Abstract
It has been 35 y since Carl Woese reported in PNAS how sequencing ribosomal RNA genes could be used to distinguish the three domains of life on Earth. During the past decade, 16S rDNA sequencing has enabled the now frequent enumeration of bacterial communities that populate the bodies of humans representing different ages, cultural traditions, and health states. A challenge going forward is to quantify the contributions of community members to wellness, disease risk, and disease pathogenesis. Here, we explore a theoretical framework for studies of the inheritance of bacterial strains and discuss the advantages and disadvantages of various study designs for assessing the contribution of strains to complex diseases.
Keywords: microbial inheritance, strain-resolution human microbial ecology analyses, effector strains, health, disease
In a 1977 issue of PNAS, Woese and Fox defined the 16S rRNA gene as a molecular and evolutionary marker to distinguish members of the domain Archaea from those belonging to Bacteria (1). This report profoundly changed the course of microbiology and opened the door for culture-independent studies (2). With the advent of next-generation sequencing, there has been an explosive increase in the use of this phylogenetic marker to characterize microbial communities in the environment and those associated with the body habitats of myriad animal species, including our own. The increasing ease and economy of defining human microbial ecology within and across individuals has spawned many studies that have sought to correlate community membership with health status (3–9). These studies have highlighted the variation in microbial diversity between body habitats in a given individual and intrapersonal variation in community membership within a given habitat over time, as well as substantial interpersonal differences. They have also revealed how a given species-level phylogenetic group in a given body habitat’s microbial community (microbiota) can be composed of multiple strains of that species, each harboring a set of shared genes, as well as genes that are variably represented across strains (10–12). This variation in a species “pan-genome” has yet to be systematically characterized for a large number of taxa recovered from different habitats of a given individual, or between large numbers of individuals. Defining this strain-level diversity and its biological significance requires moving beyond the 16S rRNA gene to genome-wide analyses.
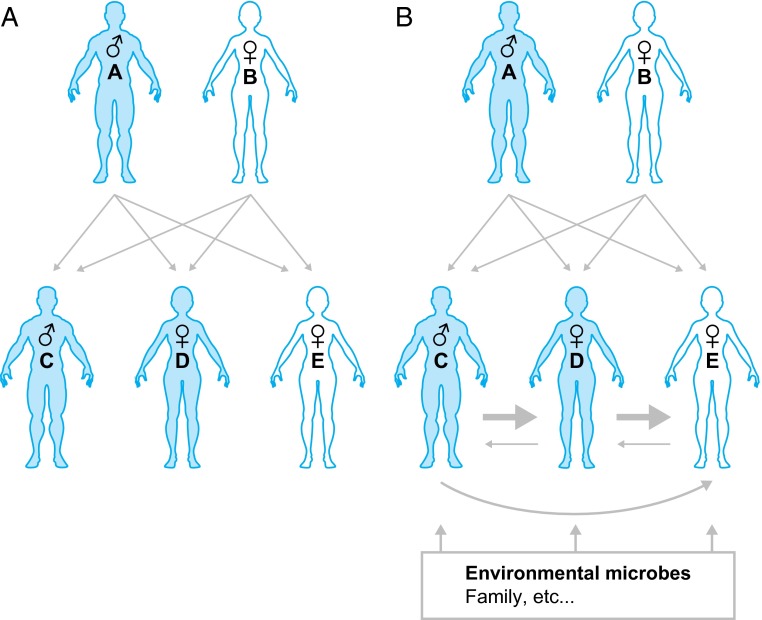
Identifying the “effector” strains represented in a microbiota that are responsible for shaping specific facets of our human biology is a daunting challenge because there are a large number of possible combinations of component organisms that could operate to produce a given phenotype. In this Perspective, we discuss the proposal that characterizing acquisition, retention, and passing of strains (“microbial inheritance”) will be a useful element in the quest to identify effector taxa associated with health or disease (Fig. 1). Using the human gut microbiota and its bacterial constituents as an illustration, we consider this proposal from the perspective of four concepts: (i) The majority of resident strains are acquired during the first three years of life; (ii) once acquired, the majority of strains are retained in an individual for decades; (iii) strain sharing is biased toward proximal microbe-rich sources (e.g., “family members”) with very little to no strain overlap in the microbiota of “nonfamily” members; and (iv) the effect of a strain’s residency may take decades to manifest itself.
Fig. 1.
Human genetic inheritance and microbial inheritance. (A) In human genetic inheritance, information transmission is vertical, with 50% of each child’s genome content inherited from the father and 50% from the mother. Note that birth order is from left to right. (B) In microbial inheritance, transmission occurs largely in the first 3 y of life from proximal microbe-rich sources, notably family members, and in particular between near-birth siblings who provide a higher degree of access to their microbes. However, microbial inheritance is not limited to family members and can include other environmental sources, including family friends, nearby surfaces, and the local water supply.
