Abstract
Cancer is one of the deadliest diseases worldwide, accounting for about 8 million deaths a year. For solid tumors, cancer patients die as a result of the metastatic spread of the tumor to the rest of the body. Therefore, there is a clinical need for understanding the molecular and cellular basis of metastasis, identifying patients whose tumors are more likely to metastasize, and developing effective therapies against metastatic progression. Over the years, Raf kinase inhibitory protein (RKIP) has emerged as a natural suppressor of the metastatic process, constituting a tool for studying metastasis and its clinical outcomes. Here, we review RKIP’s role as a metastasis suppressor and the signaling networks and genes regulated by RKIP in metastatic, triple-negative breast cancer. We also highlight the clinical implications and power of building gene signatures based on RKIP-regulated signaling modules in identifying cancer patients that are at higher risk for metastases. Finally, we highlight the potential of RKIP as a tool for developing new therapeutic strategies in cancer treatment.
Keywords: BACH-1, gene signature, HMGA-2, metastasis, Raf-1, RKIP, metastasis, suppressor, triple-negative breast cancer
I. INTRODUCTION
Metastasis is the process by which tumor cells leave their primary site and spread to the rest of the body. In most cases, multiple organ dysfunction or failure due to the aggressive metastatic spread of the tumor is the cause of lethality in cancer patients.1 When cancer patients present with a late-stage metastatic tumor at the clinic, their prognosis is generally poor regardless of the type of cancer. Moreover, the metastatic phenotype correlates with resistance to treatment and recurrence, which can partially be attributed to tumor cells’ ability to escape and seed in new parts of the body. Metastasis is a dynamic process by which tumor cells continually move in and out of tissues that can even result in seeding of tumor cells back in their primary site long after the primary tumor has been removed.2 Developing therapies that block the metastatic process are, therefore, an urgent clinical need. Blocking metastasis, however, is challenging because it involves multiple steps, each of which is mediated by diverse signaling events within the tumor cell as well as its micro- environment.3 Reaching a comprehensive understanding of how metastasis works and elucidating the key signaling mechanisms and cell types that mediate this process is essential for development of effective anti-metastatic therapies.
How do we study such a complex event? One effective way of investigating metastasis is by studying the natural inhibitors of this process—metastasis suppressors. Metastasis suppressors are proteins that can block the spread of tumor cells without affecting the growth properties of the primary tumor.4 Metastasis involves a multitude of steps including migration, invasion through tissue, intravasation into blood or lymph vessels, extravasation from circulatory vessels, and colonization at new tissue sites 5 Each step involves different cell-cell interactions and signaling pathways, rendering it difficult to sort out the mechanisms regulating metastasis. Metastasis suppressor proteins can interfere with different stages of the metastatic process such as intravasation or colonization. Understanding the function of these metastasis suppressors and the signaling events that they govern can yield important mechanistic insights into the complexity of the metastatic process.
Raf kinase inhibitory protein (RKIP), a modulator of kinase activity and a cellular homeostasis factor, also functions as a metastasis suppressor in multiple solid tumor types such as prostate and breast cancer.6,7 We will focus here on kinase signaling events and downstream gene expression changes induced by RKIP to block metastasis of breast tumor cells. Building RKIP-regulated signaling networks allows us not only to get a comprehensive view of the metastatic process at a cellular and molecular level, but also to generate pathway-based gene signatures that can identify subgroups of patients with the most aggressive tumors that are more likely to metastasize.
RKIP’s role as a metastasis suppressor was initially described in prostate cancers. Keller and colleagues demonstrated with their comparison of metastatic prostate cancer cell lines to non-metastatic cell lines that expression levels of RKIP are a determinant factor for the metastatic phenotype.6 RKIP also acts as a metastasis suppressor in breast cancers, particularly in triple negative breast cancers (TNBCs). RKIP expression is usually lost in these poor-prognosis tumors, correlating with the highly metastatic phenotype.8–11 In both in vitro and in vivo models of TNBC, the Rosner lab demonstrated that RKIP blocks multiple steps of the metastatic process. In vitro, RKIP expression results in significant reduction in invasive potential of these cells without any effect on the growth properties.7 In xenograft models, RKIP blocks intravasation of tumor cells as well as later stages of metastasis such as extravasation and colonization. Understanding how RKIP blocks metastasis in TNBCs is particularly important because there are currently no targeted therapies available for TNBC patients. This review will focus on signaling pathways and genes that mediate RKIP function in cancers, with a focus on TNBCs, and describe how these signaling networks can be used clinically to predict patient survival outcomes as well as to develop therapeutic strategies.
