SUMMARY
The inhibitory receptor Tim-3 has emerged as a critical regulator of the T cell dysfunction that develops in chronic viral infections and cancers. However, little is known regarding the signaling pathways that drive Tim-3 expression. Here, we demonstrate that IL-27 induces NFIL3, which promotes permissive chromatin remodeling of the Tim-3 locus and induces Tim-3 expression together with the immunosuppressive cytokine IL-10. We further show that the IL-27/NFIL3 signaling axis is crucial for the induction of Tim-3 in vivo. IL-27-conditioned Th1 cells exhibit reduced effector function and are poor mediators of intestinal inflammation. This inhibitory effect is NFIL3 dependent. In contrast, tumor-infiltrating lymphocytes (TILs) from IL-27R−/− mice exhibit reduced NFIL3, less Tim-3 expression and failure to develop dysfunctional phenotype, resulting in better tumor growth control. Thus, our data identify an IL-27/NFIL3 signaling axis as a key regulator of effector T cell responses via induction of Tim-3, IL-10, and T cell dysfunction.
Introduction
T cell immunoglobulin and mucin domain-3 (Tim-3) was initially identified as an inhibitory receptor expressed on IFN-γ-producing CD4+ (Th1) and CD8+ T (Tc1) cells 1. Interaction between Tim-3 and its ligand, galectin-9, was shown to suppress effector T cell function resulting in Tim-3-dependent cell death during autoimmune tissue inflammation 2. Exciting new research has demonstrated that Tim-3 is a key regulator of the exhausted antigen-specific CD4+ and CD8+ T cells that arise in both humans and mice during chronic viral infections such as HIV, HCV, HBV and LCMV 3–5 and in cancer 6–8. Exhaustion refers to a state of dysfunction that typically arises in a hierarchical fashion whereby effector T cells first lose the ability to proliferate and be cytotoxic in response to antigen stimulation. This is then followed by the loss of IL-2 secretion, which is followed by a gradual loss of TNF and IFN-γ and increased production of the immunosuppressive cytokine IL-10. Accordingly, exhausted T cells pose a significant barrier to the induction of productive anti-viral or anti-tumor immunity. In contrast, one could envisage that in autoimmune diseases, the induction of T cell exhaustion would be beneficial. While primarily studied in CD8+ T cells, exhaustion also occurs in CD4+ T cells 3.
Exhausted T cells are characterized by their sustained expression of inhibitory receptors. Programmed death-1 (PD-1) was the first such molecule to be identified; its inhibitory function is essential for the induction of T cell exhaustion during chronic LCMV infection in mice, and during chronic HIV infection in humans 9–12. It is now appreciated that co-expression of PD-1 with other inhibitory receptors, such as Tim-3, contributes to the induction of T cell exhaustion and as such defines T cells with more deeply exhausted phenotype 5. Importantly, simultaneous blockade of the Tim-3 and PD-1 signaling pathways restores CTL function and cytokine production, while blockade of the PD-1 pathway alone is less effective. Thus, targeting Tim-3 on exhausted T cells provides a potential therapeutic avenue for treating multiple chronic viral infections and cancers. On the other hand, increasing Tim-3 expression would be beneficial for autoimmunity as reduced amounts of Tim-3 expression have been associated with a number of human autoimmune diseases 13. In spite of the increasing data linking Tim-3 to the suppression of T cell immunity, little is known about the signals by which its expression is induced on T cells. It was therefore important to identify the cytokines and pathways that induce the expression of this inhibitory molecule.
In this study, we demonstrate that IL-27, an immunosuppressive cytokine, is a potent inducer of Tim-3 expression on T cells. IL-27 strongly induces the expression of the transcription factor nuclear factor, interleukin 3 regulated (NFIL3), which cooperates with T-bet, to induce the expression of Tim-3 and IL-10. In addition, IL-27-conditioned Th1 cells exhibited poor effector function and are poor mediators of intestinal inflammation in an NFIL3-dependent manner. We show that IL-27 signaling is required for the induction of Tim-3+ exhausted T cells and promotion of tumor growth. Thus, we have uncovered that an IL-27/NFIL3 signaling axis drives inhibition of effector T cells via the induction of Tim-3, IL-10, and dysfunctional T cell phenotype.
