key regulators to produce dynamic profiles of gene expression.
Keywords: super enhancers, enhancer RNAs, transcription factors, GRO-Seq, macrophage
Abstract
Enhancers are critical genomic elements that define cellular and functional identity through the spatial and temporal regulation of gene expression. Recent studies suggest that key genes regulating cell type-specific functions reside in enhancer-dense genomic regions (i.e., super enhancers, stretch enhancers). Here we report that enhancer RNAs (eRNAs) identified by global nuclear run-on sequencing are extensively transcribed within super enhancers and are dynamically regulated in response to cellular signaling. Using Toll-like receptor 4 (TLR4) signaling in macrophages as a model system, we find that transcription of super enhancer-associated eRNAs is dynamically induced at most of the key genes driving innate immunity and inflammation. Unexpectedly, genes repressed by TLR4 signaling are also associated with super enhancer domains and accompanied by massive repression of eRNA transcription. Furthermore, we find each super enhancer acts as a single regulatory unit within which eRNA and genic transcripts are coordinately regulated. The key regulatory activity of these domains is further supported by the finding that super enhancer-associated transcription factor binding is twice as likely to be conserved between human and mouse than typical enhancer sites. Our study suggests that transcriptional activities at super enhancers are critical components to understand the dynamic gene regulatory network.
Enhancers are cis-acting regulatory elements located distantly from transcription start sites (TSSs) that are regulated by the binding of sequence-specific transcription factors (TFs) (1, 2). In addition to core promoters that recruit general transcriptional machinery to modulate gene expression, enhancers play a critical role in transcriptional regulation of tissue- and cell type-specific gene expression (2–4). Recent studies exploring how enhancers exert their functions in gene regulation have revealed a variety of enhancer properties including chromatin looping between enhancers and target gene promoters, enhancer-specific histone modification such as histone H3 mono- and di-methyl lysine 4 and H3 lysine 27 acetylation modifications, histone variants, coactivator binding, and an open chromatin architecture (3, 5–11). More recent studies have indicated that enhancers are the key elements in cell-type specificity and have shown that there are regions where enhancers are clustered together near key genes that are involved in determining cell identity, called “super enhancers” (SEs) or “stretch enhancers” (12–15). It has been shown that these enhancer-dense regions are distinct from the typical enhancers (TEs) in their ability to activate cell-type and tissue-specific genes and result in a higher susceptibility for disease when mutated (12, 13, 15, 16). Interestingly, previous genomic studies have shown that active enhancers overlap with RNA Pol II loading, which generates active bidirectional transcripts called enhancer RNAs (eRNAs) (10, 17–19). Although the role of enhancer transcription remains unknown, it is considered a hallmark of functionally active enhancers (20–24).
Previous studies of eRNAs that are associated with the binding of p53 and nuclear receptors such as estrogen receptor (ER), androgen receptor (AR), and Rev-erbs have demonstrated eRNA knockdown can lead to parallel changes in gene expression (20, 23, 25–27). These studies suggest that active enhancers generate at least some eRNAs that are required for transcriptional activation. Other studies suggest that eRNAs may promote enhancer–promoter looping or recruit TFs to specific enhancers (5, 8, 21, 23, 28). However, most studies focused mainly on the functions of enhancers and eRNAs in transcriptional activation, whereas signal-dependent transcriptional repression has not been directly addressed in the context of enhancer function. Recent studies have demonstrated that signaling through nuclear receptors such as ER and peroxisome proliferator-activated receptor can cause release of coactivators at enhancers, resulting in repression of gene expression (14, 29, 30). However, the contribution of eRNA repression, if any, to this process remains unknown.
In macrophages, inflammatory signaling triggers immediate and dramatic changes at the level of gene transcription in response to external stimuli (3, 31, 32). Although the contribution of lineage-determining factors such as ccaat-enhancer–binding proteins (C/EBP) and PU.1 in macrophage identity has been studied, macrophage SEs as targets of signal-dependent control have just begun to be explored. A recent study has demonstrated the importance of NF-κB–dependent redistribution of bromodomain containing 4 (BRD4) within SEs to exert rapid inflammatory transcriptional responses (33). Although the mechanistic details of the phenomena remain to be explored, the study provides insight into how SEs play critical roles in inflammatory responses. It has also been strongly implicated that eRNAs and enhancer functions are involved in the regulation of inflammatory transcription networks (22, 34–37); however, it remains to be solved how, and if at all, SE-associated eRNAs (seRNAs) contribute to the regulatory landscape. Using global nuclear run-on sequencing (GRO-Seq) to map the location and orientation of all active RNA polymerases genome-wide (38), we found eRNAs extensively transcribed within the macrophage SE subset. We found that SEs, although only 3% of the enhancer network, are strongly enriched near genes that are either induced or repressed in response to TLR4 signaling, suggesting that they may play a key role in both signal response and developmental transcriptional programs. The key regulatory activity of these domains is further supported by the finding that SE-associated TF binding is twice as likely to be conserved between human and mouse than TE sites. Notably, we found that each SE acts as a single regulatory unit within which transcription of eRNAs and their genic transcripts are coordinately regulated. Induction or repression of transcription within each SE appears to be largely dictated by the cumulative binding of specific TFs. Collectively, these results suggest that each SE and its associated eRNAs function as a type of molecular rheostat with a high dynamic range to control gene expression.
