Significance
MicroRNAs limit gene expression by recruiting a large protein complex known as the RNA-induced silencing complex (RISC) to target mRNAs. While attempting to understand physiological regulation of RISC assembly, we found that most healthy adult tissues retain a reserve of microRNAs not stably associated with target mRNA. Recruitment of microRNAs to large mRNA-containing complexes was accompanied by an increase in their ability to repress targets and was regulated in part by phosphoinositide-3 kinase–RAC-alpha serine/threonine-protein kinase–mechanistic target of rapamycin pathway-dependent enhancement of the glycine-tryptophan protein of 182 kDa protein expression. Data presented here suggest that in vivo, many expressed microRNAs exist in an inactive reserve, allowing resting cells to use microRNAs to dynamically regulate the translation of target mRNAs in their environment.
Keywords: microRNA, Argonaute, GW182, T cells, mTOR
Abstract
MicroRNAs repress mRNA translation by guiding Argonaute proteins to partially complementary binding sites, primarily within the 3′ untranslated region (UTR) of target mRNAs. In cell lines, Argonaute-bound microRNAs exist mainly in high molecular weight RNA-induced silencing complexes (HMW-RISC) associated with target mRNA. Here we demonstrate that most adult tissues contain reservoirs of microRNAs in low molecular weight RISC (LMW-RISC) not bound to mRNA, suggesting that these microRNAs are not actively engaged in target repression. Consistent with this observation, the majority of individual microRNAs in primary T cells were enriched in LMW-RISC. During T-cell activation, signal transduction through the phosphoinositide-3 kinase–RAC-alpha serine/threonine-protein kinase–mechanistic target of rapamycin pathway increased the assembly of microRNAs into HMW-RISC, enhanced expression of the glycine-tryptophan protein of 182 kDa, an essential component of HMW-RISC, and improved the ability of microRNAs to repress partially complementary reporters, even when expression of targeting microRNAs did not increase. Overall, data presented here demonstrate that microRNA-mediated target repression in nontransformed cells depends not only on abundance of specific microRNAs, but also on regulation of RISC assembly by intracellular signaling.
MicroRNAs are ∼22 nucleotide (nt) single-stranded RNAs that regulate gene expression by repressing translation of target mRNAs and/or inducing their degradation (1). Because the majority of expressed mRNAs are predicted to be microRNA targets (2), microRNA-mediated gene regulation is thought to contribute to most physiological and pathological processes. Nevertheless, elimination of an individual microRNA is rarely sufficient to trigger an overt phenotype in adult animals, yet following stress, phenotypes are often seen (3, 4), suggesting that microRNA utilization changes in response to external stimuli.
In mammals, most microRNAs are generated from long, RNA polymerase II transcripts, which are reduced to an ∼22-nt double-stranded RNA fragment by two steps of cleavage, one mediated by the endoribonuclease Drosha in the nucleus and one by the endoribonuclease Dicer in the cytoplasm. One of the two strands, known as the guide strand, is incorporated into one of the four Argonaute proteins (Ago1–4), creating the minimal unit of the RNA-induced silencing complex (RISC) (5). Argonaute-bound microRNAs are capable of endonucleolytic cleavage of perfectly complementary targets through the small interfering RNA (siRNA) pathway (6) or translational repression and degradation of mRNA targets with partial complementarity through the microRNA pathway (7). Repression of partially complementary target mRNAs requires interaction of microRNA-containing Argonaute proteins with one of three functionally redundant TNRC6 proteins, TNRC6A/the glycine-tryptophan protein of 182 kDa, (hereafter GW182), TNRC6B, and TNRC6C (8, 9). These proteins bind one or more Argonaute proteins simultaneously and act as docking factors for the recruitment of enzymes involved in translational repression and/or target degradation (10).
Regulation of endogenous microRNA targets in mammals occurs almost exclusively through the microRNA pathway, which relies on assembly of large ribonucleoprotein complexes. As such, Argonaute-bound microRNAs are found in distinct complexes, ranging from low molecular weight RISC (LMW-RISC) composed of an Ago protein complexed to an 18- to 25-nt microRNA (∼100 kDa), to high molecular weight RISC (HMW-RISC), larger than 2 MDa (11–13). However, the biological significance of these complexes and what determines distribution of microRNAs between them is largely unknown.
We have previously shown that accumulation of LMW-RISC can be induced in vitro by long-term depletion of nutrients or growth factors from the medium of apoptosis-resistant cells (13). Under these conditions, LMW-RISC is a stable complex that retains bound microRNAs, is able to reassemble into HMW-RISC, and can induce target repression upon restoration of growth conditions. These observations prompted us to examine the distribution of microRNA–Argonaute complexes between HMW-RISC and LMW-RISC in vivo. We found that, whereas proliferating cells maintain most microRNAs in HMW-RISC bound to mRNA, many adult tissues keep a large portion of their microRNA repertoire in LMW-RISC, not engaged in target repression. Distribution between LMW-RISC and HMW-RISC varied among microRNA species, as individual microRNAs showed characteristic distributions between the two complexes. Furthermore, the ability to assemble microRNAs into HMW-RISC and enhance target repression was regulated, at least in part, by induction of GW182 protein downstream of the phosphoinositide-3 kinase (PI3K)–RAC-alpha serine/threonine-protein kinase (Akt)–mechanistic target of rapamycin (mTOR) signaling pathway. Taken together, these data suggest that assembly of functional RISC is regulated globally by availability of RISC proteins and specifically by interactions between individual microRNAs and target mRNAs.