RESULTS
IL-27 is a potent inducer of Tim-3 in naïve CD4+ T cells
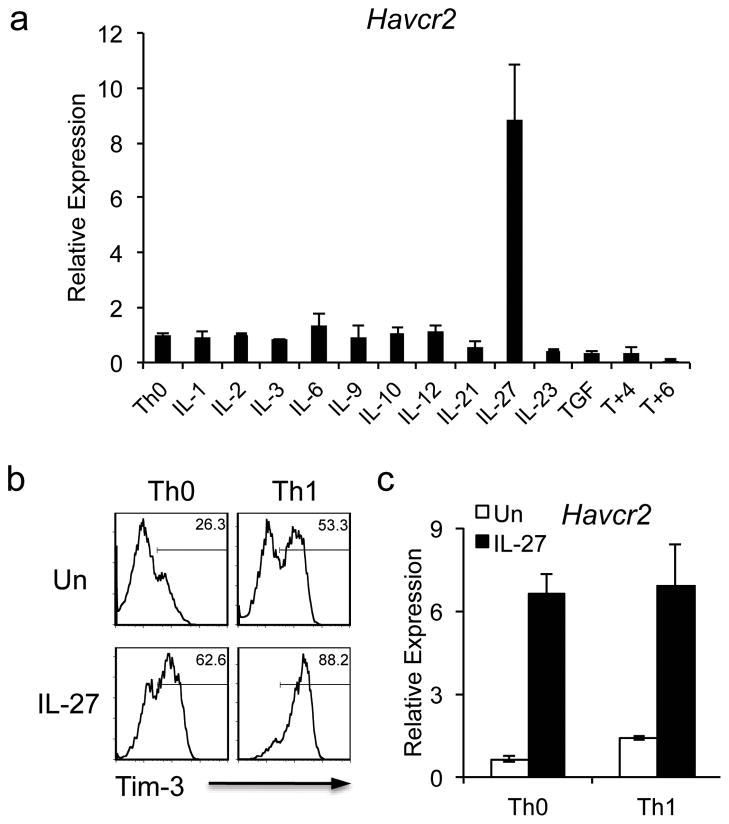
Our previous study indicated that T-bet is more functionally critical than STAT4 in the induction of Tim-3 expression on Th1 cells 14. The modest reduction of Tim-3 expression in IL-12-polarized STAT4−/− Th1 cells indicated that Tim-3 expression is not completely dependent on IL-12 signaling. To further explore other cytokines with potential to induce Tim-3, we tested a panel of cytokines for their ability to induce Tim-3 expression on naïve CD4+ T cells. After analyzing Tim-3 transcription by real time PCR, we observed that IL-27 was the most potent inducer of Tim-3 transcription (Fig. 1a). Indeed, IL-27 was more potent than IL-12, which only slightly increased Tim-3 transcription over that observed in the control neutral (Th0) condition (Fig. 1a).
Figure 1.
IL-27 induces Tim-3 expression. (a) Naïve CD4 T cells were activated by anti-CD3 and anti-CD8 antibodies in the presence of different cytokines. 72 hours after activation, cells were harvested for quantitative PCR analysis of Tim-3 (Havcr2) transcription. Havcr2 expression was normalized to that of β-actin. (b) and (c) Naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 antibodies under neutral (Th0) or Th1 (IL-12 treatment) conditions with or without IL-27. To analyze Tim-3 protein expression, the cells were restimulated with anti-CD3 and anti-CD28 antibodies on day 5 for 24 hours before examination of Tim-3 expression by flow cytometry (c). The transcription of Tim-3 (Havcr2) was examined by quantitative PCR (qPCR) 72 hours after activation (b). Data are representative of at least 3 independent experiments with similar results (a, b: mean ± s.d.).
Since Tim-3 expression is up-regulated during Th1 differentiation 1, we asked whether IL-27 further enhances Tim-3 expression on differentiating Th1 cells. We therefore activated naïve CD4+ T cells under Th0 or Th1 (with IL-12) conditions in the presence or absence of IL-27, and observed that IL-27-treated T cells (Th0+IL-27) exhibited higher Tim-3 protein expression than cells cultured with IL-12 alone (Th1 condition). Importantly, addition of IL-27 to differentiating Th1 cells (Th1+IL-27) exhibited an additive effect and induced the most Tim-3 protein expression (Fig. 1b). Similarly, we found that IL-27 had a dominant effect on Tim-3 mRNA induction in both conditions (Fig. 1c).
