Significance
Computationally predicting drug resistance mutations early in the discovery phase would be an important breakthrough in drug development. The most meaningful predictions of target mutations will show reduced affinity for the drug while maintaining viability in the complex context of a cell. Here, the protein design algorithm K* in Osprey was used to predict a single-nucleotide polymorphism in the target dihydrofolate reductase that confers resistance to an experimental antifolate in the preclinical discovery phase. Excitingly, the mutation was also selected in bacteria under antifolate pressure, confirming the prediction of a viable molecular response to external stress.
Keywords: drug resistance, antifolate, protein design, DHFR, MRSA
Abstract
Methods to accurately predict potential drug target mutations in response to early-stage leads could drive the design of more resilient first generation drug candidates. In this study, a structure-based protein design algorithm (K* in the OSPREY suite) was used to prospectively identify single-nucleotide polymorphisms that confer resistance to an experimental inhibitor effective against dihydrofolate reductase (DHFR) from Staphylococcus aureus. Four of the top-ranked mutations in DHFR were found to be catalytically competent and resistant to the inhibitor. Selection of resistant bacteria in vitro reveals that two of the predicted mutations arise in the background of a compensatory mutation. Using enzyme kinetics, microbiology, and crystal structures of the complexes, we determined the fitness of the mutant enzymes and strains, the structural basis of resistance, and the compensatory relationship of the mutations. To our knowledge, this work illustrates the first application of protein design algorithms to prospectively predict viable resistance mutations that arise in bacteria under antibiotic pressure.
Effectively treating infectious disease has become increasingly complicated by the prevalence of strains that are resistant to multiple classes of antimicrobial agents. Ideally, the lifetime of newly introduced drugs could be extended by prospectively predicting and overcoming potential resistance during the discovery cycle. For example, mutations to a drug target in response to an experimental candidate could be predicted in silico, and the results could be used to design a second generation of compounds that is active against the WT and mutated enzymes. Minimally, a successful algorithm would predict resistance mutations that maintain enzyme function and reduce inhibitor affinity. However, a more powerful algorithm would also predict mutations that maintain the fitness of the pathogen and are, therefore, likely to be selected in vitro or in vivo. Predicting fit mutations is a significant challenge, because the variables that contribute to fitness in an organism are complex and often unknown. Additionally, a successful algorithm would predict novel mutations that are responsive to novel compounds. A prospective strategy such as this would be especially effective in the discovery of therapeutics for which it is difficult to generate resistant cells in vitro.
In a previous work (1), we used the structure-based K* algorithm in the OSPREY protein design suite (2, 3) to predict double mutations in dihydrofolate reductase (DHFR) from methicillin-resistant Staphylococcus aureus (MRSA) that confer resistance to the novel propargyl-linked antifolates. S. aureus DHFR (SaDHFR) is an ideal model system for these predictions because the development of a single amino acid mutation results in trimethoprim resistance; higher levels of resistance are conferred by double mutations (4). Although the propargyl-linked antifolates exhibit greater affinity for the mutant enzymes and are active against MRSA strains resistant to trimethoprim-sulfamethoxazole (5), it would be useful to predict mutations that may arise for this new class of antifolates. Using ratios of positive design scores that predict binding of the substrate dihydrofolate and negative design scores that predict binding of the inhibitor, OSPREY/K* (2, 3) identified catalytically competent resistance mutations.
Additional previous attempts to predict mutational drug resistance have been reported; a recent review summarizes efforts using computational and structural methodologies (6). In contrast to the approach reported here, these other attempts have been retrospective, correlating computational results with approved therapeutics and known mutations in the target (7–10). Because these studies are retrospective analyses of known mutations that arise under pressure from known drugs, they do not address the problem of prospectively predicting a fit mutation.
Herein, we report the application of the structure-based protein design algorithm K* in OSPREY to identify prospective single-nucleotide polymorphisms (SNPs) that confer resistance to one of the propargyl-linked antifolates. For these studies, we required the algorithm to identify an SNP that conferred resistance and would, therefore, be more likely to be selected in the bacteria. From a ranked list of potential SNPs, we created and evaluated four of the mutant enzymes and found that all four conferred resistance (2- to 58-fold) at the enzyme level while maintaining native function. In parallel, we selected mutants of S. aureus in vitro using applied drug pressure with the antifolate to simulate a natural course for resistance to occur. Excitingly, we discovered that two of the predicted mutations (V31L and V31G) arise in the background of a compensating F98Y mutation. These results show that protein design algorithms are capable of prospectively identifying resistance mutations that are both biochemically validated and viable in the bacteria. Using bacterial fitness and enzyme kinetic experiments along with the determination of a high-resolution crystal structure of the mutant enzyme, we clarify the structural and biochemical bases of the resistance, including an explanation for the compensatory relationship of the two mutations. Overall, this work shows that reductionist biophysical algorithms can prospectively predict a molecular response to specific stress factors.