II. RKIP-REGULATED SIGNALING MODULES
A. Raf-MEK-ERK Module
RKIP is an inhibitor of Raf-stimulated MAP kinase (MAPK) signaling. A yeast two-hybrid assay that utilized the kinase domain of Raf-1 as bait originally identified RKIP as a Raf binding protein.12 High RKIP concentrations are able to interfere with MEK binding, preventing MEK activation.13 However, RKIP depletion in cells revealed that RKIP blocks Raf-1 activation under physiological conditions. RKIP inhibits Raf-1 subsequent to Raf-1 membrane translocation but prior to phosphorylation of downstream targets such as MEK and ERK.14 RKIP binding to subdomains I and II of the Raf-1 kinase domain blocks phosphorylation of residues Ser338 by PAK and Tyr340/341 by Src, both of which are required for activation of Raf-1 on growth factor stimulation.15 Enhanced Raf-1 activation in RKIP-depleted cells enabled increased DNA synthesis and cell proliferation at lower EGF concentrations, effectively enhancing the sensitivity of the system to stimuli.14
Interestingly, RKIP-mediated inhibition of Raf activation is isoform specific. RKIP depletion studies show that RKIP inhibits the phosphorylation and activation of only Raf-1, and not B-Raf.14,16 Some residues targeted by RKIP are constitutively active in B-Raf, which may explain why RKIP does not inhibit B-Raf activation. Consistent with these findings, RKIP knockout has no effect on ERK stimulation in MEFs that are largely dependent on B-Raf signaling.17 However, when B-Raf expression or activity is compromised in MEFs, then subsequent RKIP depletion can rescue MAPK signaling through activation of Raf-1.17 RKIP may also regulate B-Raf signaling indirectly through Raf-1. Thus, in some cell types, Raf-1 and B-Raf form heterodimers to activate downstream MAPK signaling, and the intensity of heterodimer-mediated signal is larger than that mediated by monomers of these MAPK activators individually.16,18 Finally, high overexpression could explain the observation that exogenous RKIP inhibits B-Raf activity in melanoma cells.19 Taken together, these studies suggest that RKIP regulation of B-Raf can vary with context and cell type.
RKIP also plays a role in protein kinase C (PKC)-induced Raf activation. The PKC family of serine/threonine kinases is required for stimulation of MAP kinase signaling by growth factors.20 However, no direct mechanism has been identified since PKC phosphorylation of Raf-1 does not appear to be necessary for Raf-1 activation. An alternative mechanism involves functional inactivation of RKIP by PKC. Rosner and colleagues showed that PKC phosphorylates RKIP at serine residue 153 (S153) in neuronal cells, and this phosphorylation leads to dissociation of RKIP from Raf-1 and subsequent activation of MEK and ERK.21 Similar findings were later reported for regulation of RKIP by PKC in cardiac myocytes.22 The role of S153 phosphorylation in regulating RKIP inhibition of Raf/MAPK signaling has now been demonstrated in a variety of cell types.7,23
B. Regulation of Spindle Checkpoint and Cell Cycle by RKIP
Multiple members of the MAPK cascade have been implicated in cell cycle entry and exit. Raf-1 is a known regulator of mitotic cell cycle progression, and phosphorylation of S338 on Raf-1 by PAK was suggested to influence this progression in mammalian cells.24,25 Activated ERK1/2 localizes with important structures of the cell cycle process such as kinetochores, spindle poles, and the midbody during different stages of cell cycle in a dynamic fashion.26,27 As discussed above, RKIP regulates phosphorylation of Raf-1 and the activation of downstream effectors MEK and ERK, raising the possibility that RKIP also influences cell cycle dynamics and mitotic progression.