Results and Discussion
Detecting Highly Active Transcription at SEs.
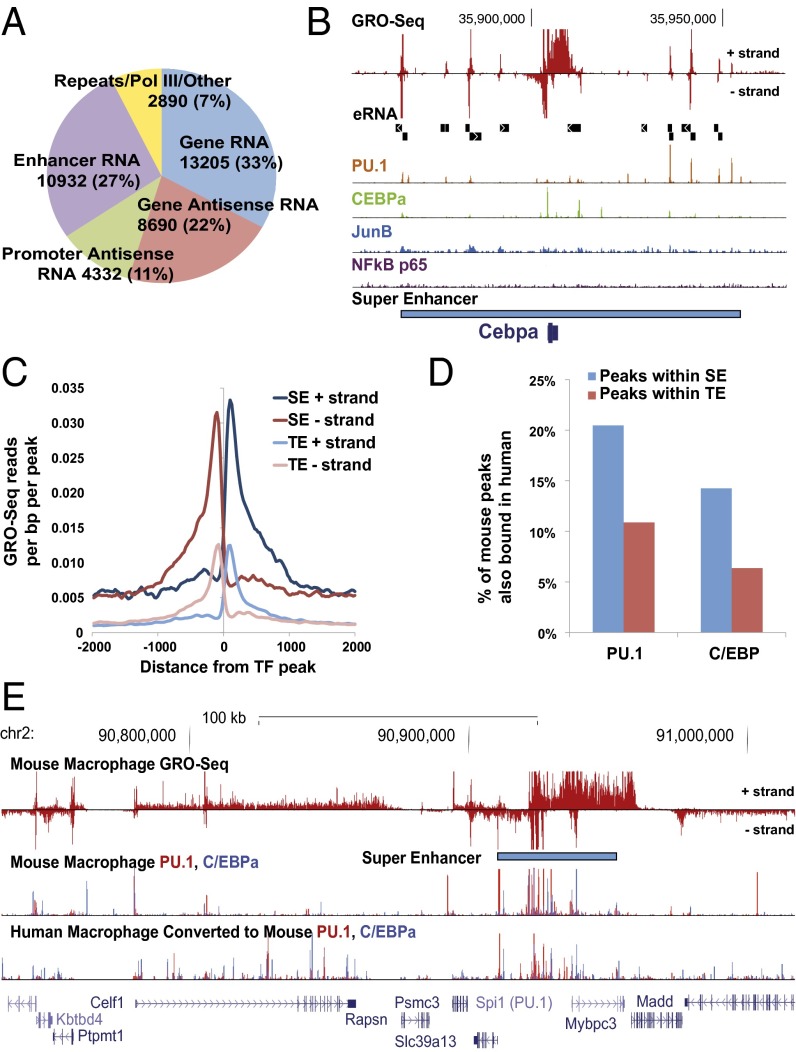
To investigate the transcriptional response of macrophages to TLR4 signaling, we generated nascent RNA transcriptomes using GRO-Seq from mouse primary macrophages in response to lipopolysaccharide (LPS) at 0, 20, 60, and 180 min. We defined de novo transcription units to characterize the macrophage response in an unbiased manner. The transcripts were classified into different groups based on their location relative to known gene annotations (Fig. 1A). Nearly 27% of transcripts were classified as eRNAs, found in intergenic regions, closely associated with TFs and epigenetic modifications. We observed that eRNAs were typically clustered at key genes within enhancer-dense regions, resembling sites that were recently described as SEs (12–14). To explore the relationship between eRNAs and SEs more closely, we analyzed ChIP-Seq data for key macrophage TFs, PU.1, C/EBPA, JUNB, and NF-κB p65 (RELA), and defined SEs based on the binding of these key factors (Dataset S1). To ensure that the SEs that are identified based on the key TFs are comparable to those that are defined based on histone modifications in macrophage, we first used H3K27Ac data to find SEs in both vehicle- and LPS-treated conditions (34). Two independently identified SEs showed statistically significant overlaps in both vehicle and LPS treatment (Fig. S1A). To further validate our SE discovery, we compared SEs reported in Brown et al. (33) to SEs identified using Hypergeometric Optimization of Motif Enrichment (HOMER) with the same mouse macrophage H4K12ac ChIP-Seq dataset (33). Over 60% of the SEs are identical between methods, with most of those that differed found just shy of the SE threshold set by the other method (Fig. S1 B and C). We independently assessed the functional quality of differential SE calls by measuring the eRNA production from intergenic TF binding sites (TFBSs) that were specific to Brown et al. or HOMER SEs, and find their transcriptional activity to be similar and quite distinct from TE levels (Fig. S1D). As exemplified by the Cebpa locus, SEs are characterized by the presence of high transcriptional activity and multiple eRNAs (Fig. 1B). We found that the number of eRNAs in the vicinity of a gene was predictive of the presence of a nearby SE, with over 60% of genes with seven or more adjacent eRNAs also containing an SE (Fig. S2 and Table S1). We refer to these SE-associated eRNAs as seRNAs herein. Interestingly, when we compared the fraction of eRNAs that overlaps with TEs or SEs, relatively few typical intergenic enhancers overlap with an eRNA (30.6%), whereas nearly all SEs contain eRNAs (93.3%) within intergenic regions. This implies that the frequency and presence of eRNAs can be used to mark SEs and these SEs are likely to be an assembly of very active and functional enhancers. Furthermore, the transcriptional activity at individual intergenic TFBSs was much higher in SEs than at sites in TEs, with about a 3.3-fold increase in the area under the curve (Fig. 1C). The fact that SEs tend to cluster at lineage-defining gene loci (12, 13) suggests that regulatory elements in these regions may be conserved between species. To