Results
RISC Molecular Weight Profile in Vivo.
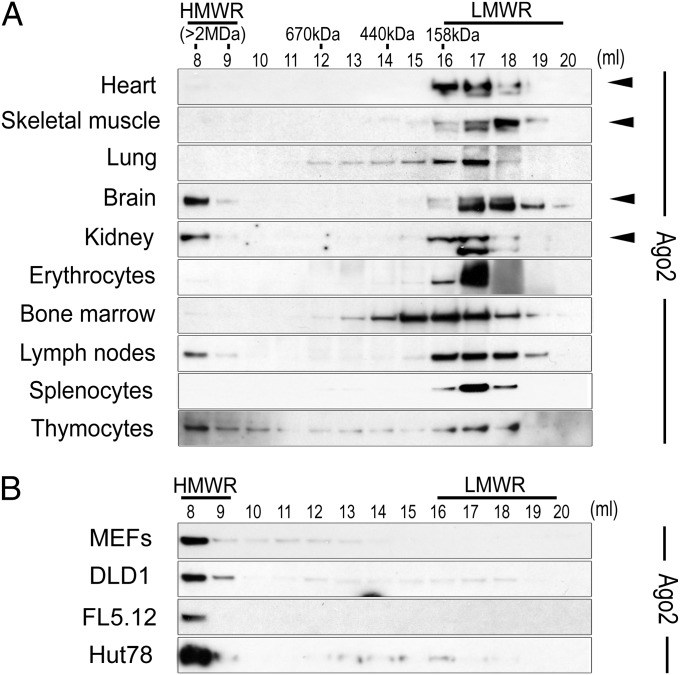
Assembly and function of the RISC has been extensively studied in immortalized cells grown in tissue culture (14). Our previous data demonstrated that RISC assembly and function could be modulated in cultured cells subjected to mitogen and/or nutrient deprivation and stimulation (13). Whether these observations could be extended to physiological, in vivo situations remained unknown. To investigate RISC assembly in vivo, we determined molecular weight profiles of Argonaute (Ago) proteins in tissues from healthy adult mice using Superose 6-based size exclusion chromatography. Strikingly, Ago proteins were greatly enriched in LMW-RISC in most adult tissues (Fig. 1A and SI Appendix, Figs. S1 and S2). In contrast, Ago proteins were found almost exclusively in HMW-RISC in cell lines in log-phase growth, irrespective of species or tissue of origin (Fig. 1B and SI Appendix, Figs. S1 and S2). This enrichment in HMW-RISC was not solely dependent on availability of nutrients and mitogens in tissue culture medium because Ago proteins were found enriched in HMW-RISC before in vitro differentiation of 3T3-L1 cells and in LMW-RISC following their differentiation to adipocytes (SI Appendix, Fig. S3).
Fig. 1.
In vivo distribution of Ago2 protein between HMWR and LMWR. Ago2 elution profiles determined by Superose 6 size exclusion chromatography followed by Western blot using extracts of (A) healthy adult mouse tissues versus (B) cell lines in log-phase growth. Arrows indicate specific Ago2 band, as a lower molecular weight nonspecific band was observed in some tissues. SI Appendix, Fig. S2 provides quantification of replicate experiments.
HMW-RISC Fractions Contain MicroRNA–Argonaute Complexes Stably Bound to mRNA.
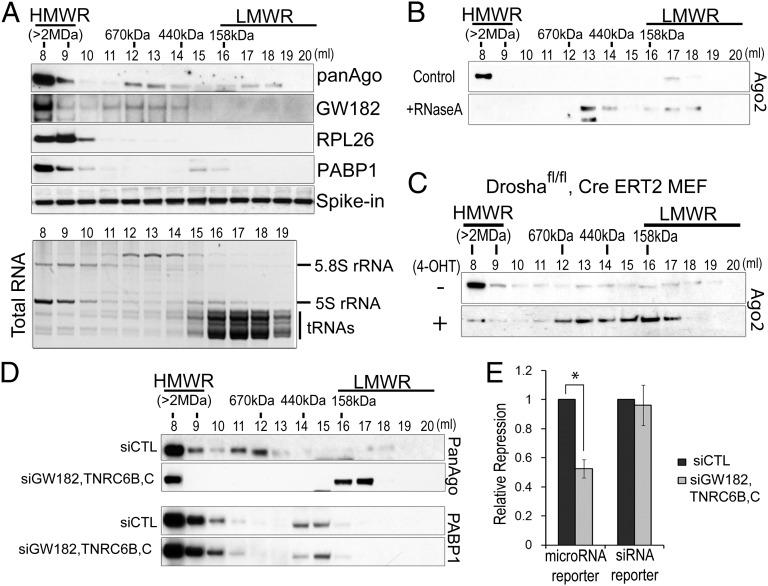
To better understand mechanisms that regulate distribution of Ago proteins between HMW-RISC and LMW-RISC, we first examined biochemical properties that govern Ago protein distribution in immortalized mouse embryonic fibroblasts (MEFs). Consistent with HMW-RISC containing mRNA-bound Ago–microRNA complexes associated with polysomes (15) and/or other mRNA binding proteins (16), Ago proteins coeluted with GW182, poly(A)-binding protein-1 (PABP1), and ribosomes in high molecular weight fractions 8 and 9 (Fig. 2A). To test this model, we assessed the requirement of RNA and protein components of RISC for stable assembly of HMW-RISC. Depletion of mRNA by treatment of extracts with RNaseA before fractionation led to complete dissolution of HMW-RISC (Fig. 2B). Similarly, knockout of Drosha (Fig. 2C and SI Appendix, Fig. S4) or Dicer (SI Appendix, Fig. S5) resulted in a marked reduction of HMW-RISC.