The gut microbiota of humans is dominated by relatively few bacterial phyla compared with other body habitats (13); a notable feature of this community is its great strain-level diversity (10, 12, 13). The difficulty in identifying causative strains for complex diseases may relate to the fact that they represent species that are common in the gut microbiota of healthy individuals. With the advent of methods for culturing a significant amount of the bacterial diversity present in a person’s fecal microbiota (14, 15), performing whole-genome sequencing of members of clonally arrayed culture collections provides one way to identify the strain-level variation that an individual harbors as a function of time, health status, and other covariates (6, 10). Although “traditional” 16S rRNA sequencing lacks the resolution and depth to track the representation and persistence of bacterial strains within a microbiota, a method known as low error amplicon sequencing (LEA-Seq) uses redundant sequencing (∼10× coverage per 16S rRNA gene amplicon) to achieve this goal with high resolution in samples collected over time from subjects (10). For example, when applied to 37 healthy adults sampled for up to 5 y, LEA-Seq revealed that each person harbored ∼100 bacterial species and ∼200 bacterial strains in their fecal microbiota. On average 60% of strains were retained in each subject during the study period. Stability followed a power law that, when extrapolated, suggests that the majority of gut strains in individuals are residents for most of their lives. Thus, early gut colonizers, once established, have the potential to exert their biological effects on our health for most and perhaps all of our adult lives (10).
A cross-sectional bacterial 16S rRNA study of the fecal microbiota of healthy children and adults belonging to families representing Guahibo Amerindians and Malawians living in rural villages, and families from metropolitan areas within the United States, revealed that the composition of the gut microbiota reached an adult-like composition by 2–3 y of age in all three populations (16). We recently found that this assembly process can be predictive of disease risk. Bacterial species whose proportional representation defines a healthy gut microbiota as it assembles during this period have been identified in a birth cohort of children living in an urban slum of Dhaka, Bangladesh. Using a sparse Random Forests-based model of these age-discriminatory bacterial taxa, a “relative microbiota maturity index” and “microbiota-for-age Z-score” was used to compare assembly (“maturation”) of a given child’s microbiota relative to the reference set of healthy children of similar chronologic age. Severe acute undernutrition is associated with significant relative microbiota immaturity that is only partially ameliorated following commonly used nutritional interventions. This immaturity is also evident in less severe forms of undernutrition and correlates with anthropometric measurements (17). These findings provide a microbial measurement of postnatal development that can be applied to individuals who are, or who are not, biologically related to one another and suggest that healthy growth is dependent upon normal maturation of the gut community.
A Model of Microbial Inheritance
Although the logic and dynamics of microbial inheritance will require experimental and theoretical refinement, a simple probabilistic model of host colonization provides a structure for understanding key variables and the effect of modifications to these variables. In this simplified model, we do not account for effects of host genotype on selection of particular taxa. This exclusion is not to imply that host genetics play no role. Well-controlled quantitative trait loci mapping studies in mice indicate that microbiota composition is affected by host genetic factors, including those involved in immune responses, as well as by environmental exposures (18, 19). Although studies of dizygotic and monozygotic twins suggest that the representation of members of the gut community is impacted by genotype (11, 20), genome-wide surveys are needed to address the important question of the contributions of host genetics versus environment on microbiota structure and function. In our simplified model, we assume the probability that the (gut) microbiota of any individual is colonized with strain i (i.e., P(colonizedi) has the following form
where P(transmissioni) is the probability of transmission of strain i, P(accessi) is the probability of access to strain i, and P(resistancei) is the resistance of the host to colonization by strain i. The probability of transmission for a particular gut strain will depend on factors such as its ability to survive environmental insults (e.g., aerotolerance, spore-forming capacity) and its competitiveness (ability to stably establish itself in one or more available niches). Access will depend on the microbe’s abundance in the source community as well as the hygiene, frequency of contact with, and the health status of people in near proximity to the recipient. Resistance will depend on the current composition of the potential recipient’s microbiota, niche availability in the recipient’s microbiota for strain i, and the hygiene of the recipient, as well as other factors including the recipient’s immune status and genotype (e.g., features of their innate immune system).
According to our model, microbial inheritance occurs preferentially between family members relative to unrelated individuals because P(accessi) is far higher for microbes harbored by individuals we most frequently encounter and because P(resistancei) is lower during the first 2–3 y of postnatal life when microbial community assembly is occurring (Figs. 1 and 2). For example, comparison of draft genomes of strains of (i) a prominent saccharolytic bacterium in the human gut microbiota, Bacteroides thetaiotaomicron (10), and (ii) the principal human gut archaeon, Methanobrevibacter smithii, isolated from the fecal microbiota of several families (11) disclosed that the same strains were shared between a majority of adult family members. Viewed in this light, microbial inheritance, where a proportion of our microbial inhabitants and their genes are passed between family members and once acquired are retained throughout life, parallels to a degree the inheritance of our human genes. However, even within a family, the simple probabilistic model suggests how differences could occur in the way strains are passed between family members.
Fig. 2.
Consequences of microbial inheritance. (A) As we age, the likelihood of transmitting and receiving microbial strains changes. The increased incidence of vomiting, diarrhea, and acute respiratory illnesses at early ages, combined with less established hygiene practices, likely enables young children to provide higher access to their microbial inhabitants. In addition, the less established community of microbes harbored in and on their bodies provides less resistance to invasion and establishment of new organisms. (B) The consequences of these age-associated changes in access and resistance are that the probability that a young child is colonized by a given microbial strain i [i.e., P(colonizedi)] is higher when both parents harbor the strain than when only one parent does [i.e., P(accessi) is higher because there are two low-access reservoirs of the strain rather than one], and higher if a near-birth sibling and a parent are colonized by the strain than if both parents are colonized [i.e., P(accessi) is higher because there is one low-access reservoir and one high-access reservoir of the strain rather than two low-access reservoirs]. (C) In the context of multiple siblings, access is highest in near-birth siblings.