Rosner and colleagues demonstrated that RKIP can regulate the spindle checkpoint during cell cycle progression.28 RKIP depletion causes downregulation of aurora kinase B activity, overrides the mitotic spindle checkpoint, and pushes cells into rapid mitotic progression in the presence of spindle poisons. These functions are all mediated at least partially by activation of the Raf-1-MEK-ERK signaling, since inhibitors of these kinases can rescue the RKIP-depletion phenotype. All of these data are consistent with the observation that RKIP potentiates apoptosis and enhances cell cycle arrest when cells are treated with chemotherapeutic agents that target microtubules, such as Taxol.29 The cells that survive RKIP loss slowly accumulate chromosomal abnormalities, potentially introducing mutations and translocations into DNA that are more likely to lead to disease states than catastrophic mitotic events.
C. Identification of an RKIP Signaling Network that Regulates Metastasis
Since RKIP functions as a metastasis suppressor, elucidating signaling events that are regulated by RKIP can reveal new therapeutic strategies that target mediators of tumor metastasis. To determine whether RKIP suppresses metastasis of triple negative breast cancer (TNBC) cells through inhibition of Raf/MEK/ERK signaling, Rosner and colleagues mapped out a signaling pathway by which RKIP prevents invasion through the extracellular matrix, entry of tumor cells into the blood, and colonization at distant organ sites, The let-7/miR-98 family of microRNAs has been shown to suppress Ras and downregulate subsequent MAPK signaling, which is a function similar to that of RKIP.30
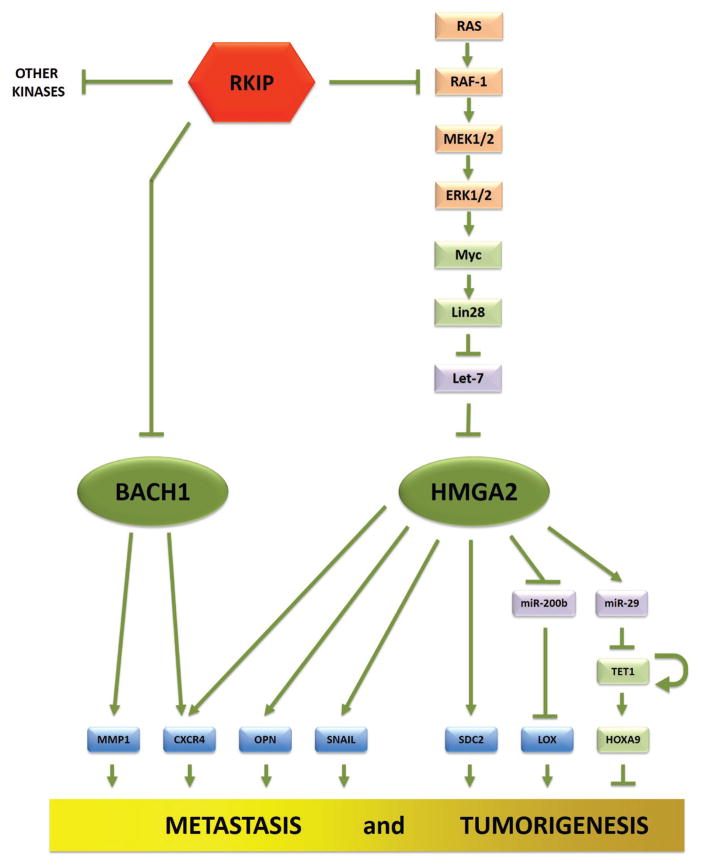
Both in vitro and in vivo work showed that RKIP-mediated downregulation of invasion and metastasis indeed involves let-7 and HMGA2. Let-7 and HMGA2 have been implicated in a variety of cancers.31–35 HMGA2 is a chromatin remodeling factor that promotes EMT and invasion by inducing transcription factors such as Snail, Slug, and Twist.7,36 These findings unraveled a downstream mechanism through which RKIP inhibits invasion, but did not reveal how RKIP actually induced let-7 expression. To address this question, Rosner and colleagues demonstrated a role for LIN28, a let-7 regulator. RKIP downregulates LIN28 by decreasing the occupancy of Myc at the LIN28 promoter region, which connects LIN28 expression to the major RKIP-regulated signaling module Raf-MEK-ERK-Myc (Fig. 1). This work demonstrated for the first time that let-7 can be regulated by a metastasis suppressor, RKIP, and showed that let-7 is a new member of a larger group of microRNAs37,38 that influence breast cancer metastasis.