Fig. 2.
HMW-RISC requires mRNA, microRNAs, and TNRC6 proteins. (A) Western blot analysis of proteins enriched in HMW-RISC fractions of Superose 6 elution profiles from MEFs in log-phase growth (Top). A total of 5 ng of recombinant GFP protein (spike-in) was spiked into each fraction to control for precipitation efficiency. Ethidium bromide staining of total RNA contained in each fraction separated on a 12% urea–PAGE gel is shown (Bottom). (B) Ago2 elution profile following RNaseA (2 µg/mL) treatment of lysates as in A. (C) Ago2 elution profile from extracts of Droshafl/fl MEFs stably expressing CreERT2 treated with 500 nM 4-hydroxytamoxifen (4-OHT) to induce Drosha deletion or left untreated for 2 wk. (D) Western blot showing elution profile of Ago proteins in MEFs 48 h after cotransfection of siRNA against GW182, Tnrc6b, and Tnrc6c or control siRNA (siCTL). Western blots for polyA-binding protein 1 (PABP1) are shown to illustrate the specific effect on Ago proteins. (E) Repression of microRNA or siRNA reporter constructs by an oligonucleotide duplex transfected into MEFs as in D. Bars represent average reporter repression ±SD of three biological replicates (*P = 0.006 as determined by two-tailed Student's t test).
Additionally, siRNA-mediated depletion of all three TNRC6 proteins (GW182, TNRC6B, and TNRC6C) in MEFs led to decreased HMW-RISC and accumulation of LMW-RISC (Fig. 2D and SI Appendix, Fig. S6). Similar results were obtained using the murine pro-B-cell line FL5.12. As TNRC6 proteins are necessary and sufficient for microRNA-mediated target repression (17), decreased HMW-RISC abundance upon knockdown of all three TNRC6 proteins was accompanied by a significant decrease in microRNA function (Fig. 2E), as assessed by the ability of a transfected oligonucleotide duplex to repress a microRNA reporter construct. Interestingly, depletion of TNRC6 proteins did not significantly affect repression of a siRNA reporter targeted by the same oligonucleotide duplex (Fig. 2E), supporting previous evidence that the microRNA and siRNA pathways are differentially dependent on TNRC6 proteins (10).
Distribution and Function Varies Between MicroRNA Species Independent of Their Expression.
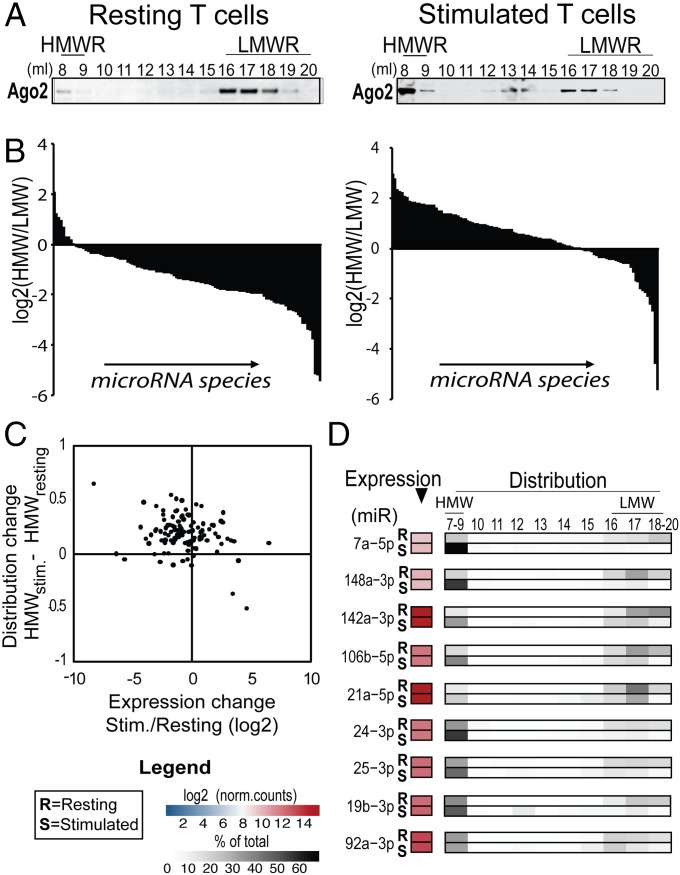
As mature microRNAs exist primarily bound to Ago proteins (18), distribution of Ago proteins between HMW-RISC and LMW-RISC should predict distribution of mature microRNAs. To test this hypothesis, we sequenced small RNAs contained in each Superose 6 fraction of lysates from freshly isolated, resting T cells that contain predominantly LMW-RISC and lysates from ex vivo stimulated T cells that contain predominantly HMW-RISC (Fig. 3A) (13). As expected, most microRNA species were enriched in LMW-RISC in resting T cells and in HMW-RISC in stimulated T cells (Fig. 3B and Dataset S1), regardless of their level of expression (SI Appendix, Fig. S7). Closer examination of these data revealed that redistribution to HMW-RISC following stimulation was distinctive for each microRNA, in agreement with previous reports (19). This shift from LMW-RISC to HMW-RISC did not significantly correlate with a change in microRNA expression (Fig. 3C and SI Appendix, Fig. S8). Analysis of individual microRNAs whose total expression did not change upon stimulation, but that redistributed between HMW-RISC and LMW-RISC, clearly demonstrated this principle (Fig. 3D).