If P(accessi) is dependent on shedding of strain i by the donor, as well as the hygiene and frequency of contact between donor and recipient, one could assume that P(accessi) is highest between similarly aged siblings early in life at a time when P(resistancei) is low (Fig. 2). Helicobacter pylori is a common member of human gastric microbiota that is first acquired in childhood. H. pylori is also a pathobiont that in some hosts causes disease, notably peptic ulcers. It is most easily spread during episodes of active shedding (e.g., during gastroenteritis) (21, 22). Studies have shown that later born siblings from large sibships are the most likely to acquire H. pylori (23): the odds of colonization with H. pylori increase with the number of 2- to 9-y-old siblings, and children born within 4 y of each other are four times more likely to be infected than those born 10 or more years apart. Moreover, the total number of siblings positive for H. pylori is a better predictor of colonization than simply the total number of siblings (23). Extrapolating from the microbial inheritance model in Fig. 2, the latter findings can be interpreted as a case where P(accessi) is high and P(resistancei) is low early in life, leading to an enrichment in shared colonization in larger sibships and near-birth siblings. A more expansive illustration of this point, and one that does not focus on a pathobiont comes from two culture-independent studies of 56 families containing monozygotic and dizygotic twin pairs and their mothers (24), and a large family composed of a father, mother, and six children (25); the results showed that siblings’ gut communities are significantly more similar to each other than to their parents (24, 25). It is important to emphasize that the definition of “family” varies between, as well as within, various human populations representing different cultural traditions, socioeconomic conditions, and other factors. P(accessi) needs to be considered in light of knowledge of infant care practices, the degree to which children and older individuals who are, and are not, related by biology share common dwellings, and other anthropologic/sociologic parameters. Future strain-level studies of microbial inheritance in large families should permit more precise quantification of time windows for optimal transmission of different microbes and should be complemented by analyses of factors that define niche and determine fitness and persistence (26–28).
Tracking microbes through families where members have a disease that is not monogenic, and where an intersection of host genetic as well as environmental factors are posited to contribute to pathogenesis, offers an opportunity to identify important microbial contributors to risk, prevention, and/or treatment of that “complex disease.” In an illustrative hypothetical example, consider gastric cancer, a disease correlated with the decades-long presence of some strains of H. pylori. In our example, monozygotic twins have a father who harbors an H. pylori strain containing the cag pathogenicity island (cag+) and a mother who is colonized with a cag− strain. During the first 2 y of life, each sibling may acquire an H. pylori strain that retains its membership in their gastric microbiota for decades, or for the remainder of their lives. The strain they acquire is critically important because acquisition of the cag+ strain will increase their risk of gastric cancer roughly twofold (29). The probabilistic model emphasizes the importance of which strain is transmitted first to one of the cotwins; the increased number of reservoirs, combined with the increased contact between siblings who are members of a twin pair, will increase the chance that the second twin will be colonized by the same strain harbored by the first cotwin to be colonized (Fig. 2), and thus help determine their concordance for this disease.
Microbial inheritance could involve acquisition of one or a number of effector strains that individually or collectively modulate the risk of developing complex diseases. Just as polygenic disorders are far more common than monogenic disorders, we posit that complex diseases are more likely to reflect the contributions of multiple microbial strains that both mediate and mitigate pathogenesis in the context of a specific host genotype. Thus, the dynamics of their exchange (inheritance) would be more complex than in the simplified example presented above and, in the case of twins, help define the degree of concordance or discordance for disease. Crohn’s disease, rheumatoid arthritis, and multiple sclerosis are examples of complex diseases where microbial agents have been hypothesized to have etiologic roles (30–36). Concordance for disease in monozygotic twins is below 40% in all three diseases (37–39). Genome-wide association studies have identified overlapping loci between the three diseases, including loci involved in immune regulation as well as microbial recognition and defense (40–42). Antibiotics are used to treat all three diseases (43–45) but with limited efficacy. Mouse models mimicking features of these diseases have shown a significantly lower incidence of pathology (or no pathology) when they are reared germ-free compared with their conventionally raised counterparts (i.e., mice that acquire a microbiota from their environment beginning at birth) (46–48).
Family Studies
Transmission Among Spouses/Cohabitating Adults.
The lack of transmission of complex disease between spouses is cited as evidence against microbial agents playing a role in pathogenesis or at least as evidence that a shared environment later in life is not an important etiologic factor. Although we are unaware of any evidence beyond case reports of an increase in relative risk for the spouse of an individual with multiple sclerosis, a small but significant increase risk of rheumatoid arthritis has been observed (49). In an analysis of two cohorts living in Belgium and France, the frequency of Crohn’s disease after marriage to an individual with this disease was significantly increased relative to what would be expected by chance alone given the population size studied (50). In both rheumatoid arthritis and inflammatory bowel disease, relative risk was extremely small (1.17 and 1.82, respectively). These small effect sizes may be related to P(resistancei) being high in adulthood either because existing members of the microbiota prevent new strains from establishing themselves or because the partner has already developed immune mechanisms to eliminate the strains(s).
Although increased relative risk upon cohabitation with an affected spouse may be small, the relative risk of Crohn’s disease (51), rheumatoid arthritis (49), or multiple sclerosis (52) is highly increased when a sibling is affected (6.3, 4.6, and 7.1, respectively). In the context of microbial inheritance, the shared risk factors are stably colonized microbial strains inherited between family members that increase (or decrease) the probability of complex disease over a lifetime.
