Significance
We report that during a recent period of hybridization between two major African malaria mosquitoes, Anopheles gambiae and Anopheles coluzzii, an island of divergence on chromosome 2 introgressed from the A. gambiae into the A. coluzzii genome and its frequency subsequently increased. This introgression was coincident with the start of a major insecticide-treated bed net campaigns in Mali. These observations suggest that increased insecticide exposure acted as a selective force sufficient to drive introgression of an entire genomic island of divergence across the reproductive barrier separating these two species. This study provides a rare example of adaptive introgression in an animal species and elucidates the dynamics of how insecticide resistance evolved in A. coluzzii.
Keywords: hybridization, gene flow, Anopheles gambiae, kdr, knockdown resistance
Abstract
Animal species adapt to changes in their environment, including man-made changes such as the introduction of insecticides, through selection for advantageous genes already present in populations or newly arisen through mutation. A possible alternative mechanism is the acquisition of adaptive genes from related species via a process known as adaptive introgression. Differing levels of insecticide resistance between two African malaria vectors, Anopheles coluzzii and Anopheles gambiae, have been attributed to assortative mating between the two species. In a previous study, we reported two bouts of hybridization observed in the town of Selinkenyi, Mali in 2002 and 2006. These hybridization events did not appear to be directly associated with insecticide-resistance genes. We demonstrate that during a brief breakdown in assortative mating in 2006, A. coluzzii inherited the entire A. gambiae-associated 2L divergence island, which includes a suite of insecticide-resistance alleles. In this case, introgression was coincident with the start of a major insecticide-treated bed net distribution campaign in Mali. This suggests that insecticide exposure altered the fitness landscape, favoring the survival of A. coluzzii/A. gambiae hybrids, and provided selection pressure that swept the 2L divergence island through A. coluzzii populations in Mali. We propose that the work described herein presents a unique description of the temporal dynamics of adaptive introgression in an animal species and represents a mechanism for the rapid evolution of insecticide resistance in this important vector of human malaria in Africa.
The major African malaria vector, Anopheles gambiae, has long been known to exist in nature as two distinct and sympatric populations. Initially described as the M and S molecular forms, they are now recognized species, designated Anopheles coluzzii and Anopheles gambiae, respectively (1). The two species exhibit varying degrees of reproductive isolation (RI) across their range (2–6), and recent studies provide strong evidence for reduced hybrid fitness in nature (5).
Previous studies comparing the genomes of A. gambiae and A. coluzzii revealed that divergence, as measured by FST values, is highest at three discrete pericentromeric regions occurring on each of chromosomes X, 2, and 3 and collectively comprise approximately 3% of the genome. These regions have been described as islands of speciation (7, 8). In a reanalysis of these data, Cruickshank and Hahn (9) rejected the islands of speciation concept, claiming that the observed islands are an artifact of the application of FST used to identify them, and concluded that the islands are related to reduced diversity in the regions of the genome in which they reside, and not to reduced gene flow. A panel of 15 species-specific SNP markers contained within these genomic regions was developed to facilitate the analysis of gene flow and introgression in natural populations (10).
A recent large-scale study using this SNP panel demonstrated the widespread occurrence of F1 and backcross hybrids (5), clearly demonstrating gene flow between A. coluzzii and A. gambiae, despite any issues arising from the application of FST. A longitudinal survey conducted as part of that study revealed temporal as well as spatial heterogeneity, with long periods of strong RI interrupted by episodes of hybridization (i.e., abrupt appearance of F1 genotypes), followed by the disappearance of hybrid genotypes and reestablishment of disequilibrium. A different pattern emerged after an episode of hybridization that occurred in 2006, when SNPs on the X and third chromosome did return to high linkage disequilibrium (LD; R2 > 0.8), but chromosome 2 SNPs, previously fixed in A. gambiae, introgressed into the A. coluzzii genome (5).
An important phenotypic difference between A. coluzzii and A. gambiae lies in their susceptibility to the insecticides widely used for malaria control. The knockdown resistance SNP, L1014F (11), in the para voltage-gated sodium channel gene (kdr) is located ∼2.4 Mbp from the 2L centromere and within the introgressed region identified by Lee et al. (5). The L1014F SNP is thought to have arisen in West African A. gambiae populations (12) and confers target site resistance to dichlorodiphenyltrichloroethane (DDT) and pyrethroid insecticides (11, 13). Recent and intense pyrethroid-based control programs are thought to have strongly selected for L1014F in A. gambiae (14, 15). L1014F has been present in some A. gambiae populations for at least 20 y (14), but was found to be absent in sympatric A. coluzzii populations (16–18). The first report of L1014F in A. coluzzii came in 2000 from samples collected in southern Benin. Based on intron sequencing, L1014F was thought to have emerged in A. coluzzii via introgression from sympatric A. gambiae (12). A recent detailed genome sequencing study clearly described how the L1014F mutation and the entire 2L island of divergence has introgressed from A. gambiae into A. coluzzii in southern Ghana. The authors reported that RI was maintained, and thus concluded that the 2L island is not critical for RI.
We undertook the present study to determine whether introgression of the 2L island, including L1014F, can be related to the recently expanded insecticide-treated bed net (ITN) campaign in Africa. We explored the role of L1014F in the breakdown of assortative mating that result in the periodic increase in hybrids, as well as the potential role of the 2L island in speciation. Finally, we wished to further characterize the dynamics of this rare example of adaptive introgression in an animal species.