IL-27-induced NFIL3 and T-bet regulate both Tim-3 and IL-10
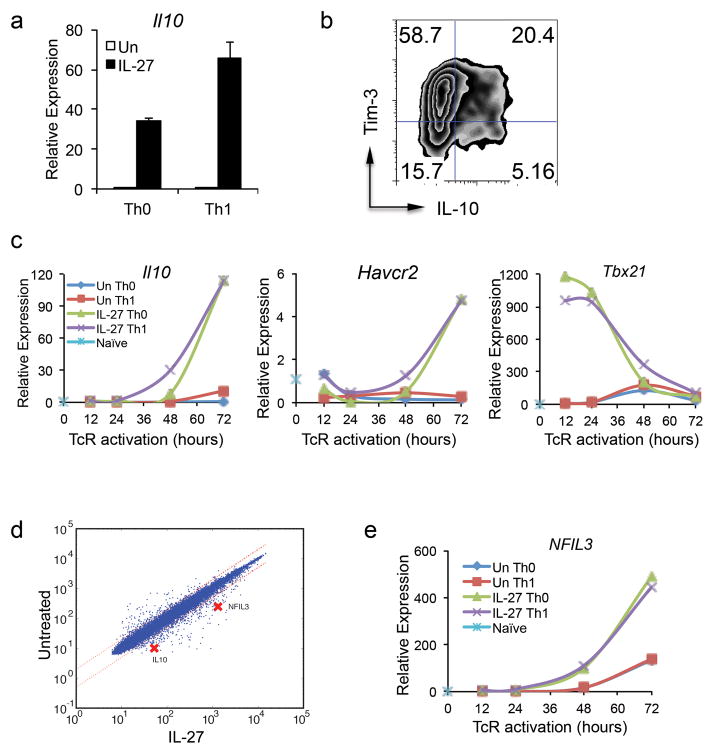
IL-27 has been shown to induce IL-10-producing Tr1 cells, which are a key component of IL-27-mediated immunosuppression 15,16. Accordingly, we examined whether IL-27 would induce IL-10 together with Tim-3 in activated T cells. We therefore examined IL-10 expression in Th0 and Th1 cells treated with IL-27 and found that IL-27 potentiated the induction of IL-10 under both conditions (Fig. 2a). We further found that IL-27 induced IL-10 together with Tim-3 as a significant fraction of IL-27-treated cells co-expressed Tim-3 and IL-10 (Fig. 2b). Similarly, IL-27 could induce Tim-3 and IL-10 expression in CD8+ T cells in both neutral (Tc0) and Tc1 (with IL-12) conditions (Supplementary Fig. 1). Examination of the kinetics of Tim-3 and IL-10 expression during IL-27 stimulation revealed that both Tim-3 and IL-10 transcription are induced with similar kinetics (Fig. 2c), suggesting a common transcriptional program for their induction.
Figure 2.
An IL-27-induced NFIL3 and T-bet regulate the expression of both Tim-3 and IL-10. Naïve CD4+ T cells were differentiated under Th0 or Th1 (by IL-12 treatment) condition with or without IL-27. (a) IL-10 (Il10) mRNA expression was detected by qPCR 72 hours after TcR activation (mean± s.d.). (b) To detect Tim-3 and IL-10 co-expression in IL-27-treated Th1 cells, the cells were restimulated with anti-CD3 and anti-CD28 antibodies on day 5 after TcR activation for 24 hours before subjected to flow cytometry analysis. (c) and (e) Naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 antibodies under Th0 or Th1 culture condition with or without IL-27. RNA was isolated at 12, 24, 48, and 72 hours after TcR activation for qPCR to determine the expression kinetics of IL-10, Tim-3, T-bet (Tbx21), and NFIL3. Data are representative of at least 3 independent experiments with similar results. (d) Gene profile studies. Naïve CD4+ T cells from C57BL/6 mice were stimulated with anti-CD3 and anti-CD28 antibodies in the presence of IL-27 for 60 hours. The cells were subjected to gene profile analysis. The scatter plot represents comparative transcriptome analysis between cells under neutral culture condition and cells treated with IL-27.