Fig. 3.
Distribution of microRNAs between HMW-RISC and LMW-RISC in resting and stimulated T cells. (A) Ago2 elution profile from freshly isolated (resting) T cells versus T cells stimulated for 3 d with beads coated with αCD3 and αCD28 antibodies in the presence of rIL2 (25 units/mL). (B) Ratio of microRNAs found in HMW-RISC (fractions 8 and 9) to LMW-RISC (fractions 16–20) in T cells as in A. Bars represent each of the 110 microRNAs expressed in resting and/or stimulated T cells as determined by small RNA sequencing from two independent experiments. (C) Change in expression of individual microRNA species (x axis) plotted against redistribution to HMW-RISC (y axis) following T-cell stimulation. Each dot represents a microRNA species. (D) Elution profiles determined by small RNA sequencing of microRNA species whose expression did not change [false discovery rate, >10%, abs (log2 fold change) <0.5] upon stimulation illustrate a shift to HMW-RISC in the absence of expression change. The elution profiles of representative microRNAs whose expression did not change with mitogenic stimulation are shown.
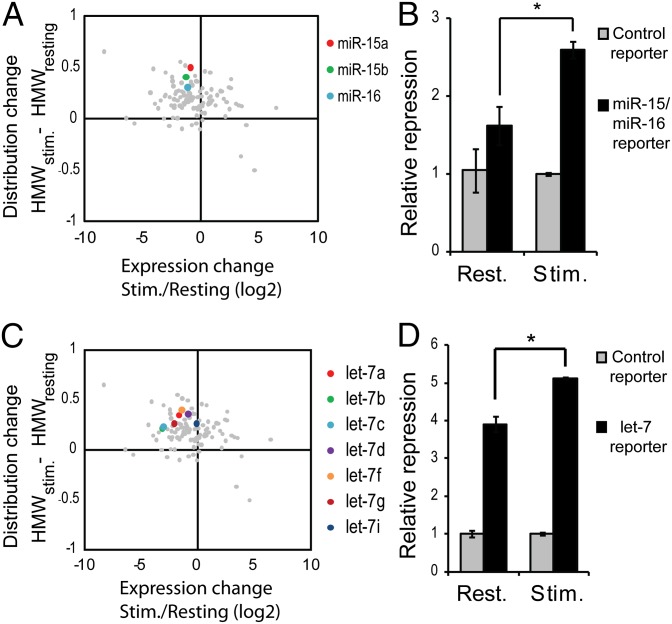
As expression and enrichment in target-bound RISC of several microRNA species were independent of one another, we hypothesized that function of these microRNAs may not be determined exclusively by their expression, but also by their enrichment in HMW-RISC. To test this hypothesis, the ability of two endogenous microRNA families to repress luciferase reporters was examined in resting and stimulated T cells. Expression of members of the miR-15/16 seed family (miR-15a, miR-15b, and miR-16) decreased in stimulated T cells, yet showed a marked enhancement in HMW-RISC (Fig. 4A and SI Appendix, Fig. S9A) accompanied by enhanced repression of a reporter with target sites complementary to the miR-15/16 seed sequence (AGCAGCA; Fig. 4B). Similarly, expression of let-7 seed family microRNAs decreased following T-cell stimulation, whereas their HMW-RISC occupancy and ability to repress a reporter with target sites complementary to the let-7 seed sequence (GAGGUAG) increased (Fig. 4 C and D and SI Appendix, Fig. S9B).
Fig. 4.
Ability of endogenous microRNAs to repress reporters can be independent of their expression. (A) Expression change plotted against distribution change of miR-15/16 seed family microRNAs (colored) following ex vivo stimulation of T cells. Gray dots represent all other expressed microRNAs. Change in expression of miR-15a/b and miR-16 was confirmed by qRT-PCR (SI Appendix, Fig. S9A). (B) Repression of a Firefly luciferase reporter containing the miR-15/16 seed target sequence cloned into its 3′ UTR in resting and stimulated T cells. Bars represent average reporter repression ±SD of a representative experiment performed in quadruplicate (*P = 0.003). (C) Expression change plotted against distribution change of let-7 seed family microRNAs (colored) following ex vivo stimulation of T cells. Gray dots represent all other expressed microRNAs. Change in expression of let-7a and let-7f were confirmed by qRT-PCR (SI Appendix, Fig. S9B). (D) Repression of a Renilla luciferase reporter containing the let-7 seed target sequence cloned into its 3′ UTR in resting and stimulated T cells. Bars represent average reporter repression ±SD of a representative experiment performed in quadruplicate (*P = 0.005 as determined by two-tailed Student's t test).
GW182 Expression Predicts HMW-RISC Assembly and MicroRNA Function in Hematopoietic Cells.