Results
A total of 1,076 A. gambiae and A. coluzzii specimens were genotyped for three SNPs that are diagnostic for species (10) and the 1,014th codon variants TTA (leucine) and TTT (phenylalanine) of the kdr gene (SI Appendix, Table S1). Samples were collected before, during, and after the 2006 hybridization event that occurred in the town of Selinkenyi (n = 632) in southwestern Mali, as well as from two other villages in Mali, Sidarebougou (n = 334) and Tissana (n = 110) (SI Appendix, Figs. S1 and S2).
Mating Structure of Sympatric Populations.
To determine whether the breakdown in assortative mating observed in 2006 is associated with the L1014F SNP itself, we tested the null hypothesis that mating is random with respect to the L1014F genotype. All A. coluzzii individuals before 2006 (n = 226) were susceptible homozygotes (+/+). The A. gambiae population included individuals with all three genotypes at the following frequencies: +/+, 0.33; +/r, 0.41; and r/r, 0.26. The observed frequency of L1014F genotypes among the 17 F1 hybrids collected in 2006 did not differ from those expected assuming random mating (Table 1); therefore, mating does not appear to have been directly influenced by the L1014F genotype.
Table 1.
kdr and 2L SNP genotype frequencies
| kdr | 2L SNP | |||||
| r/r | r/+ | +/+ | g/g | c/g | c/c | |
| A. gambiae | ||||||
| Pre-2006 observed | 2 | 5 | 3 | 11 | 0 | 0 |
| 2006 observed | 7 | 8 | 8 | 24 | 0 | 0 |
| Post-2006 observed | 57 | 3 | 3 | 62 | 0 | 0 |
| F1 | ||||||
| 2006 observed | 0 | 7 | 10 | — | — | — |
| 2006 expected | 0 | 7.9NS | 9.1NS | — | — | — |
| A. coluzzii | ||||||
| Pre-2006 observed | 0 | 0 | 226 | 0 | 0 | 133 |
| 2006 observed | 0 | 7 | 70 | 0 | 12 | 69 |
| Post-2006 observed | 94 | 127 | 79 | 87 | 125 | 86 |
| Post-2006 expected* | ||||||
| Lower bound† | 0.6 | 25.8 | 273.6 | 1.6 | 40.8 | 255.5 |
| Upper bound‡ | 1.6 | 41.1 | 257.2 | 6.5 | 75.1 | 216.3 |
The susceptible allele for kdr is denoted by “+,” and the resistance allele is denoted by “r.” For the 2L island SNP, “g” denotes the A. gambiae-specific SNP and “c” denotes the A. coluzzii-specific SNP. Observed F1 kdr genotype frequencies do not depart from H-W expectations, indicating that mating is random with respect to kdr, and that the kdr gene is not directly related to the breakdown in assortative mating. Both kdr + and r alleles were present in A. gambiae populations before 2006, but absent in A. coluzzii. A highly significant excess of resistance genotypes in post-2006 A. coluzzii suggests strong selection for kdr-r. P values were calculated from G tests comparing post-2006 observed and expected genotype frequencies.
P < 2.2e-16.
Lower bound: assumes no hybridization after 2006; thus, mating only occurred within A. coluzzii with a minimum allele frequency of kdrr and the A. gambiae 2L island SNP.
Upper bound: assumes asymmetric hybridization after 2006, with F1 individuals preferentially mating back with A. coluzzii, providing a maximum allele frequency of the resistance allele for kdrr and the A. gambiae 2L island SNP.
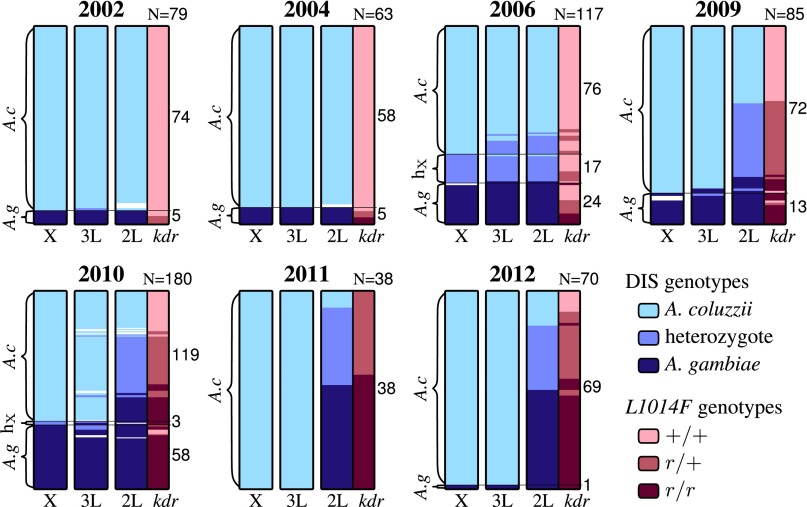
A. coluzzii and A. gambiae are defined on the basis of alternate and fixed SNPs on the X chromosome (1), and backcrossed individuals can be distinguished by their mixed genotypes for species-specific SNPs on the autosomes (10). The frequency of F1 hybrids was 14.5% in 2006 (n = 117), but dropped to zero in subsequent years (2009, 2010, and 2012; n = 335), indicating reestablishment of assortative mating after 2006 (Fig. 1 and Table 1). Backcross individuals were common in post-2006 populations and were composed mainly of individuals with the A. gambiae-associated chromosome 2L SNP in an otherwise A. coluzzii genome (Fig. 1). The frequency of backcrossed genotypes was substantially higher in post-2006 A. coluzzii populations than in A. gambiae (71.1% vs. 6.1%; Fig. 1). The 2L island and L1014F were found to be in very tight LD in post-2006 A. coluzzii populations (R2 = 0.65–0.96), with the resistance allele nearly always co-occurring with the A. gambiae-specific 2L SNP (SI Appendix, Table S2). Therefore, gene flow in backcrossed generations (2009, 2010, 2011, and 2012) appears to have been strongly asymmetric, with hybrids successfully mating with A. coluzzii but only occasionally with A. gambiae. In this case, introgressive hybridization mostly involved the transfer of the L1014F from A. gambiae to A. coluzzii.