address this hypothesis, we compared binding of PU.1 and C/EBPA, using ChIP-Seq data from human and mouse macrophages. Interestingly, mouse PU.1 and C/EBPA binding peaks that are located in SEs are more likely to bind conserved regions in human homologs compared with the peaks at TEs (∼twofold enrichment) (Fig. 1D). As an example, PU.1- and C/EBP-bound peaks near the Spi (PU.1) gene locus showed more conserved binding at the defined SE regions between human and mouse compared to other binding sites (Fig. 1E), indicating the possibility that this conservation of SEs is linked to their conserved roles in gene regulation in different species.
Fig. 1.
SEs are highly transcriptionally active and conserved. (A) De novo transcription units are defined in an unbiased manner based on GRO-Seq reads. All of the transcripts are further classified into different classes based on their location relative to known gene annotations that are publically available. (B) Browser representations of GRO-Seq data, eRNA transcript unit (black box), ChIP-Seq data of key TFs (PU.1, C/EBPA, NF-κB p65, and JUNB), and SE unit (blue box) at Cebpa locus. (C) Analyses of average GRO-Seq reads per bp per ChIP-Seq peak within either SEs or TEs. (D) Comparison of TF (PU.1 and C/EBPA) binding conservation at SEs or TEs between human and mouse. (E) Genome browser view of Spi1 (PU.1) gene loci where PU.1 and C/EBPA binding are conserved between human and mouse.
Regulation of SEs and seRNAs upon Activation of TLR4.
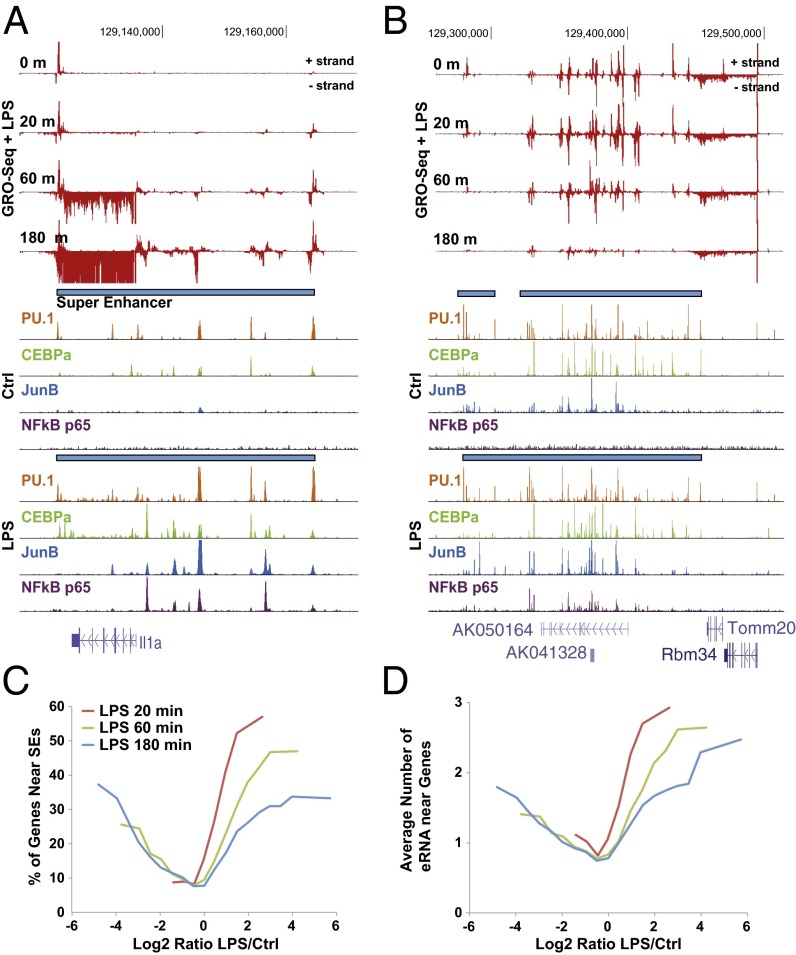
Previous genome-wide studies have demonstrated that SEs are closely correlated with key developmental genes (12, 13, 39). To explore SEs and their associated genes as targets of ligand signaling, we examined the regulation of seRNAs following LPS activation of the TLR4 pathway. This revealed that genes activated by LPS were often found in close proximity to SEs and seRNAs (Fig. 2A). Unexpectedly, genes repressed by LPS were also found in the vicinity of SEs and seRNAs (Fig. 2B), indicating that functional roles of SEs are correlated not only to activation of nearby genes but also with gene repression. Additionally, we have investigated Pol II levels at actively up- and down-regulated SEs that are associated with gene expression. In aggregate, GRO-Seq provides a higher sensitivity readout for active Pol II than Pol II ChIP-Seq at enhancer regions, although the results are highly comparable (Fig. S3). We also compared the regulation of seRNAs with previous GRO-Seq data generated in the presence of Kdo2-LipidA (KLA), the active moiety of LPS (34). Both sets of results showed highly similar regulation of seRNAs as expected (Fig. S4). In analyzing the physical association between SEs and gene transcription, we found that genes undergoing greater fold regulation were highly correlated to SE proximity (Fig. 2C). For instance, upon 180 min of LPS stimulation, ∼35% of genes near SEs exhibited ∼16-fold up- or down-regulation. This suggests that SEs similarly impact not only TLR4-elicited activation but also repression. Additionally, genes with high fold changes show parallel changes in nearby eRNA expression (Fig. 2D), demonstrating the close association between the proximity of SEs, seRNAs, and gene control.