FIG. 1.
Network summarizing RKIP regulation of metastatic cascades in breast cancer. This scheme highlights novel signaling pathways and potential drug targets. See text for further explanation of approaches and data supporting this scheme.
The RKIP-Myc-LIN28-let-7 signaling cascade was further expanded by Rosner and colleagues, who identified biologically and clinically relevant pro-metastatic factors that are downstream of let-7.39 To generate novel signaling networks, they developed an integrated experimental and bioinformatics approach based on clinical gene expression data and cell line verification that enabled both hypothesis building and testing as well as clinical validation.40 Data from over 1200 patients with heterogeneous tumor subtypes were analyzed. The clinical significance of this and subsequent studies from the Rosner group lies in the use of large expression data sets from breast cancer patients for identification of novel signaling networks as well as independent cohorts of breast cancer patients for validation.
Let-7 expression cannot be directly interrogated in the majority of databases because it is a microRNA. Therefore, Rosner and colleagues rationalized that some of the predicted let-7 targets should also be regulated by RKIP. Comparing genes that are downregulated when RKIP is overexpressed to genes predicted to be let-7 targets should identify common genes that are potentially downstream players of the RKIP-let-7 cascade. With this rationale, Yun et al. identified the BTB and CNC homology 1 (BACH1) gene as a novel target of let-7 that regulates metastasis of breast cancers along with HMGA2.39 A similar approach, based on an inverse correlation between RKIP and a ~100 gene bone metastasis signature,41 identified additional downstream regulators of metastasis. Finally, in vitro and in vivo experiments demonstrated that the RKIP-let-7 module regulates CXCR4, MMP1, and OPN via the identified let-7 targets HMGA2 and BACH1 (Fig. 1).
Gene expression and microRNA expression arrays using TNBC cell lines further extended the RKIP signaling cascade to new microRNAs and extracellular matrix target genes that are involved in metastatic signaling. These analyses identified three additional downstream targets of RKIP and HMGA2: miR-200, lysine oxidase (LOX), and syn-decan 2 (SDC2).42 miR-200 has been implicated in breast tumor cell initiation and the epithelial-mesenchymal transition that leads to cell invasion.32 LOX is a known collagen and elastin cross-linker that helps invasion and metastasis.43 SDC2 is a transmembrane heparan sulfate proteoglycan that mediates cell adhesion, cell-matrix interactions, and signaling, but its function in breast cancer is not known.44 Experimental data demonstrated that RKIP regulates LOX and SDC2 expression, and this regulation is mediated by inhibition of HMGA2. Reduction of HMGA2 expression by RKIP results in upregulation of miR-200, which in turn downregulates its direct target LOX. SDC2, on the other hand, was not a direct miR-200 target, but was still induced by HMGA2, suggesting that SDC2 contributes to RKIP function independently of the HMGA2-miR-200 module (Fig. 1). Depletion of SDC2 was shown to suppress breast cancer cell growth, invasion, and metastasis both in vitro and in vivo.
Another target of HMGA2 is the epigenetic regulator ten-eleven translocation 1 (TET1). TET proteins are known to initiate demethylation of DNA by converting methylcytosine in hydroxymethylcytosine. Methylation of gene promoter regions are thought to suppress transcription of the gene, and demethylation enables transcriptional activation. HMGA2 depletion in breast cancer cells results in suppression of miR29 and induction of the miR29 target, TET1 (data now shown).45 TET1, in turn, demethylates promoters for the homeobox A (HOXA) genes and activates their transcription (Fig. 1). HOXA9 expression, in particular, dramatically increases after exogenous expression of TET1. TET1 also demethylates its own promoter as part of a positive feedback loop. TET1 and HOXA9 expression suppress cancer cell invasion, tumor growth, intravasation, extravasation, and metastasis of TNBC cells under in vitro as well as in vivo conditions. TET1, as a downstream mediator of RKIP signaling, represents a global epigenetic mechanism that can account in part for the role of RKIP as a master regulator of complex processes such as metastasis.