Fig. 1.
SNP evidence for introgression of the 2L speciation island and kdr from A. gambiae into A. coluzzii. Genotypes for the three divergence island SNPs (DIS) and the L1014F SNP (kdr resistance) in Selenkenyi/Kela populations over 10 y. Columns represent SNPs (X divergence island, 3L island, 2L island, L1014F), individual mosquitoes are represented by colored horizontal lines, with individuals stacked vertically. Light blue indicates homozygous for A. coluzzii-associated alleles; dark blue, homozygous for A. gambiae-associated alleles; purple, heterozygous; white, missing data. L1014F genotypes are shown in dark red for homozygous resistant (r/r), salmon for heterozygous (r/+), and light pink for homozygous-susceptible (+/+). Population assignments [A. coluzzii, hybrid (hx), and A. gambiae] are indicated by brackets on the left of each heat map. Sample sizes are listed on top right of each figure, and sample size per population is provided along the right side. A “burst” of hybridization in 2006 is evident from the presence of individuals that are heterozygous across all (F1) or multiple (backcrossed, designated hx) DIS SNPs. There is no obvious association between these hybrids and L1014F. By 2009, individuals heterozygous at the X and 3L DIS are virtually absent, but more than 50% of A. coluzzii carry both the A. gambiae typical 2L DIS and L1014F with an almost perfect association between these two SNPs. As time progresses, the frequency L1014F continues to increase, but the linkage weakens. By 2011, there are numerous individuals with L1014F and the A. coluzzii typical 2L DIS, and by 2012, the frequency of the A. gambiae typical 2L DIS begins to decrease in A. coluzzii.
Selection for Insecticide Resistance in Post-2006 A. coluzzii Populations.
It appears that A. coluzzii hybrid individuals carrying the resistance SNP were strongly selected for and backcrossed with parental populations (2009–2012 in Fig. 1). In Selenkenyi, only 2 out of 280 backcrossed individuals were homozygous-susceptible (+/+), a significant deficiency relative to expected (Nexp = 24.4; P < 0.0001 using randomization goodness of fit with 10,000 replicates). Lee et al. (5) showed that an earlier hybridization episode in 2002, before the introduction of ITNs in Selenkenyi, was followed by strong selection against hybrids, presumably related to ecological maladaptation or decreased mating success. Thus, it appears that the introduction of L1014F in the presence of insecticide pressure was a key factor that allowed hybrids to overcome an intrinsic selective disadvantage and backcross with the A. coluzzii parental population, contributing to gene flow between the two species.
Interestingly, the A. gambiae-associated 2L SNP and linked L1014F were detected in an allopatric A. coluzzii population collected in 2011 from the village of Tissana, nearly 400 km north of Selenkenyi (SI Appendix, Fig. S2). This is a dry region in which A. gambiae is absent, suggesting that once L1014F emerged in A. coluzzii, it spread rapidly via gene flow between geographically distant A. coluzzii populations.
Selection for L1014F was 10-fold stronger in A. coluzzii than in sympatric A. gambiae (A. coluzzii: s = 0.13–0.14; A. gambiae: s = 0.065–0.014) (SI Appendix, Table S3), owing to either (i) the need for the resistance allele in the presence of insecticide to overcome the selective disadvantage inherent in being a hybrid or (ii) phenotypic penetrance of L1014F differing in the A. gambiae genetic background.
Patterns of Genomic Introgression and Selection.