Fig. 2.
Regulation of SEs and seRNAs upon LPS stimulation. Genome browser representations of SE loci that are (A) up-regulated and (B) down-regulated upon activation of TLR4 signaling by LPS. (C) Relationship between gene regulation (log2 ratio, x axis) and presence of SEs (±50 kb from the TSS), presented as a moving average. (D) Relationship between gene regulation (log2 ratio, x axis) and the number of eRNAs within ±50 kb from the TSS, presented as a moving average.
Signal-Dependent SEs.
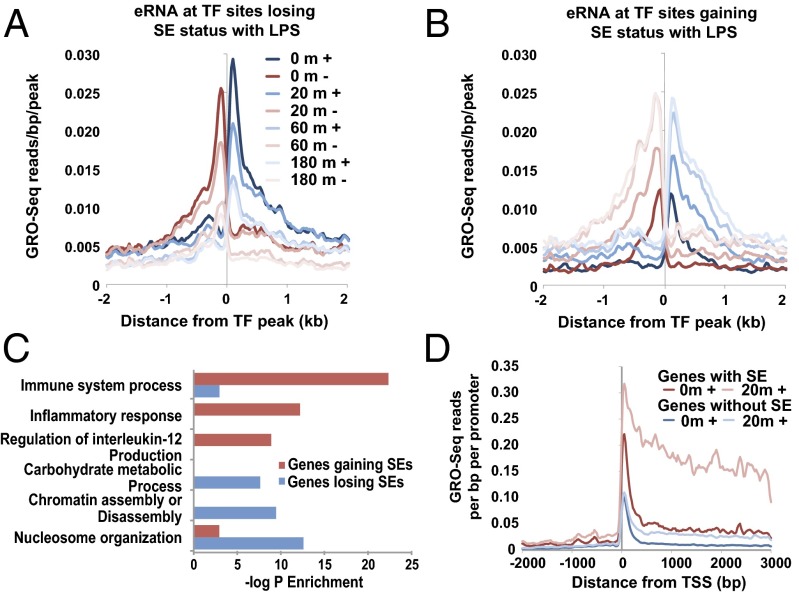
LPS treatment provides the opportunity to screen for “facultative” or “signal-dependent” SEs that can be “gained” or “lost” during macrophage activation (34, 36). Indeed, within minutes of LPS addition, 80 new SEs materialize, whereas 277 SEs recede within minutes of LPS addition (Fig. S5). These transitory SEs reflect gained or lost seRNAs that directly center over regions of previously identified TFs (Fig. 3 A and B). Interestingly, genes that gain SEs appear to be involved in immune processes and inflammatory responses (Fig. 3B), whereas the genes that lose SEs upon LPS stimulation tend to have more general functions in cellular metabolism and nuclear organization (e.g., chromatin assembly/disassembly, nucleosome organization) (Fig. 3C). This suggests that SEs are involved in regulating not only cellular identity at the basal level by marking the subset of lineage-restricted genes but also functional identity by concurrently robustly activating immune response genes and repressing cellular maintenance genes. It is possible that LPS maximizes inflammatory responses by redistributing TFs that are required to transcribe genes involved in normal cellular metabolism in a resting condition to SEs that mark functionally relevant proinflammatory genes. Thus, the decrease in transcriptional levels might be due to either a loss or redistribution of these TFs or, as previously shown in other systems, a sequestering of TFs (14, 29, 30, 33). Further studies are needed to address this issue. It is notable that genes that respond within 20 min of LPS stimulation are marked by nearby SEs and show a clear bias toward activation (Fig. 2 C and D). Thus, new seRNA transcripts appear to be the immediate early products of TLR4 signaling. In exploring the mechanism of this rapid transcriptional response, we found that this SE subset has higher levels of paused Pol II at basal levels compared with genes without SEs. Furthermore, genes with LPS-induced SEs show enhanced release of Pol II into the gene body and increased recruitment of Pol II at their promoters (Fig. 3D). This suggests that SEs mark those genes selected to rapidly respond to LPS stimulation. It is possible that the presence of signal-dependent SE subsets allows scalable communication, to allow precise temporal control of activated and repressed gene expression networks.