Before identification of H. pylori as the etiologic agent, familial aggregation was cited as evidence supporting genetic factors over environmental factors in the pathogenesis of peptic ulcer disease. In a 1950 analysis of familial risk, Doll and Buch (53) stated that “the possibility that similar environments may have been responsible for the results cannot be excluded. It is unlikely, as an excess of ulcers occurred in two generations of the relatives. For if environmental influences were responsible they would have to have operated in childhood, and it is difficult to believe that the fathers have been equally subjected to the same influences in their childhood”. In light of our current knowledge of the persistence and familial transmission of H. pylori (23, 54–57), an increased incidence in peptic ulcer disease across two generations can just as easily be ascribed to transmission of a pathogenic strain as it can to the genetic origins suggested by Doll and Buch.
Transmission Among Siblings.
Given that P(accessi) for effector strains will be higher when proximal environmental sources have less developed hygiene practices, more episodes of shedding, increased number of reservoirs (e.g., multiple sibling carriers), and increased contact with the recipient (Fig. 2), one would also expect an increased relative risk of disease in the context of affected siblings and clustering of cases between consecutively born children. Blaser et al. found a statistically significant correlation between higher birth order and gastric ulcer (58). In the context of cag+ H. pylori strains, the risk of developing gastric cancer was twice as high for individuals in sibships of more than seven individuals compared with individuals in sibships of one to three individuals (59). In a study of 102 Crohn’s disease sibships with at least two affected siblings and one healthy sibling, we observed a statistically significant increase in affected siblings in consecutive births relative to nonconsecutive births (60). In a study of multiple sclerosis in 370 Canadian twin pairs, a trend toward increased disease susceptibility was observed with dizygotic twins, with a proband-wise concordance of 5.4 ± 2.8% compared with 2.9 ± 0.6 for their nontwin siblings (37).
Spatial Clusters
Spatial clusters characterized by a statistically significant increase in disease prevalence in a particular geographic region provide compelling evidence for an environmental contribution, including a microbial one. Although such clusters are often disputed as statistical outliers, or dismissed for other reasons, the abundance of observed clusters in different geographic regions can cumulatively provide evidence for a shared environmental risk factor in disease pathogenesis. Disease clusters and outbreaks are systematically tracked for many viruses and foodborne illnesses whereas clusters of complex disease are more commonly identified from large cohorts (61) or through citizens and local physicians who notice and report a surprising number of cases in a given locale (62, 63).
In the probabilistic model of microbial inheritance, a disease cluster would form when P(transmissioni) is high and P(accessi) is high across a geographic region. Although most of our inherited microbes come from other humans with whom we interact, broad access to an effector strain or strains would likely require a nonhuman source. Public water supplies, swimming pools, rivers, lakes, and ponds are often hypothesized to be the source of regional clusters. These clusters also imply effector strains that can survive (and ideally multiply) outside of a human host, which is uncommon for the large proportion of taxa colonizing the gut. Although we are unaware of any documented geographic clusters of rheumatoid arthritis, numerous clusters of Crohn’s disease have been identified throughout the world, including Mankato, MN (64) and Blockley Parish, Gloucestershire, England (65). With large-cohort databases, geographic clusters can be detected systematically in an unbiased manner using spatial scan statistics that search large regions for variations in disease incidence and reduce the chance of spurious clusters due to sampling biases (66). We recently applied these statistical methods to the Registre Epidemiologique des Maladies de l”Appareil Digestif inflammatory bowel disease (IBD) cohort and identified 18 significant Crohn’s disease geographic clusters, the largest with 61 cases among a population of only 5,703 (10 times higher than expected) (61). Spatial techniques also identified geographic clusters of IBD in Manitoba, Canada (67). Multiple sclerosis has a large number of proposed disease clusters, including the Faroe Islands (36) and Key West, FL (63). Advances in spatial epidemiology plus geographical information systems and global positioning systems are already being applied to the study of infectious diseases (68), and their forthcoming application to chronic diseases should enable more rigorous and sensitive identification of spatial variation in complex diseases (62, 69).
Microbial Inheritance as a Tool to Identify Effector Strains
Several study designs and strategies can be envisioned for quantifying the risk associated with stably colonized and transmitted members of the (gut) microbiota. The key to such studies is to generate a “microbe type” equivalent to a genotype. This microbe type requires high-resolution tools to identify the unique set of microbes in each individual at the strain level.
An initial challenge with microbe typing is that the organisms in a community are present at different abundances and that their genomes have differing degrees of homology to one another. Therefore, a sample of that community needs to be assayed to a sufficient depth to detect all of the relevant organisms, and the assay must be of sufficient resolution to dereplicate the sequences into the unique set of strains present [high-abundance organisms will be assayed many orders of magnitude more frequently than low-abundance organisms, and assay noise (e.g., sequencing errors) could overinflate the microbe type with false positives]. The most accurate method to microbe type would be to acquire the whole genome sequence of each member of the community, but this is not practical at the present time. Currently, high-throughput whole-genome microbe typing is largely limited to culturable organisms. For example, using this approach, we recently sequenced over 500 microbial genomes, generating microbe types for six individuals (10). By quantifying the fraction of DNA alignment between each pair of bacterial genomes (coverage score) (70), we found taxa from the same genus and species had coverage scores of 0.30 ± 0.20 and 0.77 ± 0.12 (mean ± SD), respectively. Strains of bacteria isolated from individuals over time and those shared between family members had a coverage score of >0.96. These findings led to a provisional proposal that a threshold for dereplication at the strain level would be to cluster isolates with genome coverage scores of >0.96 (10).