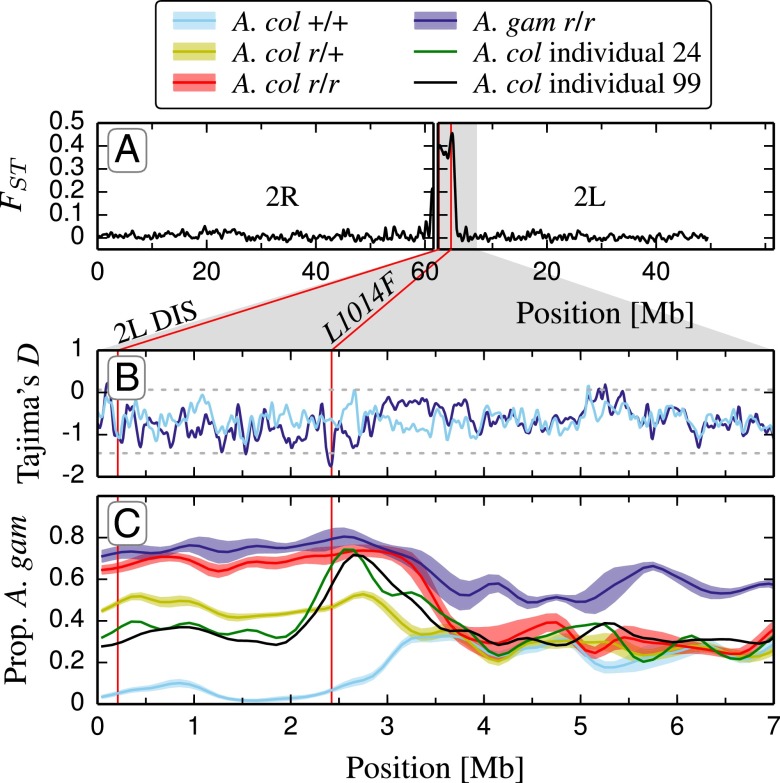
To explore the chromosomal extent of introgression, we compared individual mosquito whole-genome sequences of pre-2006 and post-2006 A. coluzzii and A. gambiae. FST values between pre-2006 and post-2006 A. coluzzii were elevated throughout a 3-Mbp region near the 2L centromere, where FST peaked at 0.45, around the kdr gene (Fig. 2A). Tajima’s D statistic provides evidence consistent with a strong selective sweep at the location of kdr in the A. gambiae genome that is not present in pre-2006 A. coluzzii (Fig. 2B). Calculation of the proportion of A. gambiae “ancestry” for this region showed that among backcrossed A. coluzzii individuals, those homozygous for the resistance allele (r/r) were genetically more similar to A. gambiae in a region from 0 to 3 Mbp, whereas heterozygotes had intermediate ancestry (Fig. 2C). This indicates that the A. gambiae 2L pericentromeric region was inherited as a single, independently assorting unit. No other major introgressed regions were detected (SI Appendix, Fig. S3), in agreement with the results reported by Clarkson et al. (19).
Fig. 2.
Genomic evidence of 2L introgression from A. gambiae into A. coluzzii. (A) FST between nonintrogressed A. coluzzii (pre-2006, +/+) and A. coluzzii carrying the A. gambiae typical 2L DIS (post-2006, r/+ and r/r) across chromosome 2. Vertical red lines denote the positions of the 2L DIS and L1014F SNPs. The high FST value from the 2L centromere to just past L1014F suggests introgression. Gray shading indicates zoomed-in region for B and C. (B) Tajima’s D in nonintrogressed, kdr +/+ A. coluzzii (light blue) and A. gambiae, kdr r/r (dark blue), suggesting selection near kdr. Computed in 5-kb windows, with gray dashed lines showing two SDs from the overall mean. Lines are smoothed with a 9-point Gaussian window. (C) Proportion of A. gambiae “ancestry” for A. gambiae (purple), preintrogression A. coluzzii (blue), homozygous introgressed A. coluzzii (red), and heterozygous introgressed A. coluzzii (yellow). Computed in 100-kb windows. Lines are smoothed with a 9-point Gaussian window, and widths indicate the SE among individuals. Two additional individuals (solid black and green lines) collected in 2012 illustrate the breakdown of linkage between the 2L DIS and L1014F, where the 2-Mb centromeric region shows a return to A. coluzzii ancestry.
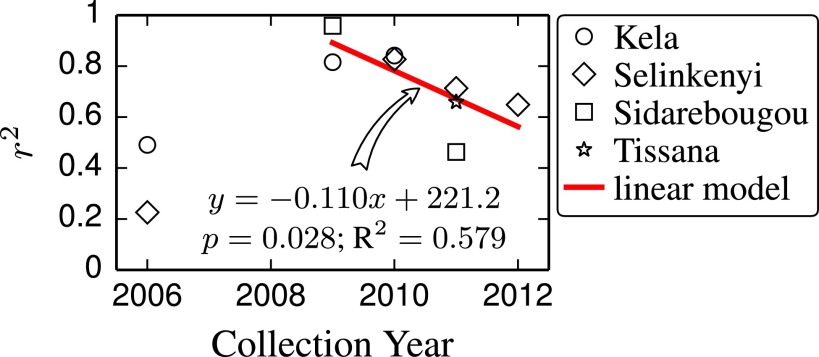
Individual mosquito whole-genome sequencing revealed two recombinant individuals (samples 24 and 99) collected in 2012, both of which were r/r while reestablishing the A. coluzzii sequence in the first 2 Mb of chromosome 2L (Fig. 2C). This observation is consistent with a general decline in LD over time between the 2L SNP and L1014F (Fig. 3). Taken together, these observations suggest that it is kdr, and not neighboring genes, that is under sufficiently strong selection to drive introgression in this case. However, selection may be driving the return of A. coluzzii genotypes in 2L outside the L1014F locus.
Fig. 3.
Breakdown of linkage between the 2L island and L1014F in introgressed A. coluzzii. LD (R2) between the 2L DIS and L1014F SNPs (2.2 Mb apart) in A. coluzzii over time is shown for four sites. Post-2006 points are fit to a linear model.