Fig. 3.
Dynamics of SEs and seRNAs. (A and B) GRO-Seq read density analyses of eRNA changes at TF peaks within SEs that are lost and gained upon LPS treatment. (C) Functional enrichment analyses on genes that gain SEs or that lose SEs. (D) A comparison of promoter pausing and elongation of Pol II between LPS-induced genes with and without SEs.
Coordinate Regulation of SEs and seRNAs.
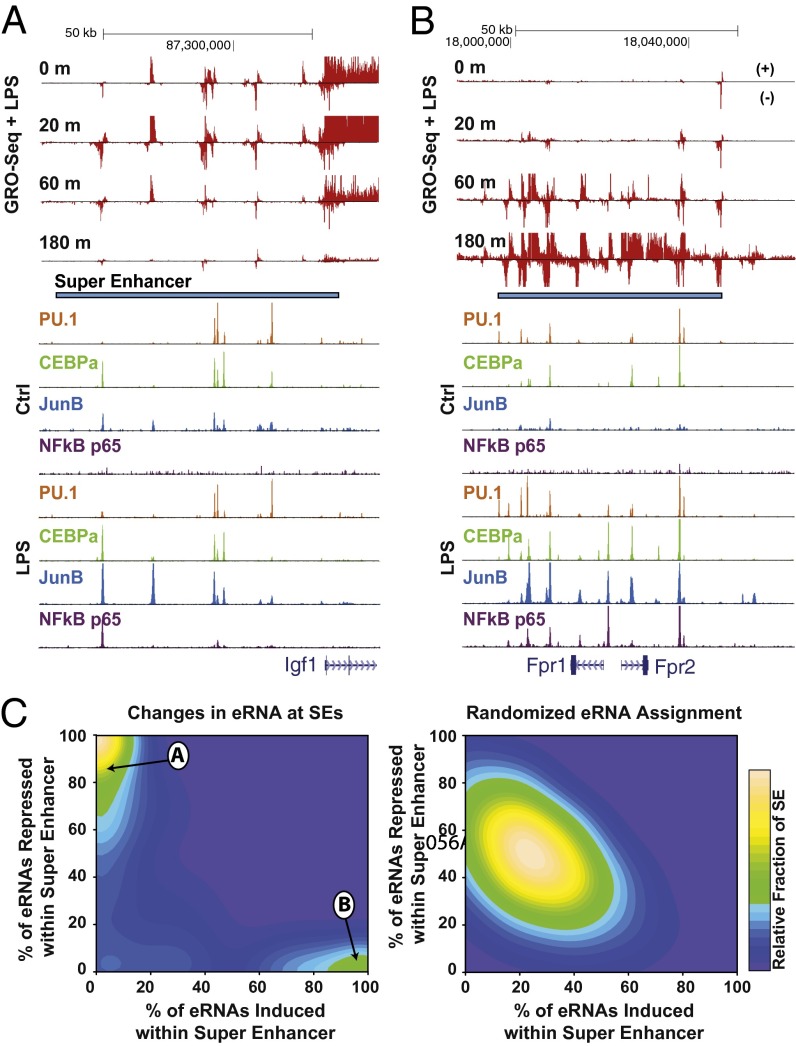
Inspection of several LPS-regulated genes revealed that the seRNAs within a given SE were often coordinately regulated with the target gene. For example, all seRNAs found in the vicinity of Fpr1 and Fpr2 genes are induced during LPS induction (Fig. 4A). Surprisingly, the same concerted trend was observed at LPS-repressed loci, such that all eRNA transcripts within the SE domain located in the upstream of Igf1 are coordinately repressed (Fig. 4B). This observation is at odds with the hypothesis that the transcriptional activity of an individual enhancer is defined by the specific bound TFs, as all SEs seem to contain PU.1, C/EBPA, p65, and JUNB sites irrespective of being activated or repressed. To assess the degree to which the dynamics of eRNA expression are coupled within an SE, we calculated the fraction of eRNAs that are either induced or repressed by LPS in each SE relative to a randomized control. Strikingly, we found genome wide that eRNAs within an SE are uniformly either up- or down-regulated rather than individually regulated (Fig. 4C). It is possible that stretches of densely packed enhancers form functional modules within which concerted regulatory activity is propagated through enhancer cross-talk. These functional modules may be associated with the formation of a topological domain where the regulations within the domain are uniformly regulated (40), a point that requires further investigation.
Fig. 4.