Identification of Resistance Mutations
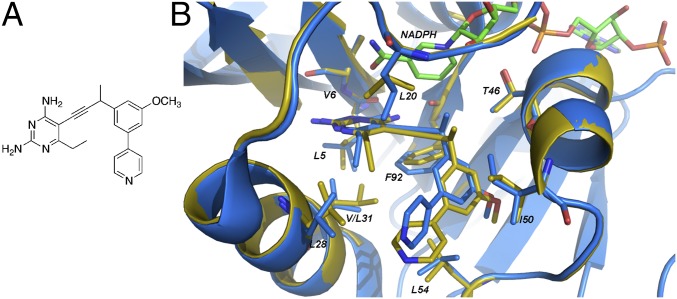
The protein design algorithm K* in OSPREY (2) was used to identify mutations characterized by SNPs that maintain binding to the substrate dihydrofolate while conferring resistance to the propargyl-linked antifolates. Nine active-site residues (Leu-5, Val-6, Leu-20, Leu-28, Val-31, Thr-46, Ile-50, Leu-54, and Phe-92) were allowed to either maintain their WT identity or mutate to a restricted group of residues that involves only an SNP (given in Materials and Methods and Fig. 1).
Fig. 1.
Modeled flexible residues in SaDHFR with NADPH and compound 1. (A) Compound 1. (B) Comparison of the structures of WT SaDHFR bound to NADPH and compound 1 (PDB ID code 3SGY; blue) with the K*-predicted lowest energy structure of SaDHFR(V31L) (yellow). All residues allowed to be flexible and mutate during the K* prediction are shown in yellow stick form. The cofactor NADPH is shown in green.
K* searches were performed on both the substrate (DHFR:NADPH:dihydrofolate) and the inhibitor (DHFR:NADPH:1) ternary complexes. K* scores approximate the binding affinity (Ka) and are computed as a ratio of Boltzmann-weighted partition functions for rotamer-based conformational ensembles of the bound protein–inhibitor complex, free protein, and free ligand. Because higher K* scores predict greater affinity, the ideal mutation would have a high score for dihydrofolate and a low or zero score for compound 1. The WT sequence was ranked 18th, with a positive-to-negative design ratio of 1.95 × 106 (Table 1). Mutants that had both (i) higher ranking than the WT and (ii) a good predicted binding affinity to dihydrofolate (i.e., positive design score) were considered as resistant mutants. Four mutations (V31L, V31I, L5I, and L5V) exhibiting high ratios for the scores representing positive (binding to DHF) and negative (binding to inhibitor) designs are shown in Table 1 (the complete ranking of all mutants is shown in Table S1). One mutation (L20F) was ranked sixth because of its high K* ratio (Table S1). However, its positive design score was over 20 orders of magnitude below the WT, predicting that L20F would lose affinity for dihydrofolate and not be a viable mutant. Therefore, L20F was discarded from consideration.
Table 1.
Characterization of WT and mutant enzymes
| Enzyme | K* ratio rank* | K* positive-to-negative design ratio | kcat/Km† | Fold loss† (Kimut/Kiwt) compound 1 |
| Sa(WT)DHFR | 18 | 1.96 E + 06 | 6.1 ± 0.3 | n/a |
| Sa(V31L)DHFR | 1 | 7.11 E + 21 | 1.60 ± 0.06 | 58 |
| Sa(V31I)DHFR | 2 | 5.95 E + 21 | 1.74 ± 0.07 | 36 |
| Sa(L5I)DHFR | 3 | 1.71 E + 15 | 2.24 ± 0.1 | 4.4 |
| Sa(L5V)DHFR | 4 | 1.16 E + 14 | 1.8 ± 0.1 | 1.9 |
n/a, not applicable.
In silico rank according to the ratio of K* scores shown in the subsequent column.
Experimentally determined.
The lowest energy-predicted conformations of the top-ranked mutations, SaV31L and SaV31I, were compared with the crystal structure of WT SaDHFR:NADPH:1 (5) to understand the basis of the prediction of resistance. Carbons Cδ1 and Cδ2 in Leu-31 are predicted to displace Phe-92, most likely to avoid steric hindrance with the phenyl side chain (Fig. 1 and Fig. S1). In addition, these carbons are observed to sterically interfere with the pyrimidine ring, displacing it from its corresponding position in the WT structure. Both of these steric interactions are predicted to significantly weaken inhibitor binding.
Biochemical and Microbiological Validation
To validate the positive and negative designs from K*, we used site-directed mutagenesis to create the top four mutant enzymes possessing SNPs resulting in V31L, V31I, L5I, and L5V. Michaelis–Menten kinetics (Table 1) reveal that the enzymes are catalytically competent, with only minor losses in kcat/Km. The activity of the mutant enzymes validates the success of the positive design component of the computational search. Sensitivity of the enzyme to inhibitor is drastically decreased with the addition of the point mutations, proving the negative design component. Resistance ranged from 2- to 58-fold, with the top-ranked mutants (V31L and V31I) conferring the greatest level of resistance. In fact, strikingly, the K* ratio rank correlated directly with the experimental ranking of inhibition constants.