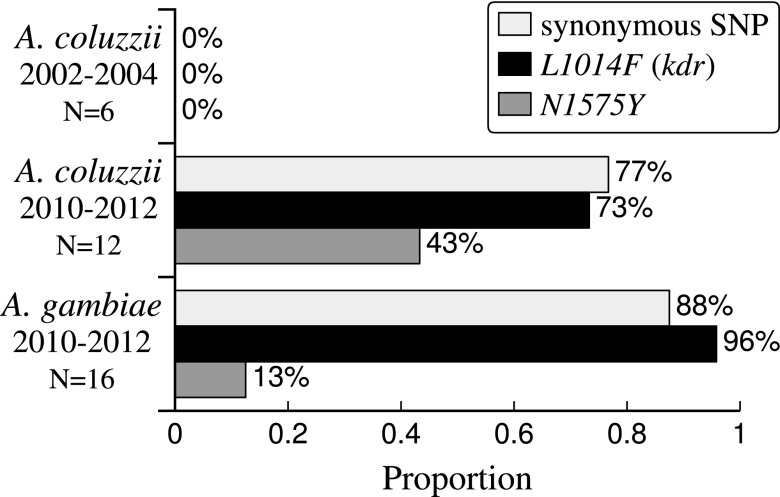
Adaptive introgression often involves the transfer of complex adaptations via “cassettes” of multiple, linked mutations (20). Previous work has shown that L1014F-based resistance to pyrethroids and DDT can be modulated by other mutations in the kdr gene (21). Using the whole-genome sequencing data, we investigated two additional kdr gene SNPs that have been shown to affect pyrethroid resistance (SI Appendix, Fig. S4). The N1575Y SNP is associated with increased pyrethroid resistance and is found only on a resistant genetic background of L1014F (22). A synonymous SNP located 4,974 bp upstream of the L1014F was detected in association mapping for pyrethroid resistance (21). Both of these mutations were absent in pre-2006 A. coluzzii, but present in more than 43% of post-2006 A. coluzzii (Fig. 4).
Fig. 4.
Cointrogression of L1014F and nearby resistance SNPs. Genotype frequencies for the minor-effect SNPs that have cointrogressed within the L1014F, identified in whole genome sequenced samples. L1014F is at 2L: 2,422,652. A synonymous/intronic [C/T] SNP located 4,974 bp upstream of L1014F was identified by Weetman et al. (20) as significantly correlated with permethrin resistance. The N1575Y [A/T] SNP located 7,093 bp downstream from L1014F was identified by Jones et al. (50), and has been shown to increase phenotypic pyrethroid resistance in an L1014F genotypic background. All three SNPs were absent in pre-2006 A. coluzzii collections but present in both post-2010 A. coluzzii and A. gambiae. Pre-2006 A. gambiae data were not available for these SNPs, but their presence is presumed.
Discussion
Where A. coluzzii and A. gambiae occur in sympatry, RI maintained by assortative mating occasionally breaks down, resulting in the appearance of F1 hybrids. Factors resulting in this breakdown remain unclear. Occasionally backcross genotypes are subsequently observed, but before 2006 these ultimately disappeared from populations, including those with an introgressed 2L island of divergence, presumably because they have reduced fitness (5). In the town of Selinkenyi, Mali, a breakdown in assortative mating occurred during the summer of 2006, resulting in F1 hybrids at a frequency of 14.5%. In this case, A. coluzzii individuals with A. gambiae chromosome 2L genotypes persisted, and their frequency increased between 2010 and 2012 (Fig. 1); however, A. coluzzii individuals carrying A. gambiae-specific chromosome 3 and X islands of divergence disappeared. Introgression of the 2L region included an insecticide-resistance allele present in A. gambiae but previously absent from sympatric A. coluzzii.
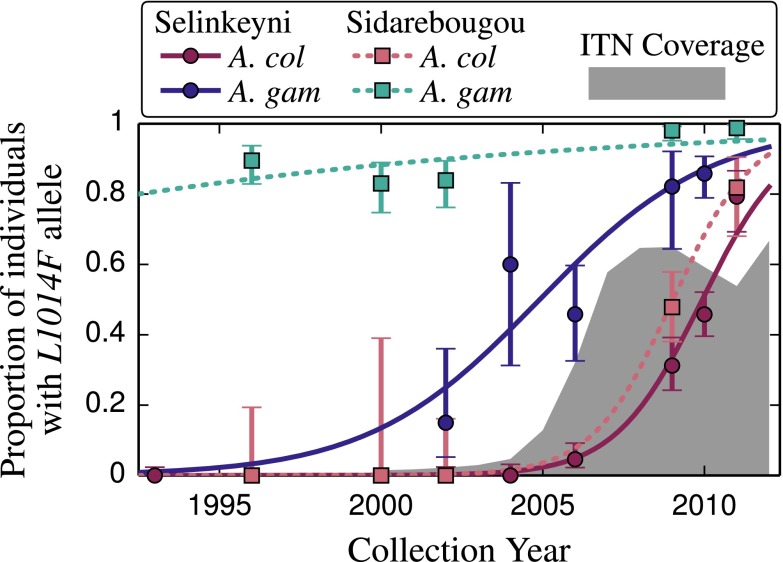
The survival and increase in frequency of backcrossed hybrids is likely related to the widespread and organized implementation of ITNs in Mali starting in 2005 (23–25), resulting in strong selection for insecticide-resistant individuals (Fig. 5). Supporting this idea is the observation that in 2002, before ITN distribution in Mali, a breakdown of assortative mating at Selinkenyi resulted in hybrid individuals reaching a frequency of 12%, but resampling of the same site in 2004 revealed no evidence of 2L introgression (5), along with a zero frequency of the kdr resistance allele (Table 1).