Microbe typing by bacterial isolation (culture) and genome sequencing is laborious and expensive. Therefore, finding alternative methods is desirable. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, a culture-dependent method that identifies microbes from their protein spectra, provides a cost-effective and rapid alternative for dereplication at the species level and perhaps at the strain level (assuming further algorithmic and sample extraction optimizations in the future) (71, 72). Sequencing 16S rRNA amplicons and shotgun sequencing of total community DNA currently represent the two most commonly used culture-independent metagenomic technologies but suffer from low resolution and difficult dereplication, respectively.
One highly anticipated outcome of microbe-typing improvements is a critical assessment of how well the definition of “strain” [based on a highly accurate 16S rDNA (or larger rRNA locus) amplicon marker sequence, or other methods including MALDI-TOF] is indicative of a given gene content, or whether cultured isolates with the same marker sequence have varying gene content (73). Improvements in read length, read number (sampling depth), and read quality could help address this challenge by (i) enabling sequencing of several components of or the entire 16S rRNA-ITS-23S rRNA region with low-error amplicon methods (10, 74), and (ii) facilitating the dereplication/assembly of shotgun reads generated from community DNA. Improvements in the throughput and quality of single-cell DNA sequencing would provide an ideal solution with its culture independence and genome-wide resolution. Continued improvements in algorithms for the assembly (75) and quantitative analysis of metagenomics data (76–78), combined with improved microbiome reference databases (79, 80) and a growing understanding of pan-genome variation within species, could spawn additional methods to track strains. These microbe-typing advances will need to be validated experimentally with defined artificial microbial communities composed of collections of sequenced organisms with differing degrees of genome similarity and different abundance distributions within these “mock” communities.
Across human populations, the specific microbial genes responsible for effects on health and disease pathogenesis may be present in the genomes of different microbial species and strains. The advantage of strain tracking is that all of the genetic information is “linked” in the genome sequence of each organism being passed between individuals: by tracking the presence or absence of strains with >96% shared genome content, statistical tests can be performed to identify strains that modulate disease risk without the need to initially identify the specific genes responsible; this linkage drastically reduces the number of variables and statistical tests. Alternatively, mapping shotgun sequencing datasets generated from microbiomes to reference gene collections (80) or reference microbial genomes (79) provides the possibility of directly identifying the genes or genomic variants that explain disease variation. However, these approaches are currently complicated by our lack of knowledge of the resolution at which pathogenic effects might occur (e.g., a rare, highly penetrant, disease-specific microbial gene versus a low penetrance polymorphism in a commonly occurring horizontally transferable microbial gene).
Study Designs
As in human genetics, several study designs can be used to attempt to identify strains with significant odds ratios between healthy and affected individuals. In the case-control design (Fig. 3A), microbe types are obtained from groups of affected and unaffected individuals with the goal of identifying strains that are significantly overrepresented in one of the two groups. This design is unlikely to be effective in identifying stably colonized effector strains because, unlike our human genomes, there seems to be very little to no strain overlap in the microbiota of nonfamily members. However, this important point needs more rigorous testing: e.g. whereas we did not observe a shared microbial strain (coverage score >0.96) in surveys of 21 unrelated people living in the United States and 578 cultured isolates representing 76 species that were cultured from their fecal microbiota, surveys of more isolates from a given individual and more individuals representing different lifestyles are needed. If the observation of little to no strain overlap in unrelated individuals holds, a case-control study would require sampling a very large population, ideally from a small geographic location with little immigration.
Fig. 3.
Study designs for identifying microbes that modulate complex disease risk. Microbial inheritance patterns provide an opportunity to identify etiologic agents of disease. By delineating the set of microbial inhabitants in each individual at the strain level, microbial inheritance patterns can be compared with disease incidence to identify strains whose presence/absence explains (correlates with) disease variation. As an illustrative example, every subject in this figure harbors a set of three microbial strains identified by their individual lower and uppercase letters (e.g., “a” is one strain and “A” is another). Healthy individuals are shown with a blue outline whereas affected individuals are shown in solid blue. Microbes that significantly increase disease risk are presented in red boldface (i.e., v, X, T, x, H, M). (A) A classic case-control design is difficult to power when searching for microbes that alter disease risk because, on average, unrelated individuals are expected to share no or very few strains. Sampling a broad enough population to have replicate observations of each strain would likely be prohibitively expensive. (B) Focusing on families increases the likelihood of identifying multiple individuals that share the same strain to enable identification of microbes enriched in either affected or unaffected individuals. Powering familial studies requires large families with multiple affected and unaffected individuals. (C) Geographic disease clusters provide a study design with high potential statistical power. Unrelated individuals are not expected to share microbial strains so identifying the same strain in multiple unrelated affected individuals would be highly significant and indicative of a shared environmental source of the identified strains.
Studies of large families where multiple members manifest a complex disease provide a way to identify effector strains significantly associated with health status (Fig. 3B) because, as noted above, microbial strains are shared among family members. Powering such studies depends on the penetrance of each effector strain and the number of affected and unaffected individuals in the family; increased power requires large families with high disease frequency (e.g., for an effector strain with 80% penetrance, at least four affected and nine total members are needed to obtain P value < 0.05 by Fisher’s exact test). Although such methods could identify a single organism that best explains a condition (e.g., H. pylori in peptic ulcer disease), they should also be capable of identifying and quantifying the combined influence, both beneficial and detrimental, of multiple microbial community members. Powering studies to quantify the combined influence of multiple strains, each contributing a small amount of disease risk or prevention, requires additional sampling. However, there are statistical advantages with strain tracking compared with human genotyping; not all strains will be acquired by vertical (parental) inheritance, thereby reducing the number of candidate effector strains; transmission involves a highly unique microbial strain that is far less likely to cooccur between two unrelated individuals by chance alone compared with SNPs, which can manifest one of four nucleotide states. Identification of candidate effector strains would set the stage for development of strain-specific assays for screening larger populations (e.g., more distant relatives and family friends), for more accurate calculations of the risk associated with harboring a given strain, and for aiding identification of genetic regions in each strain that confer altered disease risk.