To show the application of these predictions to a living pathogen, we then aimed to determine whether any of these mutations would be selected by the MRSA bacteria under pressure from compound 1 (5). To our surprise, a first round of selection revealed known mutations, and one of which (F98Y, a TTT to TAT transversion mutation) (Fig. S2) has a resistance frequency of 1.21 × 10−12. The F98Y strain was then exposed to a second round of selection for mutations. Excitingly, five of nine colonies of surviving bacteria yielded the computationally predicted V31L mutation (a GTT to CTT transversion mutation) (Fig. S2) in addition to the F98Y mutation for a final frequency of 7.56 × 10−24. One of nine colonies yielded V31G (14th in the ranked list, with a 10-fold increase in the K* ratio over the WT), and three colonies possessed only F98Y. Both the single F98Y and double V31L/F98Y mutant strains were characterized to evaluate antimicrobial susceptibility and relative fitness. Trimethoprim, with a resistance profile that includes the F98Y mutation, was included as a reference (Table 2).
Table 2.
Characterization of strain susceptibility and fitness
| Strain | MIC compound 1 (µg/mL; fold loss) | MIC TMP (µg/mL; fold loss) | Relative fitness | Doubling time (min) |
| WT | 0.0781 | 0.3123 | 1 | 22.5 |
| F98Y | 2.5 (32) | 10 (32) | 0.98 | 23.2 |
| V31L/F98Y | 20 (256) | 40 (128) | 0.86 | 25.4 |
Compound 1 and TMP have minimum inhibitory concentration (MIC) values of 2.5 and 10 µg/mL, respectively, for the F98Y strain, representing a 32-fold loss compared with the WT ATCC 43300 strain. The addition of the Val31Leu mutation confers an additional 8-fold loss for compound 1 and a 4-fold loss for TMP, resulting in total 256- and 128-fold losses, respectively. As observed previously, the Sa(F98Y)DHFR mutation does not reduce the fitness of the strain (11, 12). The presence of the V31L/F98Y mutation reported here results in minimal (14%) loss of fitness. Log-phase cell growth was minimally affected by the presence of the single F98Y or double V31L/F98Y mutation, with doubling times at 22.5 min for the WT strain and 23.2 and 25.4 min for the mutants, respectively.
To understand why the V31L mutation arises only in the background of the F98Y mutation, we examined detailed inhibition and kinetic data for the enzymes. Here, we also included the F98Y mutation and data for trimethoprim as a comparator. The Sa(F98Y) mutation affects trimethoprim more significantly, resulting in 38- and 5-fold losses in Ki against trimethoprim and compound 1, respectively. However, the Sa(V31L) mutation affects compound 1 more significantly, resulting in 15- and 60-fold losses in Ki against trimethoprim and compound 1 (Table 3). The Sa(V31L/F98Y) double mutant clearly confers much greater resistance, with a 148-fold loss in Ki for trimethoprim and an 189-fold loss in Ki for compound 1.
Table 3.
Enzyme characterization
| DHFR | Ki (TMP; fold loss; μM) | Ki (compound 1; μM) | Km (DHF; μM) | Km (NADPH; μM) | kcat | kcat/Km |
| WT | 0.0035 ± 0.0005 | 0.0028 ± 0.0002 | 17.5 ± 2 | 32.6 ± 4 | 106.9 ± 2 | 6.1 ± 0.3 |
| F98Y | 0.131 ± 0.004 (38) | 0.013 ± 0.001 (5) | 8.4 ± 0.7 | 56.8 ± 5 | 44.7 ± 0.4 | 5.33 ± 0.08 |
| V31L | 0.054 ± 0.002 (15) | 0.17 ± 0.02 (60) | 42.9 ± 3 | 15.6 ± 3 | 68.8 ± 2 | 1.60 ± 0.06 |
| V31L/F98Y | 0.52 ± 0.03 (148) | 0.53 ± 0.03 (189) | 4.1 ± 0.8 | 22.3 ± 2 | 44.8 ± 2 | 10.9 ± 0.8 |
Michaelis–Menten constants were determined for the enzymes (Table 3), revealing an interesting compensatory relationship between the mutations. The Km value for DHF is reduced from the WT value (17.5 μM) by 2-fold for Sa(F98Y)DHFR to 8.4 μM, whereas the same value for Sa(V31L)DHFR is increased by 2.4-fold to 42.9 μM. In combination, the Sa(V31L/F98Y)DHFR restores the Km for DHF to 4.1 μM. Interestingly, the Km value for NADPH for Sa(F98Y) increases to 56.8 μM from the WT value of 32.6 μM, and the Km value for NADPH in Sa(V31L) decreases to 15.6 μM. The double mutant maintains the decreased Km of Sa(V31L) and has an overall lower Km relative to the WT at 22.3 μM. Taken together, the data show that, although F98Y and V31L negatively affect NADPH and DHF, respectively, the two mutations combined restore the Km values to WT levels. Additionally, the kcat/Km value for the double mutant is increased to 10.9 μM−1.