Coordinate regulation of SEs and seRNAs. (A and B) Browser views on two loci where eRNAs within SEs are all up- or down-regulated as a single regulatory unit. (C) Contour graph representations of the fraction of eRNAs that are either repressed or induced upon LPS stimulation within an SE based on the actual observations (Left) and the randomization (Right).
TF Composition in SEs Determines Regulatory Properties.
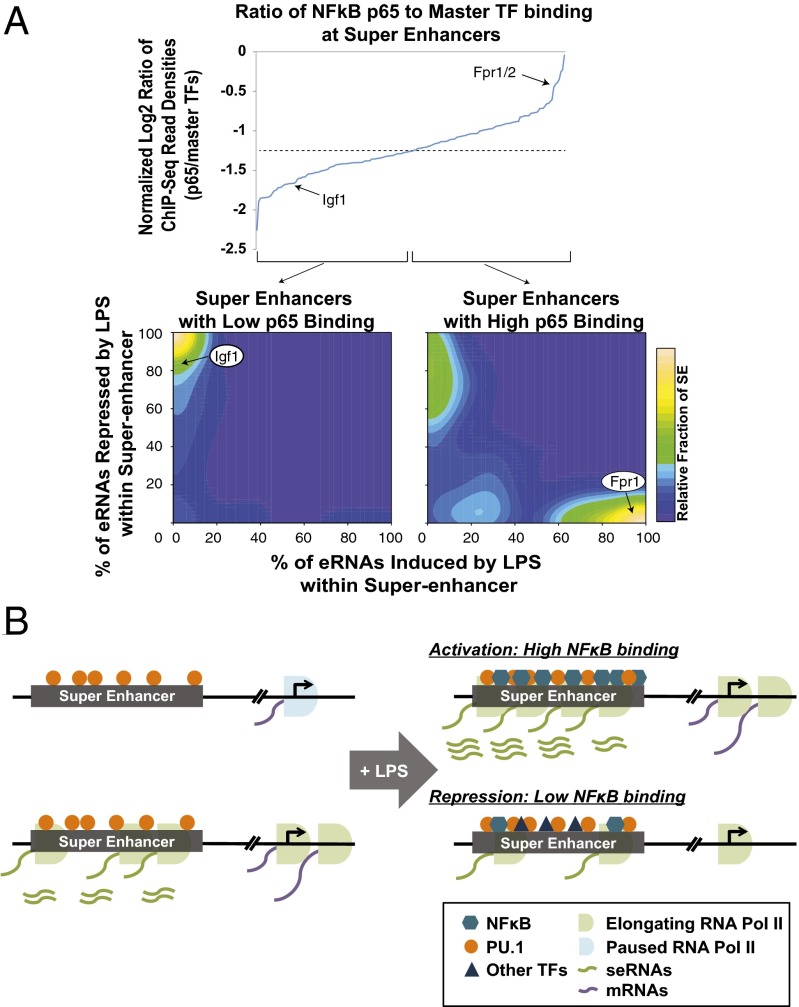
With regard to TLR4 activation, a key question is “What are the molecular determinates that direct these opposing events?” TFs activated by TLR4 signaling, such as NF-κB, are known to play a key role in inducing gene expression (31–33). A recent study has also shown the involvement of NF-κB in SE formation (33). Interestingly, ChIP-Seq for NF-κB (p65) shows the binding in both induced and repressed SEs, which is at odds with its assumed function as an activator (41). Closer inspection revealed that key inflammatory genes show more intense p65 binding within the SE relative to repressed loci. To assess whether SE regulation may be related to the quantity of binding at multiple sites within the regulatory domain, we calculated the ChIP-Seq read densities of NF-κB relative to other key master TFs that are used to define SEs and ranked them based on the ratio of NF-κB to these key TFs. We then compared this ratio to the fraction of eRNAs induced or repressed within each SE (Fig. 5A). Interestingly, SEs with strong p65 binding relative to other factors, such as Fpr1/2, exhibited high fractions of eRNA induction, whereas SEs with low p65 binding, such as Igf1, exhibited eRNA reduction (Fig. 5A), indicating that TF composition in SEs determines regulatory properties, in both eRNA activation and repression (Fig. 5B). Because it is possible that a subset of SEs requires redistribution of transcriptional cofactors such as BRD4 (33), we examined a series of seRNAs by quantitative RT-PCR in response to LPS treatment in the presence and absence of JQ1, a BRD4 inhibitor. The results show a clear reduction in transcription with LPS in the presence of JQ1 (Fig. S6), suggesting the involvement of BRD4 in seRNA transcription, at least in the SE subset examined in our system. Additionally, it is possible that these specific subsets of SEs and seRNAs serve as targets for anti-inflammatory pathways by modulating the composition of TFs and attenuating their seRNA expression, leading to reduced expression of key inflammatory genes.
Fig. 5.
Regulation of TF composition and seRNAs in SEs. (A) Relationship between the NF-κB ChIP-Seq read density relative to combined master TF ChIP-Seq read density used to define SEs (log2 ratio) and the ratio of NF-κB p65 over master TF binding ratio at SE. Igf1 represents low NF-κB p65 binding and Fpr1/2 for high NF-κB p65 binding (Upper). Contour graph representations of the fraction of LPS-stimulated eRNAs that are repressed with low NF-κB p65 binding (Lower Left) or induced with high NF-κB p65 binding (Lower Right) within an SE. (B) Model depicting LPS-dependent regulations of seRNA as well as NF-κB and TF binding changes within an SE.