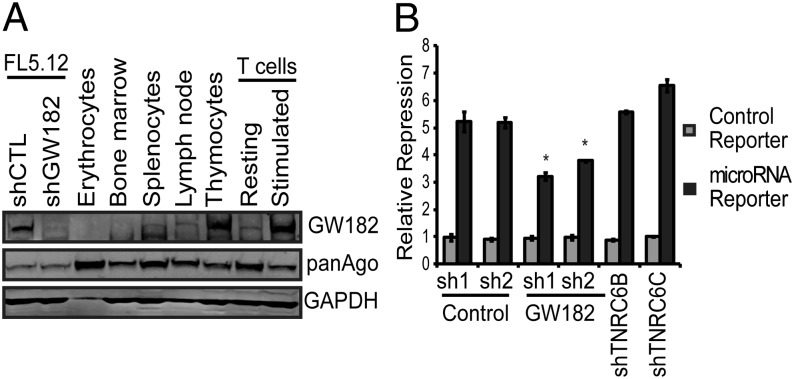
Whereas differences existed between recruitment of individual microRNAs to HMW-RISC, overall the trend was for increased occupancy in HMW-RISC of both Ago proteins and microRNA following ex vivo stimulation of primary T cells. This enhancement of HMW-RISC assembly in stimulated T cells was accompanied by increased expression of GW182 (13, 20) (Fig. 5A), one of three redundant RISC scaffold proteins. Expression of GW182 correlated with the extent to which Ago protein partitioned to HMW-RISC in hematopoietic tissues (Fig. 5A compared with Fig. 1) as well as in several solid tissues (SI Appendix, Fig. S10 compared with Fig. 1).
Fig. 5.
Expression and function of GW182 in hematopoietic cells. (A) GW182 expression in the indicated primary hematopoietic cell types assessed by Western blot. FL5.12 cells expressing a control shRNA or a shRNA targeting GW182 mRNA are shown to demonstrate specificity of the GW182 antibody used. (B) Repression of a microRNA reporter construct targeted by an oligonucleotide duplex (details in SI Appendix, SI Materials and Methods) transfected into T cells expressing the indicated shRNAs and stimulated for 2 d with beads coated with αCD3 and αCD28 antibodies in the presence of rIL2. Bars represent average reporter repression ±SD of a representative experiment performed in quadruplicate (*P = 0.003 as determined by two-tailed Student's t test).
Consistent with an important role for GW182 in assembly of HMW-RISC in hematopoietic cells, we found that increased GW182 expression following mitogenic stimulation was accompanied by a shift of an exogenously introduced synthetic microRNA from LMW-RISC to HMW-RISC and an increase in the ability of this synthetic microRNA to repress a reporter through the microRNA pathway (13) (SI Appendix, Fig. S11). Using the same synthetic microRNA/reporter system, we found that microRNA function in activated T cells could be specifically inhibited by knockdown of GW182 (Fig. 5B and SI Appendix, Fig. S12), but not by knockdown of TNRC6B or TNRC6C. This suggests that, among the three TNRC6 proteins, GW182 plays a critical role in enhanced microRNA function in hematopoietic tissues.
GW182 Expression, RISC Assembly, and MicroRNA Function Are Regulated by the PI3K-Akt-mTOR Signaling Pathway.
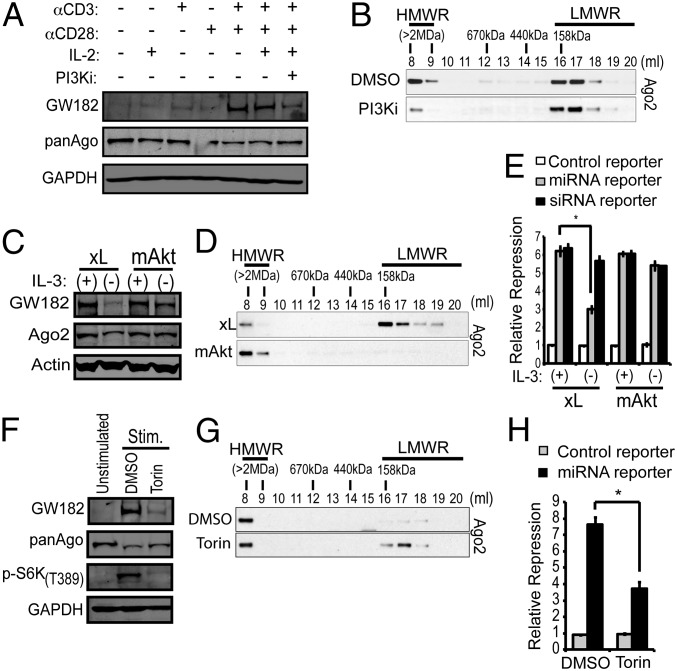
To dissect the signaling responsible for enhanced GW182 expression, RISC assembly and microRNA function, freshly isolated T cells were stimulated with microbeads coated with αCD3 and/or αCD28 antibodies in the presence or absence of interleukin-2 (IL-2). Maximal GW182 protein induction was seen following costimulation with αCD3 and αCD28 for 24 h (Fig. 6A). As the phosphatidylinositol 3-kinase (PI3K) signaling pathway is a key mediator downstream of CD3/CD28 costimulation (21), the effect of the PI3K inhibitor LY294002 on GW182 protein expression and RISC composition was examined. As shown in Fig. 6 A and B, accumulation of GW182 protein and HMW-RISC in stimulated T cells could largely be prevented by the inhibitor, suggesting that signaling through the PI3K-Akt pathway stimulates HMW-RISC assembly through induction of GW182 expression.