Structure of Sa(V31L/F98Y)DHFR
The data in Tables 1 and 3 indicate that the mutations have a direct influence on substrate and cofactor binding as well as inhibitor potency. To understand the structural effects of the mutations, we determined a crystal structure of the double-mutant enzyme. Crystals of Sa(V31L/F98Y)DHFR produced diffraction amplitudes to 2.1 Å when cocrystallized with NADPH and a propargyl-linked antifolate (statistics for data and refinement are found in Table 4, and electron density is shown in Fig. S3). The structure was solved using Fourier methods based on the model of single-mutant Sa(F98Y) bound to NADPH and a propargyl-linked antifolate [Protein Data Bank (PDB) ID code 3F0U] (13). The Sa(V31L/F98Y) structure features the standard extended form of NADPH but lacks compound 1 present during cocrystallization.
Table 4.
Crystallographic data collection and refinement statistics
| Sa(V31L/F98Y):NADPH | Sa(F98Y):NADPH | |
| PDB ID code | 4Q6A | 4Q67 |
| Space group | P6122 | P6122 |
| No. monomers in asymmetric unit | 1 | 1 |
| Unit cell (a, b, c; Å) | 79.33, 79.33, 107.53 | 79.26, 79.26, 107.42 |
| Resolution (Å) | 50.00–2.10 (2.14–2.10) | 42.30–2.04 (2.51–2.04) |
| Completeness % (last shell; %) | 99.87 (100) | 98.7 (98.5) |
| Unique reflections | 12,235 | 13,132 |
| Redundancy (last shell) | 13.7 (13.5) | 10.22 (10.44) |
| Rsym (last shell) | 0.094 (0.320) | 0.070 (0.340) |
| <I/σ> (last shell) | 31.7 (20.0) | 16.9 (5.6) |
| Rfactor/Rfree | 0.1647/0.2063 | 0.1752/0.2330 |
| No. of atoms (protein, ligands, and solvent) | 1,510 | 1,446 |
| rmsd Bond lengths (Å), angles (°) | 0.008, 1.214 | 0.007, 1.205 |
| Average B factor for protein (Å2) | 19.57 | 28.75 |
| Average B factor for ligand (NADPH; Å2) | 14.32 | 21.11 |
| Average B factor for solvent molecules (Å2) | 32.75 | 35.18 |
| Residues in most favored regions (%)* | 98.14 | 98.10 |
| Residues in additional allowed regions (%)* | 1.86 | 1.90 |
| Residues in disallowed regions (%)* | 0 | 0 |
| Collection location | BNL X4A | Rigaku Micromax-007 HF |
Ramachandran plot analysis.
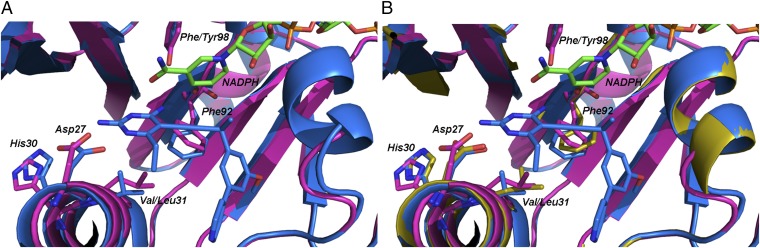
Comparisons of the Sa(V31L/F98Y) structure with the structure of the WT enzyme bound to compound 1 (Fig. 2A) reveal a structural basis of resistance indicated by significant conformational changes induced by the presence of the two mutations. A full table of interactions of the amino acids and compound 1 is presented in Dataset S1. The most significant amino acid reorientations are observed between Phe-92, Val-3, and the binding site of the diaminopyrimidine moiety of compound 1. The major change centers on Leu-31, which projects 2.0 Å farther into the active site than the WT Val-31 residue, resulting in a corresponding 2.3-Å shift in Phe-92 into the active site and a 1.4-Å shift of the backbone carbonyl (Fig. 2A). This new Phe-92 orientation restricts the side chain from adopting the position needed for ligand binding and disrupts stabilizing hydrophobic interactions with the acetylene linker. The shift of the Phe-92 carbonyl also results in the loss of a hydrogen bond to the four-amino group of the pyrimidine. Additionally, Leu-31 is 2.2 Å from the 6-ethyl substituent of the diaminopyrimidine, resulting in repulsive steric interactions.
Fig. 2.
Crystal structures of the WT and mutant enzymes show conformational changes at Phe-92 and His-30. (A) Superposition of the structures of Sa(WT):NADPH:1 from PDB ID code 3SGY (blue) with Sa(V31L/F98Y):NADPH (magenta). (B) Superposition of the structures of Sa(WT):NADPH:1 from PDB ID code 3SGY (blue) with Sa(F98Y):NADPH (yellow) and Sa(V31L/F98Y):NADPH (magenta).