Conclusion
Collectively, using macrophage activation as a model, we describe a relationship between SEs, seRNAs, and TF binding that reveals a role for SE domains and the roles of transcriptional events in those sites. In our study, we showed that SEs and seRNAs, although spread over long genomic regions, function as a single regulatory unit, which allows coordinated genic transcriptional regulation in the context of gene activation and gene repression. Previous studies have focused on the role of SEs in maintaining cellular identity (12, 13, 16, 39). However, Brown et al. (33) and our current study further investigate the regulatory dynamics of SEs upon inflammatory stimuli in endothelial cells and macrophages. Interestingly, both studies identify signal-regulated loss or gain of SE formation. Whereas Brown et al. focus on BRD4 as an enhancer output that directs SE formation and transcriptional changes of proinflammatory genes (33), our study uses eRNA production and regulation as a readout to determine enhancer activities in addition to TF binding. One of the advantages of using eRNA synthesis as a readout for determining SE is that it can allow more unbiased information of active loci throughout the genome. We also demonstrated that SEs are likely to be more conserved among species compared with TEs and can mediate dynamic signaling events such as macrophage activation. Interestingly, both activating and repressive properties of SEs and seRNAs are largely dictated by the composition of TFs within SEs. It is possible that this may not only be limited to LPS but may also apply to any signaling pathway that triggers changes in the transcriptome. For instance, different TLR ligands can regulate different compositions of IRFs in SEs, resulting in a distinctive set of gene regulations based on the stimuli. As SEs provide insights into how inflammatory responses are driven, SEs might also play a critical role in developing endotoxin tolerance in macrophages. It is possible that low-dose LPS priming is sufficient to fully induce eRNA synthesis as well as key TF binding within SEs, masking further activation of inflammatory responses in macrophages at a high-dose LPS response. Although further mechanistic understanding is required, we propose that proinflammatory SEs may be desirable genomic targets of anti-inflammatory drugs and that SEs function as molecular sensors and rheostats integrating the binding profiles of key regulators to produce dynamic profiles of gene expression in a signal-dependent fashion.
Materials and Methods
Cell Culture and Treatments.
All primary macrophages were differentiated from bone marrow isolated from an isogenic C57 background as described previously (42). The primary macrophages were plated for experiments in macrophage serum-free media (Invitrogen) before treatment with 100 ng/mL of LPS (Sigma-Aldrich) for the time indicated.
Generation of GRO-Seq Libraries.
GRO-Seq library generation was carried out as described previously (20) from two biological replicates of mouse primary macrophages treated with LPS. GRO-Seq libraries were sequenced on an Illumina HiSeq2500 sequencer to a depth of over 50 million reads per library.
Genomic Data Analysis and Visualization.
GRO-Seq analysis.
GRO-Seq sequencing reads were mapped to the mouse genome (mm9, National Center for Biotechnology Information build 37) using Bowtie2 with default parameters (v2.1.0) (43). Only reads that mapped to a single unique location were considered for further analysis. Genome browser BedGraph tracks and read density histograms were generated using HOMER (44). De novo transcript identification was performed using HOMER (findPeaks program using the “-style groseq” option), which looks for regions of continual GRO-Seq read coverage (45). Discovered transcripts were annotated as genic (overlapping known RefSeq genes, sense direction), antisense (overlapping known genes in the antisense direction), repetitive (5′ end of transcript overlapping University of California Santa Cruz (UCSC) annotated repeatmasker region), promoter antisense (5′ end emanates from promoter within 500 bp in the opposite direction from a gene), or eRNA (intergenic transcript not belonging to any of the other categories). Gene expression or transcript expression was determined using HOMER to count reads within transcript boundaries (exon + intron) for either previously annotated or de novo identified transcripts. eRNA levels at individual enhancer locations were calculated as the total GRO-Seq signal within ±500 bp relative to the peak center. Only intergenic enhancers at least 3 kb upstream of the TSS or greater than 10 kb downstream of the transcript end were used for these calculations to minimize signal from polymerase read-through from genic transcripts.
ChIP-Seq analysis.
The macrophage ChIP-Seq data used in this study came from the following Gene Expression Omnibus data accessions: GSE46494 (PU.1 and CEBPA), GSE16723 (NF-κB p65), GSE38379 (JUNB), and GSE31621 (human PU.1 and CEBPB). All ChIP-Seq data were analyzed from raw sequencing reads and aligned to either the mouse or human genomes (mm9/hg19) using Bowtie2. Genome browser tracks, ChIP-Seq peak finding (“-style factor”), and ChIP-Seq read density calculations were all performed using HOMER. To compare PU.1 and CEBP recruitment between human and mouse macrophages, the mapped coordinates of human ChIP-Seq reads and peak positions were converted to the mm9 genome using the UCSC Liftover tool.