III. POTENTIAL CLINICAL APPLICATIONS OF RKIP SIGNALING PATHWAYS
A. Building RKIP Pathway-Based Gene Signatures for Analysis of Clinical Outcomes
One of the major difficulties in cancer treatment is identifying patients who will benefit from therapy. Organ-specific metastasis gene signatures based on the gene expression profiles of tumors have been developed in order to predict the likelihood that a patient’s primary tumor will metastasize to distant organs. A few of these signatures have been in clinical use, but they are primarily effective for patients that express the estrogen or HER2 receptors.46,47
Because the metastasis signaling networks regulated by RKIP (Fig. 1) were identified using TNBCs, Rosner and colleagues generated new gene signatures based on these networks. Initially, they developed a seven-gene “RKIP pathway metastasis signature” (RPMS).39 In addition to the five key factors of the pathway (RKIP, HMGA2, MMP1, CXCR4, and OPN), it included two meta-genes, Let-7-TG (let-7 target genes) and BACH1-TG. These meta-genes are derived using a weighted average of expression values for let-7 and BACH1 target genes, respectively, based on correlations with RKIP expression. Therefore, even though the gene signature is based on a network of seven genes, it actually measures expression of 117 genes, most of which are averaged into let-7 and BACH1 meta-genes. The genes in this signature only stratify patients when they are utilized together and not analyzed individually, suggesting that the entire pathway is the critical predictor. This is confirmed by random survival forest (RSF) analysis of these genes, which showed that these genes cooperate in patient stratification, especially in low-RKIP tumors. Other gene signatures were also derived based on RKIP, HMGA2, LOX, and SDC2,42 and the HMGA2, TET1, HOXAA7, and HOXA9 networks.45 The general strategy for developing pathway gene signatures in this manner has been described previously.40
In order to improve the RPMS gene signature and increase its ability to predict patient outcome specifically in TNBC patients, Lee and colleagues developed a BACH1-pathway metastasis signature (BPMS).48 BPMS differs from previously generated metastasis gene signatures for a number of reasons including (i) the BACH1 meta-gene is defined based on experimental data as opposed to bioinformatically predicted targets; (ii) the number and choice of genes are optimized (30 genes total); (iii) the numerical cutoffs of high versus low expression for each gene are optimized; and (iv) the BPMS is a single patient predictor. With these improvements, BPMS is able to predict patient outcome related to metastasis-free survival specifically in triple negative breast cancer patients.
B. Inhibition of RKIP to Regulate Metastatic Progression
Loss of RKIP in cancers enables activation of pro-growth signals at subthreshold concentrations of growth factors, and increases the maximum effect that these factors have on cells.14 This sensitizes the tumors to extracellular stimuli and drives their progression, resulting in more aggressive and metastatic tumors. In addition, Rosner and colleagues recently identified an inverse relationship between the metastasis suppressor RKIP and the metastasis promoter BACH1 that can play a role in potentiating breast cancer progression.49 They showed that BACH1 directly inhibits RKIP as well as its own (BACH1) transcription. In combination with the indirect inhibition of BACH1 by RKIP (Fig. 1), this regulatory architecture enables single cells to generate a stable subpopulation of pro-metastatic cells without any genetic changes. Thus, stochastic fluctuations in BACH1 expression can lead to RKIP suppression and the induction of BACH1-expressing pro-tumorigenic cells.
This study also found that histone deacetylase inhibitors (HDACs) or depletion of the polycomb repressor EZH2 differentially affect BACH1 and RKIP gene expression in a cell type-dependent manner—in some cases leading to a pro-metastatic phenotype. Together, these results demonstrate that a mutually inhibitory relationship between two regulators of metastasis like RKIP and BACH1 (or Snail) has the potential to promote metastatic progression in cancer, and suggest that using HDAC or EZH2 inhibitors for treatment of breast cancer may lead to mixed outcomes.