Fig. 6.
PI3K-Akt-mTOR signaling promotes GW182 expression, HMW-RISC assembly, and microRNA function. (A) GW182 expression by Western blot in T cells stimulated for 24 h with bead-bound αCD3 antibody, αCD28 antibody, or rIL-2 (25 units/mL) alone or in combination ± the PI3K inhibitor LY294002 (10 µM). (B) Ago2 elution profile from T cells treated as in the last two lanes of A. (C) Western blot for GW182 expression in IL-3 dependent FL5.12 cells engineered to overexpress Bcl-xL (FL5.12.xL) or myristoylated Akt (FL5.12.mAkt) cultured in the presence or absence of IL-3 for 24 h. (D) Comparison of Ago2 elution profiles from FL5.12.xL versus FL5.12.mAkt cells cultured in the presence or absence of IL-3 for 72 h. (E) Repression of microRNA or siRNA reporter constructs by an oligonucleotide duplex transfected into cells cultured as in C. Bars represent average reporter repression ±SD of a representative experiment performed in quadruplicate (*P < 0.001). (F) Western blot for GW182 expression in T cells stimulated with beads coated with αCD3 and αCD28 antibodies in the presence of rIL2 ± 500 nM Torin1 for 48 h. (G) Ago2 elution profile from T cells treated as in the last two lanes of F. (H) Repression of a microRNA reporter construct by an oligonucleotide duplex transfected into T cells cultured as in the last two lanes of F. Bars represent average reporter repression ±SD of a representative experiment performed in quadruplicate (*P < 0.001 as determined by two-tailed Student's t test).
To confirm that the PI3K signaling pathway regulates HMW-RISC assembly, hematopoietic cell lines, in which mitogen-stimulated GW182 expression and HMW-RISC assembly are dependent on the presence of interleukin-3 (IL-3) (13), were used. Expression of tamoxifen-inducible PI3K P110 subunit (PI(3)K-ER) or myristoylated Akt (mAkt-ER) in BaF3 cells conferred 4-hydroxytamoxifen (4-OHT)-dependent GW182 protein expression in the absence of IL-3 (SI Appendix, Fig. S13). Similarly, constitutive expression of myristoylated Akt in FL5.12 cells was sufficient to maintain GW182 in the absence of IL-3, independent of the antiapoptotic effect of constitutive PI3K-Akt pathway activation because GW182 expression was not maintained by overexpression of the antiapoptotic protein Bcl-xL (Fig. 6C and SI Appendix, Fig. S14). Consistent with the putative role of GW182 in HMW-RISC assembly and microRNA function, IL-3 withdrawal from FL5.12 cells expressing Bcl-xL led to accumulation of Ago proteins in LMW-RISC (Fig. 6D) and decreased repression of reporters targeted by synthetic (Fig. 6E) or endogenous (SI Appendix, Fig. S15) microRNAs. Constitutive Akt activation was sufficient to maintain Ago protein occupancy in HMW-RISC (Fig. 6D) and microRNA reporter repression (Fig. 6E) in the absence of IL-3 stimulation.
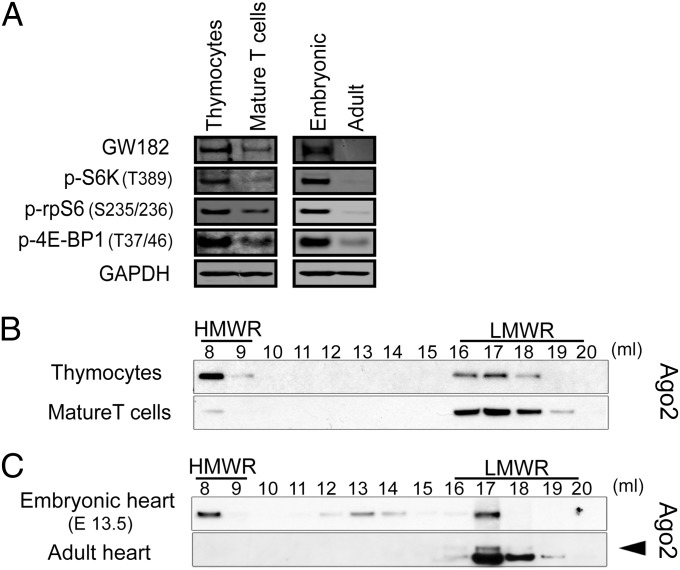
The mechanistic target of rapamycin (mTOR) is a central component of the PI3K-Akt signaling network known to influence the fate of stimulated T cells (22). Inhibition of mTOR serine/threonine kinase activity using the chemical inhibitor Torin1 resulted in decreased expression of GW182 protein in stimulated T cells (Fig. 6F) or FL5.12 cells (SI Appendix, Fig. S16A). Furthermore, Torin1 treatment inhibited HMW-RISC assembly following T-cell stimulation (Fig. 6G) and decreased repression of a reporter construct targeted by an exogenously introduced synthetic microRNA in stimulated T cells (Fig. 6H) or FL5.12 cells (SI Appendix, Fig. S16B). In vivo, mTOR is a critical component of signal transduction-induced thymic T-cell maturation (23, 24) and development of the embryonic heart (25). Consistent with these observations, we found increased phosphorylation of the mTOR targets S6, S6 kinase, and 4E-BP1 when comparing freshly isolated thymocytes to mature T cells or embryonic heart to adult heart (Fig. 7A and SI Appendix, Fig. S17). Enhanced mTOR signaling correlated with increased GW182 protein expression (Fig. 7A) and Ago2 occupancy in HMW-RISC in both thymocytes (Fig. 7B) and embryonic heart (Fig. 7C). Taken together, these data demonstrate that the relative abundance of HMW-RISC and LMW-RISC is dynamically regulated by the PI3K-Akt-mTOR pathway, at least in part, through induction of GW182 expression.