Fig. 5.
kdr resistance allele L1014F frequency over 20 y in sympatric populations of A. coluzzii and A. gambiae. Data before 2006 are from Fanello et al. (17) and Tripet et al. (13). Solid lines (Selenkenyi/Kela) and dashed lines (Sidarebougou) show selection model estimation. ITN coverage is indicated in the gray shaded area of the graph. ITN data were taken from the World Health Organization’s Mali country profile (22).
We have found no evidence suggesting that the pericentromeric region of 2L is directly involved with RI. However, with the exception of kdr and its immediate surroundings, we observed that the introgressed region reverted to the typical A. coluzzii form over time. This finding strongly suggests divergent selection and/or epistatic interactions between the pericemtromeric region of 2L and elsewhere in the genome, which drives differentiation between A. gambiae and A. coluzzii in this region. These observations suggest that divergent selection on 2L remains strong, lending support to the possibility that the 2L island indeed may represent a “speciation island” promoting divergence between these closely related species.
Selection for L1014F had been strong in the A. gambiae population in the Sidarebougou region for decades, likely related to widespread pyrethroid use in cotton agriculture in this region (13, 14). Likewise, before L1014F was present in A. gambiae, although at a lower frequency, in Selinkenyi, where intermediate usage of pyrethroid on crops has been reported (14). In 2005, the Malian Ministry of Health began providing free ITNs to children (26), and the President’s Malaria Initiative began operations in Mali shortly thereafter, providing 369,800 ITNs in 2007 (24). By 2007, 20–40% of children age <5 y slept under an ITN in the provinces where our study sites are located (27). The rapid scale-up of insecticide-based malaria control measures in Africa during the first decade of the 2000s has had measurable effects on malaria vectors, especially on A. gambiae and A. coluzzii, which are highly dependent on humans for blood and indoor resting sites. In some cases, selection pressure from these measures has been strong enough to drive A. gambiae and A. coluzzii to local extinction (28). In addition, the feeding behavior of Anopheles arabiensis, a sister species of A. gambiae and A. coluzzii, has shifted to biting outdoors and on nonhuman hosts (28), thereby avoiding contact with ITNs. A. coluzzii seems to have responded to insecticide selection pressure by co-opting pyrethroid and DDT resistance, in the form of the L1014F mutation, from A. gambiae through adaptive introgression.
Interspecific hybridization due to anthropogenic habitat disturbance has been described in plant systems (29), but well-documented examples are rare in animal taxa (30). Adaptive introgression could be rare in animals because (i) hybridization between animal species is rare (30–32) and/or (ii) hybrids normally have decreased fitness (33–35). For adaptive introgression to occur, a beneficial mutation should cross the species barrier through a rare hybridization event and overcome any fitness cost incurred from hybrid incompatibilities. The most well-characterized animal examples include warfarin resistance in European house mice (36) and melanism in North American gray wolves (37). Adaptive introgression of an insecticide-resistance gene between A. gambiae and A. coluzzii has been reported in numerous papers (12, 19, 38–41). These examples suggest that reproductive barriers between animal species break down facilitating adaptation to changes in the environment.
Our results distinguish the role of hybridization and adaptation. This longitudinal study in Selinkenyi, Mali provides a unique view of adaptive introgression by documenting its evolution before, during, and after the transfer of an adaptive genetic variant (insecticide resistance) from a donor (A. gambiae) to a recipient species (A. coluzzii). The susceptible genotypes found in F1 hybrids (Table 1) suggest that the kdr resistance SNP did not directly affect assortative mating. In other words, hybridization did not occur preferentially for A. coluzzii and A. gambiae carrying the kdr resistance SNP. Hybridization between A. coluzzii and A. gambiae is fairly common (>1%) (5, 42, 43). A drastic decline in hybrid genotypes in 2004 following a bout of hybridization in 2002 (1% F1 and 11% other hybrids) indicates that A. coluzzii/A. gambiae hybrids typically are less fit in nature (5). Backcrossed hybrids were found at fairly high frequencies (>3%) in Selinkenyi populations in all years examined (5). This finding suggests a potential for A. coluzzii to acquire the kdr resistance SNP (and the 2L divergence island containing it) from sympatric A. gambiae before 2006, but this did not occur. We propose that the introduction of ITNs in 2005–6 changed the fitness landscape and, combined with a bout of hybridization, provided an environment in which previously susceptible A. coluzzii populations acquired adaptive insecticide resistant genotypes that increased in frequency to near fixation by 2012. Thus, in this case, adaptive introgression is a byproduct of hybridization, not its cause.
Materials and Methods
Sample Collection.
A. coluzzii and A. gambiae were collected by mouth aspiration inside houses from three sites in Mali: Selenkenyi/Kela (in 2002, 2004, 2006, 2009, 2010, 2011, and 2012), Sidarebougou (in 2002, 2009, and 2011), and Tissana (in 2006 and 2011), during the rainy season (August–October). The samples from Kela, which is located 20 km from Selinkenyi, were grouped with those from Selinkenyi because of geographic proximity and species composition similarity. DNA was extracted using Biosprint (Qiagen).