Furthermore, the B helix adjacent to the active site and possessing critical amino acids for ligand binding shifts 0.4 Å away from the active site. Distances between the ligand and amino acids Asn-25, Asp-27, His-30, and Leu-34 are increased by 0.4 Å, concomitantly reducing hydrogen bonding and van der Waals interactions essential for stability and ligand binding. Moreover, the shift in the B helix results in a 1.1-Å shift in the imidazole ring of His-30. The shifted His-30 side chain extends the binding site, allowing for a glycerol molecule to displace a water molecule that typically provides stabilizing hydrogen bonds between the pyrimidine amino group of 1 and the His-30 imidazole (Fig. S4). A similar disruption of the water network has been previously shown in a crystal structure of the clinically observed resistance mutant Sa(H30N/F98Y) with NADPH and a propargyl-linked antifolate (14). Combined, the observations for Phe-92, Leu-31, and the B helix explain the lower affinity of compound 1 for the mutant enzyme.
The Sa(V31L/F98Y) enzyme maintains catalytic competency. Comparisons of the Sa(V31L/F98Y) structure with WT Sa/NADPH/DHF (PDB ID code 3FRD) (15) indicate that a shift in the Phe-92 peptide carbonyl would have little or no effect on DHF binding or turnover, because there are no direct interactions between the two groups. Any minor steric interactions between Phe-92 and the pterin ring of DHF may be compensated by the additional interactions in the glutamate tail, which remains undisturbed.
To verify which structural effects result from the presence of the mutations and which result from an enzyme lacking a bound ligand, we determined the structure of Sa(F98Y)DHFR bound only to NADPH (statistics for data and refinement are found in Table 4). Comparisons of the structures of binary Sa(F98Y):NADPH, Sa(F98Y/V31L):NADPH, and ternary Sa:NADPH:1 indicate that the reorientations of Phe-92 and His-30 are caused by the presence of the mutations. Like the Sa(F98Y/V31L):NADPH structure, the Sa(F98Y):NADPH structure features the same 0.4-Å shift of the B helix away from the active site. However, the conformation of Phe-92 in the Sa(F98Y/V31L) structure is clearly influenced by the V31L mutation, because the conformation of this residue in the Sa(F98Y) structure is ∼0.5 Å closer to that observed in the ternary structure. Similarly, the V31L mutation influences the conformation of His-30, because a comparison of the Sa(F98Y):NADPH and ternary (Sa:NADPH:1) structures shows that the His-30 conformation is the same (Fig. 2B).
Comparisons of the crystal structure of Sa(F98Y/V31L):NADPH (Fig. 2A) with the lowest energy-predicted structure of the K*-predicted single V31L mutant (Fig. 1) show a conservation of the effect of the V31L mutation on Phe-92. The predicted structure also indicates a steric interaction between Leu-31 and the C-6-ethyl substituent of the pyrimidine ring of compound 1, which matches the crystallographic results. Incidentally, when K* was used to predict resistance mutations with another propargyl-linked antifolate that maintains the same atoms as compound 1 other than possessing a methyl instead of an ethyl group at the C-6 position of the pyrimidine ring, the Val-31 mutants ranked lower than the Leu-5 mutations. These results validate that the steric interaction between Leu-31 and the ethyl group specifically contributes to resistance.
In summary, the K* algorithm in OSPREY was used to predict unique single mutations in the active site of S. aureus DHFR that confer resistance to an experimental propargyl-linked antifolate, 1. Four of the predicted mutant enzymes were created and shown to be catalytically competent and resistant to compound 1, with the top-ranked mutant having a 58-fold reduction in inhibitor potency. Excitingly, the computational predictions were shown to be not only biochemically validated but also, selected in the bacteria under antibiotic pressure, because the top-ranked mutation, V31L, was selected in the background of an F98Y mutation, which has been clinically observed. Exploration of the enzymatic fitness of this double mutant revealed a compensatory relationship between the single F98Y and V31L mutations that results in a doubly mutated enzyme with fitness comparable with the WT enzyme. Consideration of the cellular fitness revealed that the double-mutant strain only suffered a slight loss in fitness over both the progenitor strain and the previously characterized F98Y strain. Crystal structures of the double-mutant enzyme revealed the structural basis of compound resistance.
The mutation V31L emerged as the top-ranked SNP that maintained dihydrofolate binding while conferring inhibitor resistance by perturbing Phe-92 and sterically interfering with the C-6-ethyl group of the pyrimidine ring. Interestingly, there is a strong correlation between these results and those obtained in the first application of K* to identify double mutants of SaDHFR that confer resistance. In a previous study (1), the seven top-ranked mutations were variants of Val-31 and Phe-92; a crystal structure of the V31Y/F92I mutant enzyme shows that the F92I mutation reduces van der Waals interactions and that the V31Y mutation introduces destabilizing steric bulk. Overall, it is striking that the same structural effect is selected with both applications of K* whether it is applied to identify double or SNP mutations.