IV. CONCLUDING REMARKS
Metastasis is a complex multistep process that involves a multitude of cellular and molecular interactions, and disentangling them can be challenging. Studying metastasis suppressors, as exemplified here using RKIP, is an effective strategy for identifying key signaling networks and mechanisms that mediate dissemination of tumor cells, as well as developing pathway-based gene signatures such as the BPMS that predicts metastatic risk in TNBC patients. As a guide to potential therapeutic strategies, it is now possible to determine whether other identified downstream targets or effectors of RKIP such as gene methylation are differentially expressed among BPMS patients.
In addition to the Raf-1-MEK-ERK module, RKIP is a regulator of other signaling pathways within the cell. RKIP-mediated kinase regulation has been implicated in G-protein coupled receptor (GPCR), NFκB, and GSK3β signaling pathways (reviewed in Ref. 50). RKIP has also been shown to play a role in the oxidative stress response that can trigger activation of the p38 stress kinase in cells.51 Downstream of these pathways are genes that are involved in cellular growth and cell cycle progression (such as Cyclin D1 and Aurora B kinase), proliferation (such as NFκB targets), EMT and invasion (such as Snail, Slug, MMP1), and metastasis (such as CXCR4, OPN, BACH1). Finally, investigation of the RKIP metastasis suppressor network has revealed a number of other regulators of tumor progression and metastasis including let-7, LIN28, HMGA2, BACH1, MMP1, OPN, CXCR4, LOX, SDC2. TET1, and the HOXA genes (Fig. 1).7,39,42,45 Taken together, RKIP functions as an epigenetic regulator that alters many steps in metastasis from kinase activation and signaling to migration, invasion and homing to metastatic sites.
Considering that RKIP regulates key factors that play a role in growth of tumor cells, it might seem contradictory that RKIP does not have much effect on the growth of the primary tumor. Such expectations stem from the assumption that signaling pathways are linear pathways, and that downstream effector functions should be mimicked by all upstream regulators under experimental conditions. However, pathways regulated by RKIP are intertwined and subject to cross talk via both positive and negative feedback loops. It is quite possible that RKIP depletion causes changes in anti-proliferative or apoptotic factors that counteract the effect of pro-proliferative signals. Therefore, understanding the function of RKIP requires systemic identification of the extensive RKIP-regulated signaling network.
Since RKIP inhibits many kinases, as discussed in this special issue, the anti-metastatic function of RKIP is likely to be mediated by smaller changes in the activity of multiple pathways that all work in concert. Developing cancer treatments that target individual kinases using small molecule inhibitors have proven inefficient as tumors almost always develop resistance to the treatment. One example is the MEK inhibitor that can potentially be used for the treatment of breast and other cancers.52 Johnson and colleagues have demonstrated that tumor cells overcome MEK inhibition by reprogramming their kinome and activating alternative pathways,53 and the reprogramming effect is stronger with higher doses of the inhibitors. This observation argues that high-dose single agent regimens can have the contradictory effect of facilitating the aggressive disease phenotype. Therefore, alternative therapeutic strategies need to be sought. Given our current understanding of RKIP function as a kinase modulator that blocks metastatic disease progression, more effective therapies can be developed by reactivating RKIP in tumor cells where its expression is suppressed, or by mimicking RKIP’s effect on the kinome of the tumor cells using combinations of multiple kinase inhibitors at much lower doses.
Acknowledgments
We thank Casey Frankenberger for helpful comments. The work presented here was supported by National Institutes of Health grant GM087630 (to M.R.R.).