Spatial clusters, if caused by microbial inheritance, provide the sample source with the highest statistical power (Fig. 3C). Identification of shared microbial strains between affected unrelated individuals in a region would be a highly significant event. Studies of spatial clusters should include unaffected, unrelated controls to ensure that strains identified as enriched are not enriched in all humans in the community due to shared environmental sources. If shared strains are identified between affected unrelated individuals, microbe typing a significant proportion of a small town with a complex disease cluster would also provide a wealth of information about microbial transmission and the degree of sharing of the town’s collective microbiota.
Improving Tools and Resources for Microbial Inheritance Studies
Microbe typing healthy families from different regions of the world with diverse dietary and cultural traditions would provide baseline transmissibility information, detailing what proportion of the (gut) microbiota is inherited from family members versus other environmental sources and which strains are more or less transmissible. A transmissibility database would facilitate development of statistical models for identifying effector strains that are weighted by the probability that such an event would occur. A strain-level database in states or countries with complementary geographical information systems in their health departments would provide an opportunity to track potential effector strains moving through the population (68, 81, 82).
Gnotobiotic animal models could help us to understand the dynamics of strain transmission in a highly controlled manner because male and female mice colonized separately with defined cultured isolates from a human father’s microbiota and a human mother’s microbiota could be mated. Cohousing male and female animals could provide a way to study microbial transmission of different strains in the context of established adult microbial communities in animals given diets similar to that consumed by the human microbiota donors. The resulting litters or subsequent rounds of litters could be compared with estimates of the proportion of strains inherited maternally and paternally and the similarity and differences between litters from the same parents. “Humanized” gnotobiotic mouse models represent a facilitated test of transmissibility: their coprophagy ensures that P(accessi) is high and relatively constant across studies.
The equation and study designs described above aim to quantify the risk or benefit associated with strains that do or do not stably engraft in an affected population relative to a control population. Persistent colonization with an effector strain in peptic ulcer disease, gastric cancer, Whipple’s disease, leprosy, and tuberculosis provides precedent for the paradigm that a chronic progressive inflammatory condition requires chronic colonization. The epidemiologic studies described above provide evidence that such effects can be detected in population data as, for example, unexpected deviations in sibling disease risk. Thus, a static model provides the simplest starting point for understanding the microbiota’s contribution to disease risk at the strain level. However, as strain-level microbe-typing technologies improve to enable longitudinal studies, the equation could be modified by including terms for the probability of a strain’s extinction [P(extinctioni)], or to enable P(transmissioni), P(accessi), and P(resistancei) of each strain i to change as a function of time. These additions would facilitate identification of transient microbes that imprint a beneficial or deleterious trajectory on their host that manifests some time after their extirpation from a microbiota. The exact form of these models and the properties encompassed by the underlying variables will undoubtedly change as they are refined by empirical data collection. Our goal with the current model is to provide a starting point for assessing the implications of a microbiota acquired early in life and stably maintained for decades.
The search for microbial community contributions to complex human disease is increasingly focused on identifying potentially pathogenic states: i.e., a “dysbiosis.” Although numerous searches for organisms that cause complex disease have failed, we have lacked tools with sufficient resolution to determine whether, among common species in communities, there was a unique disease-associated strain or strains. Whether a given disease is mediated by a single or multiple organisms, we are still attempting to solve a community-wide problem in the sense that the goal is to identify key elements of the community, be they individual strains, metabolic states, or thus far unknown characteristics that are causally related to, rather than just the effect of, that disease. If the community state is the primary mediator of disease, the potential for a pathogenic conformation of the community is likely influenced or determined by the strain-level composition of the community. Assuming that some diseases are mediated by multiple strains in a community, we must carefully quantify what this assumption entails. In the context of microbial inheritance, the probability that unrelated individuals share a single member of their microbiota at the strain level is small. If microbes were inherited independently, even a disease that is only mediated by two strains would represent the product of two small probabilities and would be extremely rare, unless the causative organisms developed nonindependent transmission mechanisms, in which case such organisms should be identifiable by strain-level cooccurrence analyses of the microbiota of individuals with the conditions of interest. The cases of multiple organisms mediating disease could represent multiple causative genes harbored in microbes across diverse phylogenetic groups yet these genes must be common enough in the global metagenome to allow the joint probability of their occurrence in a single affected host. As we move from one causative organism or gene to two and beyond, the probabilities quickly become vanishingly small unless these organisms or genes are common or have developed mechanisms to transmit nonindependently through the human population. A major advantage of microbial inheritance, and the study designs described here, is that it is agnostic to the number of organisms and forgoes the need to initially search for causative genes by taking advantage of the linkage of genes to the strains that are being passed between hosts. Enrichment or depletion of one strain or many strains in affected individuals should be detectable.