SNP Genotyping.
A totals of 1,076 samples were analyzed with an iPLEX Gold multiplexed SNP genotyping array at five loci: one in the X divergence island (28S rDNA intergenic sequence, also used to differentiate A. coluzzii and A. gambiae), one in the 2L divergence island (8), one in the 3L divergence island (8), and L1014F [TTA→TTT; (11)] SNPs (SI Appendix, Table S1). The genotypes of the three divergence island SNPs determined species. Data were used to calculate standard molecular indices, Hardy–Weinberg equilibrium, SNP frequency, and significance of LD using Arlequin 3.1 (44). R2 values were calculated using the maximum likelihood approach implemented in the EMLD program (45, 46).
Selection Coefficient Modeling.
Selection coefficients for each site were estimated by fitting resistance allele frequency per year in the corresponding location to a recursive selection equation (47) using the leastsq option in the scipy.optimize package in Python (scipy.org). Details are provided in SI Appendix.
Whole-Genome Sequencing.
Thirty-three individual mosquitoes from Selenkenyi (12 A. gambiae, 6 pre-2006 A. coluzzii, and 15 post-2006 A. coluzzii) were whole-genome sequenced at 5–10× coverage per individual. Library preparations were made with 25–50 ng genomic DNA input, using the Nextera DNA Sample Preparation Kit and TruSeq dual indexing barcodes (Illumina). Samples were sequenced on the Illumina HiSeq2500 platform with paired-end, 100-bp reads. Reads were mapped to the PEST AgamP3 reference genome version 3.7 using Stampy (48) and BWA-mem (49). VCFtools (50) was used to call SNPs and calculate FST values, Tajima’s D statistics, and A. gambiae vs. A. coluzzii ancestry. The methodology is described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Allison Weakly and Catelyn Nieman for assistance with DNA extraction and genotyping; Julia Malvick and the University of California Davis Veterinary Genetics Laboratory for assistance with iPLEX genotyping; Dr. Sekou Traore and his team at the Malaria Research and Training Center at the University of Bamako; the QB3 Vincent J Coates Genomics Sequencing Laboratory at University of California Berkeley; Adama Sacko, Rebecca Trout-Fryxell, Stephanie Seifert, and Michelle Sanford for help with field collection; and Clare Marsden for valuable comments on the manuscript. Funding was provided by National Institutes of Health Grants T32AI074550, R01 AI 078183, and D43TW007390.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All karyotype and SNP data have been deposited in the PopI OpenProject “AgKDR” database (popi.ucdavis.edu/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418892112/-/DCSupplemental.
References
- 1.Coetzee M, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619(2):246–274. [PubMed] [Google Scholar]
- 2.della Torre A, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10(1):9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 3.Marsden CD, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20(23):4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riehle MM, et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331(6017):596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 2013;110(49):19854–19859. doi: 10.1073/pnas.1316851110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor C, et al. Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics. 2001;157(2):743–750. doi: 10.1093/genetics/157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19(5):925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23(13):3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Marsden CD, Nieman C, Lanzaro GC. A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour. 2014;14(2):297–305. doi: 10.1111/1755-0998.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Torres D, et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7(2):179–184. doi: 10.1046/j.1365-2583.1998.72062.x. [DOI] [PubMed] [Google Scholar]
- 12.Weill M, et al. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol Biol. 2000;9(5):451–455. doi: 10.1046/j.1365-2583.2000.00206.x. [DOI] [PubMed] [Google Scholar]
- 13.Reimer L, et al. Relationship between kdr mutation and resistance to pyrethroid and DDT insecticides in natural populations of Anopheles gambiae. J Med Entomol. 2008;45(2):260–266. doi: 10.1603/0022-2585(2008)45[260:rbkmar]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Tripet F, et al. Longitudinal survey of knockdown resistance to pyrethroid (kdr) in Mali, West Africa, and evidence of its emergence in the Bamako form of Anopheles gambiae s.s. Am J Trop Med Hyg. 2007;76(1):81–87. [PubMed] [Google Scholar]
- 15.Lynd A, et al. Field, genetic, and modeling approaches show strong positive selection acting upon an insecticide resistance mutation in Anopheles gambiae s.s. Mol Biol Evol. 2010;27(5):1117–1125. doi: 10.1093/molbev/msq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandre F, et al. Current distribution of a pyrethroid resistance gene (kdr) in Anopheles gambiae complex from west Africa and further evidence for reproductive isolation of the Mopti form. Parassitologia. 1999;41(1-3):319–322. [PubMed] [Google Scholar]
- 17.Awolola TS, Brooke BD, Koekemoer LL, Coetzee M. Absence of the kdr mutation in the molecular “M” form suggests different pyrethroid resistance mechanisms in the malaria vector mosquito Anopheles gambiae s.s. Trop Med Int Health. 2003;8(5):420–422. doi: 10.1046/j.1365-3156.2003.01034.x. [DOI] [PubMed] [Google Scholar]
- 18.Fanello C, et al. The pyrethroid knock-down resistance gene in the Anopheles gambiae complex in Mali and further indication of incipient speciation within An. gambiae s.s. Insect Mol Biol. 2003;12(3):241–245. doi: 10.1046/j.1365-2583.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- 19.Clarkson CS, et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5:4248. doi: 10.1038/ncomms5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26(2):229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 21.Weetman D, et al. Association mapping of insecticide resistance in wild Anopheles gambiae populations: Major variants identified in a low-linkage disequilbrium genome. PLoS ONE. 2010;5(10):e13140. doi: 10.1371/journal.pone.0013140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CM, et al. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci USA. 2012;109(17):6614–6619. doi: 10.1073/pnas.1201475109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization 2013 Country profile: Mali. Available at: www.who.int/countries/mli/en/. Accessed July 8, 2014.