ABBREVIATIONS
- BACH1
BTB and CNC homology 1, basic leucine zipper transcription factor 1
- BMS
bone metastasis signature
- CTGF
connective tissue growth factor
- CXCR4
chemokine (C-X-C motif) receptor 4
- EGF
epidermal growth factor
- EMT
epithelial mesenchymal transition
- ERK
extracellular signal-regulated kinase
- HMGA2
high mobility group AT-hook 2
- HOXA
homeobox A
- IL-11
interleukin-11
- KSR
kinase suppressor of Ras
- MEF
mouse embryonic fibroblast
- MEK
mitogen/extracellular signal-regulated kinase
- MMP1
matrix metalloproteinase 1
- MYC
v-myc avian myelocytomatosis viral oncogene homolog
- NEK1
NIMA-related kinase 1
- NFκB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- OPN
osteo-pontin
- PAK
p21-activated kinase
- PKC
protein kinase C
- RAF-1
V-Raf-1 murine leukemia viral oncogene homolog 1
- RAS
rat sarcoma
- RKIP
Raf kinase inhibitory protein
- RPMS
RKIP pathway metastasis signature
- RSF
random survival forest
- SDC2
syndecan-2
- TET1
tet methylcytosine dioxygenase 1
- TNBC
triple negative breast cancer
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Comen E, Norton L, Massague J. Clinical implications of cancer self-seeding. Nat Rev Clin Oncol. 2011;8:369–77. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 3.Brabletz T, Lyden D, Steeg PS, Werb Z. Roadblocks to translational advances on metastasis research. Nat Med. 2013;19:1104–9. doi: 10.1038/nm.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, Vivian CJ, Brinker AE, Hampton KR, Lianidou E, Welch DR. Microenvironmental Influences on Metastasis Suppressor Expression and Function during a Metastatic Cell’s Journey. Cancer Microenviron. 2014 doi: 10.1007/s12307-014-0148-4. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–21. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–89. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 7.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–58. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinho O, Pinto F, Granja S, Miranda-Goncalves V, Moreira MA, Ribeiro LF, di Loreto C, Rosner MR, Longatto-Filho A, Reis RM. RKIP inhibition in cervical cancer is associated with higher tumor aggressive behavior and resistance to cisplatin therapy. PLoS One. 2013;8:e59104. doi: 10.1371/journal.pone.0059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinho O, Granja S, Jaraquemada T, Caeiro C, Miranda-Goncalves V, Honavar M, Costa P, Damasceno M, Rosner MR, Lopes JM, Reis RM. Downregulation of RKIP is associated with poor outcome and malignant progression in gliomas. PLoS One. 2012;7:e30769. doi: 10.1371/journal.pone.0030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Mulla F, Bitar MS, Thiery JP, Zea TT, Chatterjee D, Bennett L, Park S, Edwards J, Yeung KC. Clinical implications for loss or diminution of expression of Raf-1 kinase inhibitory protein and its phosphorylated form in ductal breast cancer. Am J Cancer Res. 2013;3:446–64. [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle B, Hagan S, Al-Mulla F, Scott L, Harden S, Paul J, Mulcahy H, Murray GI, Sheahan K, O’Sullivan J, Kolch W. Raf kinase inhibitor protein expression combined with peritoneal involvement and lymphovascular invasion predicts prognosis in Dukes’ B colorectal cancer patients. Histopathology. 2013;62:505–10. doi: 10.1111/his.12014. [DOI] [PubMed] [Google Scholar]
- 12.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–7. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 13.Yeung K, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol Cell Biol. 2000;20:3079–85. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem. 2005;280:24931–40. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- 15.Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. Embo J. 1999;18:2137–48. doi: 10.1093/emboj/18.8.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–72. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng L, Ehrenreiter K, Menon J, Menard R, Kern F, Nakazawa Y, Bevilacqua E, Imamoto A, Baccarini M, Rosner MR. RKIP regulates MAP kinase signaling in cells with defective B-Raf activity. Cell Signal. 2013;25:1156–65. doi: 10.1016/j.cellsig.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–8. [PubMed] [Google Scholar]
- 19.Park S, Yeung ML, Beach S, Shields JM, Yeung KC. RKIP downregulates B-Raf kinase activity in melanoma cancer cells. Oncogene. 2005;24:3535–40. doi: 10.1038/sj.onc.1208435. [DOI] [PubMed] [Google Scholar]