S1 DHFR (from dfrA), a plasmid-acquired enzyme on transposon Tn4003 that confers high levels (>100-fold) of trimethoprim resistance, shares three of four active-site mutations (F98Y, V31I, G43A, and L5I) with the predicted K* mutations (16). Similar to our conclusions, S1 DHFR crystal structures indicate that F98Y disrupts NADPH binding. In fact, crystal structures with S1 DHFR bound to NAPDH and trimethoprim completely lack NADPH in three of six molecules in the asymmetric unit (17); related experiments show that the synergy between NADPH and trimethoprim is eliminated in dfrA. The F98Y mutation has also been shown to disrupt NADPH binding in the context of the propargyl-linked antifolates. When Sa(F98Y) is crystallized with inhibitor, the structure possesses two conformations of NADPH: the standard extended β-NADPH and one with the pyrophosphate moiety in a rotated position (13).
Having validated the mutational prediction capabilities of K*/OSPREY through bacterial selection of the predicted mutants, the algorithm could potentially be applied to many different research areas. Specifically, the computational prediction of drug resistance mutations could be valuable in cases where it is more difficult to raise mutant strains or cell lines in vitro, such as with viruses or cancer cell lines. Overall, the extension of the computational prediction of drug resistance to observations of biologically relevant mutants provides new opportunities in drug discovery, especially for those targets that are most affected by mutational resistance.
Materials and Methods
Computational Prediction of Resistance Mutations.
The K* algorithm (18, 19) within the OSPREY protein design program (2) predicted binding affinities of DHFR mutants to both DHF (positive design) and compound 1 (negative design). Drug resistance was predicted by ranking each mutant and the WT protein by the K* ratio: the K*-positive design score divided by the K*-negative design score.
The set of mutant sequences was selected by choosing all single-nucleotide mutants of the active-site residues. Nine residues in the active site were modeled as flexible and allowed to mutate by up to one nucleotide (substitution): L5{L/V/I/R/Q}, V6{V/A/L/I/F/D/G}, L20{L/V/I/F/S}, L28{L/V/M/W/F/S}, V31{V/A/I/F/L/D/G}, T46{T/A/R/I/K/S}, I50{I/V/L/M/F/N/S/T}, L54{L/R/Q/V}, and F92{F/V/L/I/Y/S/C}. This combination resulted in a total of 47 sequences. Two structures were used as input: a model of the DHF:DHFR:NADPH WT complex (for the positive design) and a model of compound 1. Because a structure of compound 1 bound to DHFR was not available at the time of the design predictions, compound 1 was modeled on the bound structure of a related DHFR inhibitor [PDB ID code 3FQC (13)]. The mutable residues were allowed to assume any conformation in the continuous conformation space (20) within 9° of the rotamers in the Richardson’s penultimate rotamer library (21); in addition, the WT rotamer of Phe-92 was added to the rotamer library. Because of observed conformations of the escape mutations to antifolate inhibitors described in ref. 11, we modeled ∼10,000 possible binding conformations of compound 1 to DHFR. These binding poses were first filtered by OSPREY’s MinDEE/A* algorithms (18) that searched for the lowest energy conformation of any of the mutants to each of the binding conformations. Conformations with predicted energies above a steric threshold where then pruned, resulting in 1,660 binding poses for compound 1. We then used the K* algorithm in OSPREY, a statistical mechanics-derived algorithm that uses the MinDEE/A* algorithms to compute Boltzmann-weighted partition functions over energy-minimized conformational ensembles and generates an approximation to Ka, the association binding constant for a given protein–substrate complex. The resulting K* scores of mutations for both DHF (positive design) and compound 1 (negative design) were used to compute a ratio of scores between positive and negative designs (Table S1).
Selection of Resistant Bacterial Colonies.
Resistant strains were selected by plating overnight culture (∼1012 cfu/mL) of the progenitor strain, ATCC 43300, or S. aureus (F98Y) on Isosensitest Agar (Oxoid) containing compound 1 at six times the MIC of the progenitor strain. After 16–18 h of growth at 37 °C, surviving colonies were harvested for characterization. Genotypic characterization was achieved using direct colony PCR to amplify the dfrB gene using sense primer (5′-ATGACTTTATCCATTCTAGTTGC-3′), antisense primer (5′-TTATTTTTTACGAATTAAATGTAG-3′), and high-fidelity Taq Polymerase (Takara) (22). Mutation frequencies were calculated based on sequencing results for resistant colonies.
Evaluation of Antibacterial Activity.
MICs were determined according to the Clinical and Laboratory Standards Institute Guideline’s Standard Microdilution broth assay using a final inoculum of 5 × 105 cfu/mL in Isosensitest Broth (Oxoid) (23). The MIC was defined as the lowest concentration of inhibitor to visually inhibit growth. Growth was monitored at A600 after 18 h of incubation at 37 °C. MICs were colorimetrically confirmed using Presto Blue (Life Technologies).