Fig. 7.
In vivo correlation between mTOR signaling pathway activation, GW182 protein expression, and enrichment of Ago proteins in HMW-RISC. (A) Western blot for phosphorylation of mTOR pathway activation and GW182 protein expression in the indicated tissues or cell types. (B) Ago2 elution profiles determined by Superose 6 size exclusion chromatography followed by Western blot from freshly isolated thymocytes versus resting splenic T cells. (C) Ago2 elution profiles determined by Superose 6 size exclusion chromatography followed by Western blot from embryonic heart (E13.5) versus adult heart. Arrow indicates specific Ago2 band.
Discussion
Altered expression of individual microRNAs has been linked to a variety of diseases, including cancer (26). As a consequence, extensive efforts have been directed to determine microRNA expression profiles (27). These studies tacitly assume that expression level of a given microRNA correlates with its ongoing repression of target mRNA. Data presented here show that differences in expression level of a given microRNA are not necessarily predictive of the ability of that microRNA to repress its cognate targets. Rather, our data suggest that changes in intracellular signaling can alter microRNA assembly on targets, and consequently function, independent of changes in expression. Moreover, microRNA–Argonaute complexes contained in most adult tissues are not associated with target mRNA (i.e., in HMW-RISC), suggesting that basal signaling required to maintain tissue homeostasis is insufficient to promote microRNA function. In light of these observations, it follows that lack of apparent phenotypes observed in many microRNA knockout animals (3, 4) may be, at least partially, explained by the fact that microRNAs are not associated with target mRNA in most differentiated adult tissues.
The endpoint of the microRNA pathway is mRNA degradation (28), a function apparently redundant to the siRNA pathway. The siRNA pathway requires, in principle, only the catalytic activity of Ago2 to induce target degradation (29), whereas the microRNA pathway requires recruitment of proteins responsible for deadenylation and degradation of mRNA targets through association of target-bound Argonaute proteins with GW proteins (GW182, TNRC6B, and TNRC6C). Data presented here suggest that regulation of this interaction, and therefore the assembly of a functional RISC, may allow cells to express microRNAs in advance of their physiologic requirement. This may provide an advantage during transition between physiological states or during stress, which could help to explain why metazoans evolved the microRNA pathway while maintaining the siRNA pathway.
Recently, it was reported that bacterial Ago proteins can restrict the acquisition of foreign nucleic acids using an siRNA-related process (30). Like mammalian Ago2, the bacterial Ago proteins have slicing activity that can degrade exogenous nucleic acids complementary to bound nucleic acids. Although metazoan microRNAs are thought to regulate gene expression primarily through translational repression, the structural conservation between eukaryotic Ago2 and its archeal and eubacterial counterparts is striking (31). Our data show that in conditions where LMW-RISC predominates, microRNA activity is compromised but siRNA activity is not (Figs. 2E, 6E, and SI Appendix, Fig. S15B). These observations raise the possibility that microRNAs stored in LMW-RISC may not only be recruited to HMW-RISC to regulate endogenous gene expression, but may also be used to cope with exogenous nucleic acids introduced by pathogens.
MicroRNA function can be regulated at the level of RISC assembly via regulation of GW182 protein expression. When comparing distribution of individual microRNAs between HMW-RISC and LMW-RISC in conditions where GW182 is not limiting (i.e., stimulated T cells), we observed differences among microRNA species. This suggests that association of individual microRNAs with target mRNAs is further regulated by factors that restrict or enhance these interactions. It is likely that differential activation of target mRNAs also contributes to differences in distribution of individual microRNAs between HMW-RISC and LMW-RISC during mitogenic activation. In future studies, the mapping of microRNA-target interactions during physiologic perturbations of primary tissues using methods such as high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation or photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation may help shed light on these issues. Furthermore, recent studies have reported additional ways in which microRNA–mRNA interactions can be altered, including changes in subcellular localization of microRNA–Ago complexes (32–34), expression of endogenous microRNA “sponges” (35), microRNA editing (36), interference by RNA-binding proteins (16, 37), changes in RNA secondary structures (38), use of alternative polyadenylation sites in target mRNAs (39), or disruption of microRNA–Ago interactions (40). All these factors collectively may contribute to the distribution of individual microRNAs between HMW-RISC and LMW-RISC in vivo.