- 24.President’s Malaria Initiative 2013 Country profile for Mali. Available at: www.pmi.gov/docs/default-source/default-p-library/country-profiles/mali_profile.pdf?sfvrsn=14. Accessed July 8, 2014.
- 25.Flaxman AD, et al. Rapid scaling up of insecticide-treated bed net coverage in Africa and its relationship with development assistance for health: A systematic synthesis of supply, distribution, and household survey data. PLoS Med. 2010;7(8):e1000328. doi: 10.1371/journal.pmed.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization . World Malaria Report 2012. World Health Organization; Geneva, Switzerland: 2012. [Google Scholar]
- 27.Noor AM, Mutheu JJ, Tatem AJ, Hay SI, Snow RW. Insecticide-treated net coverage in Africa: Mmapping progress in 2000-07. Lancet. 2009;373(9657):58–67. doi: 10.1016/S0140-6736(08)61596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govella NJ, Chaki PP, Killeen GF. Entomological surveillance of behavioural resilience and resistance in residual malaria vector populations. Malar J. 2013;12:124. doi: 10.1186/1475-2875-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crispo E, Moore JS, Lee-Yaw JA, Gray SM, Haller BC. Broken barriers: Human-induced changes to gene flow and introgression in animals. An examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays. 2011;33(7):508–518. doi: 10.1002/bies.201000154. [DOI] [PubMed] [Google Scholar]
- 30.Hedrick PW. Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 2013;22(18):4606–4618. doi: 10.1111/mec.12415. [DOI] [PubMed] [Google Scholar]
- 31.Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. [Google Scholar]
- 32.Mallet J, Dasmahapatra KK. Hybrid zones and the speciation continuum in Heliconius butterflies. Mol Ecol. 2012;21(23):5643–5645. doi: 10.1111/mec.12058. [DOI] [PubMed] [Google Scholar]
- 33.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 34.Svedin N, Wiley C, Veen T, Gustafsson L, Qvarnström A. Natural and sexual selection against hybrid flycatchers. Proc Biol Sci. 2008;275(1635):735–744. doi: 10.1098/rspb.2007.0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burke JM, Arnold ML. Genetics and the fitness of hybrids. Annu Rev Genet. 2001;35:31–52. doi: 10.1146/annurev.genet.35.102401.085719. [DOI] [PubMed] [Google Scholar]
- 36.Song Y, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21(15):1296–1301. doi: 10.1016/j.cub.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson TM, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323(5919):1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.della Torre A, Merzagora L, Powell JR, Coluzzi M. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics. 1997;146(1):239–244. doi: 10.1093/genetics/146.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diabate A, et al. KDR mutation, a genetic marker to assess events of introgression between the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in the tropical savannah area of West Africa. J Med Entomol. 2003;40(2):195–198. doi: 10.1603/0022-2585-40.2.195. [DOI] [PubMed] [Google Scholar]
- 40.Diabate A, et al. The spread of the Leu-Phe kdr mutation through Anopheles gambiae complex in Burkina Faso: Genetic introgression and de novo phenomena. Trop Med Int Health. 2004;9(12):1267–1273. doi: 10.1111/j.1365-3156.2004.01336.x. [DOI] [PubMed] [Google Scholar]
- 41.Donnelly MJ, Pinto J, Girod R, Besansky NJ, Lehmann T. Revisiting the role of introgression vs. shared ancestral polymorphisms as key processes shaping genetic diversity in the recently separated sibling species of the Anopheles gambiae complex. Heredity (Edinb) 2004;92(2):61–68. doi: 10.1038/sj.hdy.6800377. [DOI] [PubMed] [Google Scholar]
- 42.Tripet F, Dolo G, Lanzaro GC. Multilevel analyses of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting mosquito projects. Genetics. 2005;169(1):313–324. doi: 10.1534/genetics.104.026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripet F, Touré YT, Dolo G, Lanzaro GC. Frequency of multiple inseminations in field-collected Anopheles gambiae females revealed by DNA analysis of transferred sperm. Am J Trop Med Hyg. 2003;68(1):1–5. [PubMed] [Google Scholar]
- 44.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Q, Shete S, Swartz M, Amos C. Examining the effects of linkage disequilibrium on multipoint linkage analysis. BMC Genet. 2005;6(Suppl 1):S83. doi: 10.1186/1471-2156-6-S1-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbosa S, Black WC, 4th, Hastings I. Challenges in estimating insecticide selection pressures from mosquito field data. PLoS Negl Trop Dis. 2011;5(11):e1387. doi: 10.1371/journal.pntd.0001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.