- 20.Parker PJ, Murray-Rust J. PKC at a glance. J Cell Sci. 2004;117:131–2. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- 21.Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR. Activation of Raf-1 signaling by protein kinase C through a Mechanism Involving Raf kinase inhibitory protein. J Biol Chem. 2003;278:13061–8. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz K, Lohse MJ, Quitterer U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature. 2003;426:574–9. doi: 10.1038/nature02158. [DOI] [PubMed] [Google Scholar]
- 23.Granovsky AE, Clark MC, McElheny D, Heil G, Hong J, Liu X, Kim Y, Joachimiak G, Joachimiak A, Koide S, Rosner MR. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol Cell Biol. 2009;29:1306–20. doi: 10.1128/MCB.01271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziogas A, Lorenz IC, Moelling K, Radziwill G. Mitotic Raf-1 is stimulated independently of Ras and is active in the cytoplasm. J Biol Chem. 1998;273:24108–14. doi: 10.1074/jbc.273.37.24108. [DOI] [PubMed] [Google Scholar]
- 25.Hayne C, Tzivion G, Luo Z. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J Biol Chem. 2000;275:31876–82. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro PS, Vaisberg E, Hunt AJ, Tolwinski NS, Whalen AM, McIntosh JR, Ahn NG. Activation of the MKK/ERK pathway during somatic cell mitosis: direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142:1533–45. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zecevic M, Catling AD, Eblen ST, Renzi L, Hittle JC, Yen TJ, Gorbsky GJ, Weber MJ. Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–58. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell. 2006;23:561–74. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, Braastad C, Sun Y, Mukhopadhyay A, Aggarwal BB, Darnowski J, Pantazis P, Wyche J, Fu Z, Kitagwa Y, Keller ET, Sedivy JM, Yeung KC. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem. 2004;279:17515–23. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- 30.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 32.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–9. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 35.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–83. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 39.Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, Liang H, Li WH, Ishwaran H, Minn AJ, Rosner MR. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–14. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minn AJ, Bevilacqua E, Yun J, Rosner MR. Identification of novel metastasis suppressor signaling pathways for breast cancer. Cell Cycle. 2012;11:2452–7. doi: 10.4161/cc.20624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 42.Sun M, Gomes S, Chen P, Frankenberger CA, Sankarasharma D, Chung CH, Chada KK, Rosner MR. RKIP and HMGA2 regulate breast tumor survival and metastasis through lysyl oxidase and syndecan-2. Oncogene. 2013;33:3528–37. doi: 10.1038/onc.2013.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 44.Park H, Kim Y, Lim Y, Han I, Oh ES. Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J Biol Chem. 2002;277:29730–6. doi: 10.1074/jbc.M202435200. [DOI] [PubMed] [Google Scholar]
- 45.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920–5. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 47.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 48.Lee U, Frankenberger C, Yun J, Bevilacqua E, Caldas C, Chin SF, Rueda OM, Reinitz J, Rosner MR. A prognostic gene signature for metastasis-free survival of triple negative breast cancer patients. PLoS One. 2013;8:e82125. doi: 10.1371/journal.pone.0082125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Lee J, Farquhar KS, Yun J, Frankenberger CA, Bevilacqua E, Yeung K, Kim EJ, Balazsi G, Rosner MR. Network of mutually repressive metastasis regulators can promote cell heterogeneity and metastatic transitions. Proc Natl Acad Sci U S A. 2014;111:E364–73. doi: 10.1073/pnas.1304840111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Mulla F, Bitar MS, Taqi Z, Yeung KC. RKIP: much more than Raf kinase inhibitory protein. J Cell Physiol. 2013;228:1688–702. doi: 10.1002/jcp.24335. [DOI] [PubMed] [Google Scholar]
- 51.Al-Mulla F, Bitar MS, Al-Maghrebi M, Behbehani AI, Al-Ali W, Rath O, Doyle B, Tan KY, Pitt A, Kolch W. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3beta. Cancer Res. 2011;71:1334–43. doi: 10.1158/0008-5472.CAN-10-3102. [DOI] [PubMed] [Google Scholar]
- 52.Akinleye A, Furqan M, Mukhi N, Ravella P, Liu D. MEK and the inhibitors: from bench to bedside. J Hematol Oncol. 2013;6:27. doi: 10.1186/1756-8722-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan PF, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]