Previous studies, such as those describing the creation of target prediction algorithms, have focused on cell lines (41). Here we demonstrate that in cultured cell lines, the vast majority of expressed microRNAs are found in HMW-RISC, regardless of their expression level. Because HMW-RISC engages in target repression, this explains why there is such tight correlation between microRNA expression and target repression in cell lines. Whereas this correlation has allowed the effective development of algorithms to predict microRNA targets, the study of freshly isolated adult tissue reveals an additional level of complexity to how microRNAs function in vivo. Adult tissues contain high levels of microRNA/Ago complexes that exist in LMW complexes that are not engaged in target repression. Such LMW-RISC may represent a regulatory reserve that is used in vivo to allow cells to modulate gene expression in response to physiologic perturbations or in response to stress. As a corollary, it is likely that the distribution of microRNAs between HMW-RISC and LMW-RISC can provide additional information concerning the contribution of individual microRNAs to gene regulation during physiological and/or pathological conditions. Further analysis will clarify whether microRNA distribution is predictive of which microRNAs are engaged in translational repression. If so, such analyses may provide a tool to generate microRNA utilization profiles, which will complement the ongoing analysis of microRNA by expression profiling.
Materials and Methods
T-Cell Stimulation.
Splenic T cells were isolated from 6- to 12-wk-old C57/BL6 mice and activated using antibody-coated microbeads in the presence or absence of recombinant mouse IL-2 (rIL-2, Becton Dickinson).
Size Exclusion Chromatography and Assessment of MicroRNA Distribution Profiles.
Crude cell lysate was processed through Superose 6 column (GE Healthcare) loaded on an AKTA FPLC system (GE Healthcare). Small RNA was isolated from each Superose 6 fraction by miRNeasy Serum/Plasma Kit (QIAGEN). Next, RNA was ligated to a barcoded 3′ adapter, pooled, ligated to a common 5′ adapter, reverse transcribed into cDNA, PCR amplified, and sequenced on the Illumina HiSeq platform.
MicroRNA Reporters.
MicroRNA function was tested using Dual-Luciferase Reporter System (Promega). MicroRNA reporters containing specific target sequences at the 3′ end of luciferase genes were cotransfected with control reporters to normalize transfection efficiency.
A detailed description of methods and reagents can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Chao Lu (The Rockefeller University) for technical assistance with the 3T3 L1 differentiation assay; Jayanta Chaudhuri for sharing the AKTA FPLC system; and Christine Mayr, Raymond Wu, Julija Hmeljak, Ping Mu, and members of the C.B.T. laboratory for critical discussions and feedback on the manuscript. This work has been funded by the National Institutes of Health Grants R01 CA105463 (to C.B.T.), K99 CA175189 (to S.H.O.), R01 CA149707 GRANT10355609 (to A.V.), U01-CA164190 and U24-CA143840 (to C.S.L.), and Memorial Sloan-Kettering Cancer Center Support Grant P30 CA008748.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing datasets analyzed in this paper are available for download at cbio.mskcc.org/public/Leslie/PNAS2015_Thompson/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424217112/-/DCSupplemental.
References
- 1.Béthune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13(8):716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 5.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: An Ago-centric perspective. Cold Spring Harb Perspect Biol. 2011;3(10):a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17(4):438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakymiw A, et al. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7(12):1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7(12):1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11(11):1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höck J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8(11):1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14(12):2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olejniczak SH, La Rocca G, Gruber JJ, Thompson CB. Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proc Natl Acad Sci USA. 2013;110(1):157–162. doi: 10.1073/pnas.1219958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13(12):1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 16.Ho JJ, Marsden PA. Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip Rev RNA. 2014;5(1):69–86. doi: 10.1002/wrna.1197. [DOI] [PubMed] [Google Scholar]
- 17.Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013;32(7):1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burroughs AM, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8(1):158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Androsavich JR, Chau BN, Bhat B, Linsley PS, Walter NG. Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver. RNA. 2012;18(8):1510–1526. doi: 10.1261/rna.033308.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, et al. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117(Pt 23):5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 21.Frauwirth KA, Thompson CB. Activation and inhibition of lymphocytes by costimulation. J Clin Invest. 2002;109(3):295–299. doi: 10.1172/JCI14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshii T, et al. Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2014;111(10):3805–3810. doi: 10.1073/pnas.1320265111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang F, et al. A critical role for Rictor in T lymphopoiesis. J Immunol. 2012;189(4):1850–1857. doi: 10.4049/jimmunol.1201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, et al. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS ONE. 2013;8(1):e54221. doi: 10.1371/journal.pone.0054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichhorn SW, et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56(1):104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12(4):340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 30.Vogel J. Biochemistry. A bacterial seek-and-destroy system for foreign DNA. Science. 2014;344(6187):972–973. doi: 10.1126/science.1252962. [DOI] [PubMed] [Google Scholar]
- 31.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336(6084):1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 33.Gibbings D, et al. Human prion protein binds Argonaute and promotes accumulation of microRNA effector complexes. Nat Struct Mol Biol. 2012;19(5):517–524, S511. doi: 10.1038/nsmb.2273. [DOI] [PubMed] [Google Scholar]
- 34.Wu PH, Isaji M, Carthew RW. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol Cell. 2013;52(1):113–123. doi: 10.1016/j.molcel.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 36.Kume H, Hino K, Galipon J, Ui-Tei K. A-to-I editing in the miRNA seed region regulates target mRNA selection and silencing efficiency. Nucleic Acids Res. 2014;42(15):10050–10060. doi: 10.1093/nar/gku662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131(7):1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 38.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505(7485):706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King IN, et al. The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J Biol Chem. 2014;289(20):14263–14271. doi: 10.1074/jbc.M114.561902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.