Strain Fitness Determination.
Relative strain fitness was determined by pairwise competition assays (24) with trimethoprim as the selective agent. Cell growth was monitored every 20 min by A600, and the doubling time of the strains was determined by monitoring cell growth in the log phase at an absorbance of 600 nm every 15 min for a total of 180 min. Fitness was calculated using the following equation: .
Enzyme Expression and Purification.
Procedures for cloning the Sa(F98Y)DHFR construct in pET41-a(+) have been previously reported (13). A QuikChange Site-Directed Mutatgenesis Kit (Agilent Technologies) was used to mutate Val31 to Leu31 on the Sa(F98Y)DHFR construct using sense (5′-GGCACCTACCAAATGATTTGAAACAT-3′) and antisense (5′-CCTGTTGATAATTTTTTAAGATGTTTC-3′) primers. Mutagenesis was confirmed by sequencing. The recombinant Sa(V31L/F98Y) enzyme was overexpressed in Escherichia coli BL21 (DE3; Invitrogen) cells and purified using nickel affinity chromatography (5Prime). Protein was desalted using a PD-10 column (GE Healthcare) into buffer containing 20 mM Tris (pH 7.0), 20% (vol/vol) glycerol, 0.1 mM EDTA, and 2 mM DTT and stored in aliquots at −80 °C.
Enzymatic Inhibition Assays.
Enzyme inhibition assays were performed by monitoring the rate of NADPH oxidation by DHFR through absorbance at 340 nm at room temperature in assay buffer containing 20 mM TES (N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid) (pH 7.0), 50 mM KCl, 0.5 mM EDTA, 10 mM β-mercaptoethanol, and 1 mg/mL BSA using 0.1 mM NADPH and 2 μg/mL enzyme. Inhibitor, in DMSO, was added to the enzyme–NADPH mixture and allowed to incubate for 5 min before the addition of 0.1 mM DHF in 50 mM TES (pH 7.0).
Enzyme kinetics were determined by nonlinear regression analysis (GraphPad) of data generated by enzyme activity assays using 12.5, 25, 50, 75, and 100 µM DHF with 20 µM NADPH to determine the Km and Vmax for DHF or 12.5, 25, 50, 75, and 100 µM NADPH with 50 µM DHF to determine the Km and Vmax for NADPH.
Sa(V31L/F98Y)DHFR and Sa(F98Y) Crystal Structures.
Sa(V31L/F98Y)DHFR was cocrystallized with NADPH and a propargyl-linked antifolate using the hanging drop vaporization method. Sa(F98Y)DHFR was cocrystallized with NADPH only. Purified protein (20 mg/mL) was incubated with 2 mM NADPH (Sigma-Aldrich) and 1 mM inhibitor in DMSO [in the case of Sa(V31L/F98Y)DHFR] for 2 h on ice. Equal volumes of the protein–cofactor solution were mixed with an optimized crystallization solution containing 13% PEG 10,000, 0.1 M sodium acetate, 0.1–0.2 M 2-(N-morphilino)ethanesulfonic acid (pH 6.0), and 5% γ-butyrolactone. When stored at 4 °C, conditions typically yielded crystals within 7 d. Crystals were frozen in cyroprotectant buffer containing 25% glycerol. High-resolution data were collected on the X4A Beamline at Brookhaven National Laboratories for Sa(V31L/F98Y)DHFR:NADPH and the Rigaku HighFlux HomeLab Protein Crystallography X-Ray System at the University of Connecticut for Sa(F98Y):NADPH.
Data for Sa(V31L/F98Y)DHFR:NADPH and Sa(F98Y):NADPH were indexed and scaled using HKL2000 and d*TREK, respectively. Phaser (25) was used to identify molecular replacement solutions for the structures of Sa(V31L/F98Y)DHFR:NADPH and Sa(F98Y):NADPH using PDB ID code 3F0U (13) or 3FQO (13) as probe molecule, respectively. The programs Coot (26) and Phenix (27) were used for structure refinement until acceptable Rwork and Rfree were achieved. Structural geometry was evaluated by Procheck (28) and Ramachandran plots.
Inhibitors.
The synthesis and characterization of compound 1 have been described (5). Trimethoprim is commercially available (Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Dennis Wright for providing compound 1 and William Clemons, Kyle Roberts, and all members of the laboratories of B.R.D. and A.C.A. for helpful discussions. The authors gratefully acknowledge the support of National Institutes of Health (NIH) Grants GM 78031 (to B.R.D.) and AI111957 (to A.C.A.). Data for this study were measured at Beamline X4A of the National Synchrotron Light Source.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.P.-M. is a guest editor invited by the Editorial Board.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4Q6A and 4Q67).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411548112/-/DCSupplemental.