Assuming that microbial inheritance plays a role in complex disease, the largest hurdle to quantifying this influence is our ability to consistently detect the same strains in and across samples. False positives in microbe-type analyses will likely be rare, particularly if whole-genome sequencing is used. However, false negatives (the inability to detect or isolate a particular organism in an individual’s microbiota) are likely to be an unwanted source of variance until better strain-level identification methods are available. One could argue that many “patterns” could be found that correlate well with disease in large samples by simple chance alone. However, these correlations should be rare because only a fraction of strains are shared even between related individuals. An additional complication is where to search for the organisms of interest. The gut and mouth, which seem to be stable sources of microbial life (10, 83–85), represent attractive initial places to microbe type. However, effector strains that increase the probability of disease might also be found in higher abundances at local sites of disease. In addition, microbe typing should not be, and need not be, limited to bacteria because inherited effector strains could also be members of Archaea or Eukarya (or their viruses).
If disease develops from a stably colonized strain or collection of strains, or a lack of these strains, microbial inheritance provides a set of strategies to facilitate detection of such organisms. Studies of microbial inheritance can be applied to a broad range of complex diseases occurring in varied geographic and cultural settings, including, for example, undernutrition of children in low-income countries where the size of extended families can be large and spatial scan statistics may reveal previously unappreciated clusters of diseases that are long standing and common in the population or of diseases emerging coincident with Westernization. Finally, one hoped-for outcome of studies of microbial inheritance would be new strategies for disease treatment and prevention that involve addition to, or elimination of, strains from an individual’s microbiota. The anticipation is that one day we will look back at the history of diagnosing and treating complex diseases that once were progressive and refractory to cure and marvel why we were not able to see their microbial roots.
Acknowledgments
Work cited from the J.J.F., J.-F.C., and J.I.G. laboratories was supported by grants from the NIH and the Crohn’s and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article is part of the special series of PNAS 100th Anniversary articles to commemorate exceptional research published in PNAS over the last century.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane DJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 8.Taur Y, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seekatz AM, et al. Recovery of the gut microbiome following fecal microbiota transplantation. MBio. 2014;5(3):e00893–e14. doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen EE, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108(15):6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier JC, et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18(12):1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 16.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107(44):18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnite AM, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE. 2012;7(6):e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsonnet J, Shmuely H, Haggerty T. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA. 1999;282(23):2240–2245. doi: 10.1001/jama.282.23.2240. [DOI] [PubMed] [Google Scholar]
- 22.Perry S, et al. Gastroenteritis and transmission of Helicobacter pylori infection in households. Emerg Infect Dis. 2006;12(11):1701–1708. doi: 10.3201/eid1211.060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman KJ, Correa P. Transmission of Helicobacter pylori among siblings. Lancet. 2000;355(9201):358–362. doi: 10.1016/S0140-6736(99)05273-3. [DOI] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2014;2:25. doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501(7467):426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell AB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16(2):227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaser MJ, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111–2115. [PubMed] [Google Scholar]
- 30.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 31.Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15(3):314. doi: 10.1007/s11926-012-0314-y. [DOI] [PubMed] [Google Scholar]
- 32.Hyrich KL, Inman RD. Infectious agents in chronic rheumatic diseases. Curr Opin Rheumatol. 2001;13(4):300–304. doi: 10.1097/00002281-200107000-00010. [DOI] [PubMed] [Google Scholar]
- 33.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7(10):569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray TJ. The history of multiple sclerosis: The changing frame of the disease over the centuries. J Neurol Sci. 2009;277(Suppl 1):S3–S8. doi: 10.1016/S0022-510X(09)70003-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtzke JF. Epidemiologic evidence for multiple sclerosis as an infection. Clin Microbiol Rev. 1993;6(4):382–427. doi: 10.1128/cmr.6.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC. Canadian Collaborative Study Group Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci USA. 2003;100(22):12877–12882. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfvarson J. Genetics in twins with Crohn’s disease: Less pronounced than previously believed? Inflamm Bowel Dis. 2011;17(1):6–12. doi: 10.1002/ibd.21295. [DOI] [PubMed] [Google Scholar]
- 39.Svendsen AJ, et al. On the origin of rheumatoid arthritis: The impact of environment and genes—a population based twin study. PLoS ONE. 2013;8(2):e57304. doi: 10.1371/journal.pone.0057304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lettre G, Rioux JD. Autoimmune diseases: Insights from genome-wide association studies. Hum Mol Genet. 2008;17(R2):R116–R121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki A, Kochi Y, Okada Y, Yamamoto K. Insight from genome-wide association studies in rheumatoid arthritis and multiple sclerosis. FEBS Lett. 2011;585(23):3627–3632. doi: 10.1016/j.febslet.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 42.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan KJ, et al. Antibiotic therapy in inflammatory bowel disease: A systematic review and meta-analysis. Am J Gastroenterol. 2011;106(4):661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 44.Ogrendik M. Antibiotics for the treatment of rheumatoid arthritis. Int J Gen Med. 2013;7:43–47. doi: 10.2147/IJGM.S56957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, et al. The prospects of minocycline in multiple sclerosis. J Neuroimmunol. 2011;235(1-2):1–8. doi: 10.1016/j.jneuroim.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Hansen J, Sartor RB. Insights from animal models. In: Bernstein CN, editor. IBD Yearbook. Remedica; London: 2007. pp. 19–55. [Google Scholar]