Identification of SEs.
SEs were identified in a highly similar manner to the method outlined in Whyte et al. (13). First, master TFs of macrophage development and activation (PU.1, CEBPA, JUNB, p65) were combined by sampling 10 million reads from each sequencing experiment and merging the results into a single metaexperiment for untreated and LPS-treated conditions separately. Input/IgG experiments were merged in the same manner. SEs were then found by running HOMER findPeaks using the “-style super” option. This process first identifies traditional ChIP-Seq peaks by treating the metaexperiment and metainput as normal ChIP-Seq experiments and then merges identified peaks found within 12.5 kb into continuous regions. The “enhancer score” for each region is defined by the number of metaexperiment reads minus the metainput reads normalized for sequencing depth. All regions are then sorted by their score and plotted by their relative rank and score (0–1). Enhancer regions past which the slope of the line reaches 1 are considered SEs, and all remaining enhancers are considered as “typical enhancers.” Discovery of these SEs was further validated by comparing SEs found in Brown et al. (33) to SEs found using HOMER with the same mouse macrophage H4K12ac ChIP-Seq dataset (33). Differences were mostly attributed to differential identification of key peaks that might allow a given SE region to expand across 12.5-kb regions to increase signal. Independently, the functional quality of differential SE calls was assessed by measuring the eRNA production from intergenic TFBSs that were specific to Brown et al. or HOMER SEs.
The binding index of a given TF within an SE is defined as the log2 ratio of reads for the given factor in the SE relative to the total reads of the metaexperiment in the SE. In this sense, if a factor is highly bound within the SE relative to other TFs, the binding index will be high, whereas if there is minimal binding of the factor in the SE relative to other factors, the binding index will be low. Gene ontology analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID). The release of RNA Pol II from proximal promoter pausing was quantified by calculating the density of Pol II ChIP-Seq reads within 200 bp of the TSS relative to the gene body (200 bp – 3 kb relative to the TSS).
Coordinate regulation within SEs.
To detect coregulated eRNAs within SEs, individual peak regions within each SE (defined by applying peak finding to the “metaexperiment” representing the merge of master regulators) were analyzed for changes in GRO-Seq read density (±500 bp from peak center). Only peaks located in intergenic regions >3 kb from the TSS and >10 kb from transcript end locations were considered. Only peaks with detectable transcription (>10 GRO-Seq reads in either control or treated conditions) were considered. Peaks were assigned to their overlapping SE or assigned to a random SE as a control. Each SE was considered in terms of the fraction of peaks with up-regulated or down-regulated eRNA expression and then plotted as a contourplot in R using a 2D Gaussian kernel density estimator. Regulated eRNAs were defined by >twofold.
Supplementary Material
Acknowledgments
We thank L. Ong and C. Brondos for administrative assistance. N.H. was supported by a Pioneer Postdoctoral Endowment Fellowship. R.M.E. is supported by National Institutes of Health Grants DK057978, DK090962, HL088093, HL105278 and ES010337; a Stand Up to Cancer Dream Team Translational Cancer Research Grant; Program of the Entertainment Industry Foundation Grant SU2C-AACR-DT0509; the Glenn Foundation for Medical Research; the Leona M. and Harry B. Helmsley Charitable Trust; Ipsen/Biomeasure; California Institute for Regenerative Medicine; and The Ellison Medical Foundation. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE60857).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424028112/-/DCSupplemental.
References
- 1.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong CT, Corces VG. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3(1):e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111(20):7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12(11):R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natoli G, Andrau JC. Noncoding transcription at enhancers: General principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 9.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siersbæk R, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Reports. 2014;7(5):1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Parker SC, et al. NISC Comparative Sequencing Program; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors; NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai F, Shiekhattar R. Enhancer RNAs: The new molecules of transcription. Curr Opin Genet Dev. 2014;25:38–42. doi: 10.1016/j.gde.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39(4):170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Mousavi K, Zare H, Koulnis M, Sartorelli V. The emerging roles of eRNAs in transcriptional regulatory networks. RNA Biol. 2014;11(2):106–110. doi: 10.4161/rna.27950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ørom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21(2):194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guertin MJ, Zhang X, Coonrod SA, Hager GL. Transient ER binding and p300 redistribution support a squelching mechanism for E2-repressed genes. Mol Endocrinol. 2014;28(9):1522–1533. doi: 10.1210/me.2014-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Step SE, et al. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARγ-driven enhancers. Genes Dev. 2014;28(9):1018–1028. doi: 10.1101/gad.237628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smale ST. Seq-ing LPS-induced enhancers. Immunity. 2010;32(3):296–298. doi: 10.1016/j.immuni.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140(6):833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JD, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56(2):219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.IIott NE, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel B, et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014;28(14):1562–1577. doi: 10.1101/gad.242685.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin JW, Wang G. The mediator complex: A master coordinator of transcription and cell lineage development. Development. 2014;141(5):977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- 40.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baltimore D. NF-κB is 25. Nat Immunol. 2011;12(8):683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 42.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24(24):2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.