References
- 1.Frey KM, Georgiev I, Donald BR, Anderson AC. Predicting resistance mutations using protein design algorithms. Proc Natl Acad Sci USA. 2010;107(31):13707–13712. doi: 10.1073/pnas.1002162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gainza P, et al. OSPREY: Protein design with ensembles, flexibility, and provable algorithms. Methods Enzymol. 2013;523:87–107. doi: 10.1016/B978-0-12-394292-0.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CY, Georgiev I, Anderson AC, Donald BR. Computational structure-based redesign of enzyme activity. Proc Natl Acad Sci USA. 2009;106(10):3764–3769. doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale GE, et al. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J Mol Biol. 1997;266(1):23–30. doi: 10.1006/jmbi.1996.0770. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan K, et al. Toward new therapeutics for skin and soft tissue infections: Propargyl-linked antifolates are potent inhibitors of MRSA and Streptococcus pyogenes. PLoS ONE. 2012;7(2):e29434. doi: 10.1371/journal.pone.0029434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao G-F, Yang G-F, Zhan C-G. Structure-based methods for predicting target mutation-induced drug resistance and rational drug design to overcome the problem. Drug Discov Today. 2012;17(19-20):1121–1126. doi: 10.1016/j.drudis.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu S, Travis SM, Elcock AH. Accurate calculation of mutational effects on the thermodynamics of inhibitor binding to p38α MAP kinase: A combined computational and experimental study. J Chem Theory Comput. 2013;9(7):3151–3164. doi: 10.1021/ct400104x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikita H, Warshel A. Predicting drug-resistant mutations of HIV protease. Angew Chem Int Ed Engl. 2008;47(4):697–700. doi: 10.1002/anie.200704178. [DOI] [PubMed] [Google Scholar]
- 9.Hao G-F, Yang G-F, Zhan C-G. Computational mutation scanning and drug resistance mechanisms of HIV-1 protease inhibitors. J Phys Chem B. 2010;114(29):9663–9676. doi: 10.1021/jp102546s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safi M, Lilien RH. Efficient a priori identification of drug resistant mutations using Dead-End Elimination and MM-PBSA. J Chem Inf Model. 2012;52(6):1529–1541. doi: 10.1021/ci200626m. [DOI] [PubMed] [Google Scholar]
- 11.Frey KM, Viswanathan K, Wright DL, Anderson AC. Prospective screening of novel antibacterial inhibitors of dihydrofolate reductase for mutational resistance. Antimicrob Agents Chemother. 2012;56(7):3556–3562. doi: 10.1128/AAC.06263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vickers AA, Potter NJ, Fishwick CW, Chopra I, O’Neill AJ. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. J Antimicrob Chemother. 2009;63(6):1112–1117. doi: 10.1093/jac/dkp090. [DOI] [PubMed] [Google Scholar]
- 13.Frey KM, et al. Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance. J Mol Biol. 2009;387(5):1298–1308. doi: 10.1016/j.jmb.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey KM, Lombardo MN, Wright DL, Anderson AC. Towards the understanding of resistance mechanisms in clinically isolated trimethoprim-resistant, methicillin-resistant Staphylococcus aureus dihydrofolate reductase. J Struct Biol. 2010;170(1):93–97. doi: 10.1016/j.jsb.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oefner C, et al. Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity. J Antimicrob Chemother. 2009;63(4):687–698. doi: 10.1093/jac/dkp024. [DOI] [PubMed] [Google Scholar]
- 16.Dale GE, et al. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermidis ATCC 14990: The origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol. 1995;177(11):2965–2970. doi: 10.1128/jb.177.11.2965-2970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaslet H, et al. Structural comparison of chromosomal and exogenous dihydrofolate reductase from Staphylococcus aureus in complex with the potent inhibitor trimethoprim. Proteins. 2009;76(3):706–717. doi: 10.1002/prot.22383. [DOI] [PubMed] [Google Scholar]
- 18.Georgiev I, Lilien RH, Donald BR. The minimized dead-end elimination criterion and its application to protein redesign in a hybrid scoring and search algorithm for computing partition functions over molecular ensembles. J Comput Chem. 2008;29(10):1527–1542. doi: 10.1002/jcc.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilien RH, Stevens BW, Anderson AC, Donald BR. A novel ensemble-based scoring and search algorithm for protein redesign and its application to modify the substrate specificity of the gramicidin synthetase a phenylalanine adenylation enzyme. J Comput Biol. 2005;12(6):740–761. doi: 10.1089/cmb.2005.12.740. [DOI] [PubMed] [Google Scholar]
- 20.Gainza P, Roberts KE, Donald BR. Protein design using continuous rotamers. PLOS Comput Biol. 2012;8(1):e1002335. doi: 10.1371/journal.pcbi.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40(3):389–408. [PubMed] [Google Scholar]
- 22.Lorian V. Antibiotics in Laboratory Medicine. Lippincott Williams & Wilkins; Philadelphia: 2005. [Google Scholar]
- 23.Clinical Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; Sixteenth Informational Supplement. Clinical Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 24.Lenski R. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution. 1988;42(3):425–432. doi: 10.1111/j.1558-5646.1988.tb04149.x. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.