- 47.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60(3):661–668. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 50.Laharie D, et al. Inflammatory bowel disease in spouses and their offspring. Gastroenterology. 2001;120(4):816–819. doi: 10.1053/gast.2001.22574. [DOI] [PubMed] [Google Scholar]
- 51.Hemminki K, Li X, Sundquist K, Sundquist J. Familial association of inflammatory bowel diseases with other autoimmune and related diseases. Am J Gastroenterol. 2010;105(1):139–147. doi: 10.1038/ajg.2009.496. [DOI] [PubMed] [Google Scholar]
- 52.Westerlind H, et al. Modest familial risks for multiple sclerosis: A registry-based study of the population of Sweden. Brain. 2014;137(Pt 3):770–778. doi: 10.1093/brain/awt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doll R, Buch J. Hereditary factors in peptic ulcer. Ann Eugen. 1950;15(2):135–146. doi: 10.1111/j.1469-1809.1949.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 54.Giannakis M, et al. Response of gastric epithelial progenitors to Helicobacter pylori Isolates obtained from Swedish patients with chronic atrophic gastritis. J Biol Chem. 2009;284(44):30383–30394. doi: 10.1074/jbc.M109.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundin A, et al. Slow genetic divergence of Helicobacter pylori strains during long-term colonization. Infect Immun. 2005;73(8):4818–4822. doi: 10.1128/IAI.73.8.4818-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Owen RJ, Xerry J. Tracing clonality of Helicobacter pylori infecting family members from analysis of DNA sequences of three housekeeping genes (ureI, atpA and ahpC), deduced amino acid sequences, and pathogenicity-associated markers (cagA and vacA) J Med Microbiol. 2003;52(Pt 6):515–524. doi: 10.1099/jmm.0.04988-0. [DOI] [PubMed] [Google Scholar]
- 57.Suerbaum S, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95(21):12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blaser MJ, Chyou PH, Nomura A. Age at establishment of Helicobacter pylori infection and gastric carcinoma, gastric ulcer, and duodenal ulcer risk. Cancer Res. 1995;55(3):562–565. [PubMed] [Google Scholar]
- 59.Blaser MJ, Nomura A, Lee J, Stemmerman GN, Perez-Perez GI. Early-life family structure and microbially induced cancer risk. PLoS Med. 2007;4(1):e7. doi: 10.1371/journal.pmed.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hugot JP, et al. GETAID Clustering of Crohn’s disease within affected sibships. Eur J Hum Genet. 2003;11(2):179–184. doi: 10.1038/sj.ejhg.5200932. [DOI] [PubMed] [Google Scholar]
- 61.Genin M, et al. Space-time clusters of Crohn's disease in northern France. J Public Health. 2013;21:497–504. [Google Scholar]
- 62.Bureau of Environmental Health, Community Assessment Program . Review of the Prevalence of Type 1 Diabetes Among Children and Adolescents in Weston, Wellesley and Newton. Massachusett Department of Public Health; Boston, MA: 2012. [Google Scholar]
- 63.Helmick CG, et al. Multiple sclerosis in Key West, Florida. Am J Epidemiol. 1989;130(5):935–949. doi: 10.1093/oxfordjournals.aje.a115426. [DOI] [PubMed] [Google Scholar]
- 64.Van Kruiningen HJ, Freda BJ. A clustering of Crohn’s disease in Mankato, Minnesota. Inflamm Bowel Dis. 2001;7(1):27–33. doi: 10.1097/00054725-200102000-00004. [DOI] [PubMed] [Google Scholar]
- 65.Allan RN, Pease P, Ibbotson JP. Clustering of Crohn’s disease in a Cotswold village. Q J Med. 1986;59(229):473–478. [PubMed] [Google Scholar]
- 66.Kulldorff M, Nagarwalla N. Spatial disease clusters: Detection and inference. Stat Med. 1995;14(8):799–810. doi: 10.1002/sim.4780140809. [DOI] [PubMed] [Google Scholar]
- 67.Green C, Elliott L, Beaudoin C, Bernstein CN. A population-based ecologic study of inflammatory bowel disease: Searching for etiologic clues. Am J Epidemiol. 2006;164(7):615–623; discussion 624-618. doi: 10.1093/aje/kwj260. [DOI] [PubMed] [Google Scholar]
- 68.Richardson DB, et al. Medicine: Spatial turn in health research. Science. 2013;339(6126):1390–1392. doi: 10.1126/science.1232257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda ML, Casper M, Tootoo J, Schieb L. Putting chronic disease on the map: Building GIS capacity in state and local health departments. Prev Chronic Dis. 2013;10:E100. doi: 10.5888/pcd10.120321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005;21(10):2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- 71.Ghyselinck J, Van Hoorde K, Hoste B, Heylen K, De Vos P. Evaluation of MALDI-TOF MS as a tool for high-throughput dereplication. J Microbiol Methods. 2011;86(3):327–336. doi: 10.1016/j.mimet.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Sandrin TR, Goldstein JE, Schumaker S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom Rev. 2013;32(3):188–217. doi: 10.1002/mas.21359. [DOI] [PubMed] [Google Scholar]
- 73.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosher JJ, et al. Improved performance of the PacBio SMRT technology for 16S rDNA sequencing. J Microbiol Methods. 2014;104:59–60. doi: 10.1016/j.mimet.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 75.Sharon I, et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23(1):111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veiga P, et al. Changes of the human gut microbiome induced by a fermented milk product. Sci Rep. 2014;4:6328. doi: 10.1038/srep06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9(8):811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francis OE, et al. Pathoscope: Species identification and strain attribution with unassembled sequencing data. Genome Res. 2013;23(10):1721–1729. doi: 10.1101/gr.150151.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelson KE, et al. Human Microbiome Jumpstart Reference Strains Consortium A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rasko DA, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365(8):709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4(148):148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG, de Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2012;15(4):1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- 84.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]