It is known that IL-27 induces T-bet 17, a transcription factor that has a role in both Tim-3 and IL-10 expression 14,18,19. However, overexpression of T-bet alone in T cells under different experimental settings does not always increase Tim-3 expression 14,19, indicating that maximal levels of Tim-3 and IL-10 expression in IL-27-treated Th1 cells may not be due to the induction of T-bet alone and that other transcription factors must be required for optimal Tim-3 transcription. Indeed, close examination of the kinetics of IL-27-induced T-bet transcription revealed that T-bet mRNA levels peaked within 24 hours of T cell activation and gradually declined thereafter. In contrast, Tim-3 and IL-10 transcription gradually increased until peaking at 72 hours at which time T-bet transcription had declined to near basal levels similar to that observed in Th0 cells (Fig. 2c). This decline of T-bet transcription prior to Tim-3 and IL-10 transcription indicates that other transcription factor(s) must be required for sustained induction of Tim-3 and IL-10 transcription.
We therefore undertook gene-expression profiling of naïve CD4+ T cells stimulated in the presence or absence of IL-27 at 8 different time points in order to elucidate the IL-27-induced transcription network in CD4+ T cells. With this information, we built a transcriptional network that could identify transcription factors based on their kinetics and by labeling each edge with a time stamp to identify a connection between a regulator and its target 20. Through this analysis, we identified that NFIL3 was a transcription factor whose transcription was highly induced by IL-27 (Fig. 2d, 2e, and Supplementary Fig. 2). Indeed, NFIL3 has been found to be essential for IL-10 expression in Th1 cells 21. Importantly, the expression kinetics of NFIL3 mRNA paralleled that of IL-10 and Tim-3 expression (Fig. 2c and 2e), thus raising the possibility that NFIL3 may be the transcription factor that mediates IL-27-driven expression of Tim-3 and IL-10.
NFIL3 is induced in both exhausted and anergic T cells
Given the association of both Tim-3 and IL-10 expression with T cell exhaustion and the indication that NFIL3 may drive Tim-3 and IL-10 expression, we examined the expression of NFIL3 in exhausted T cells. Indeed, we found that NFIL3 is induced to much higher levels in virus-specific CD8+ T cells in mice infected with the chronic clone-13 (Clone 13) strain of LCMV compared to virus-specific CD8+ T cells in mice infected by the Armstrong (Arm) strain. In addition, we found that NFIL3 is upregulated in ionomycin-induced anergic T cells (Supplementary Fig. 3a and 3b), which also up-regulate Tim-3 (Supplementary Fig. 3c), Taken together these data support that NFIL3, a transcription factor that is highly induced by IL-27 signaling, may represent a key regulator of Tim-3 expression and the induction of T cell dysfunction.
NFIL3 is critical for the expression of Tim-3 and IL-10
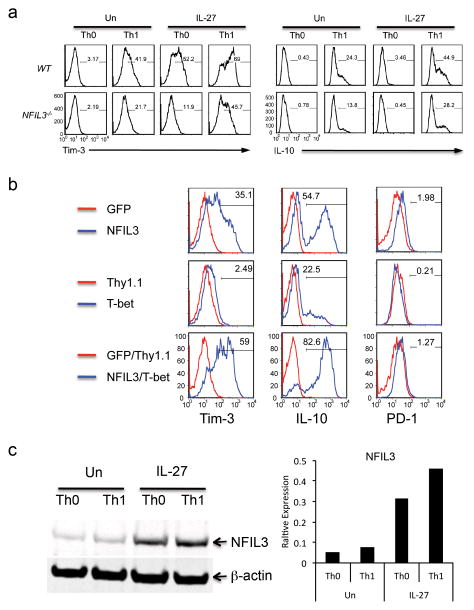
To confirm the role of NFIL3 in the regulation of Tim-3 and IL-10, we cultured naïve wild type (WT) and NFIL3−/− CD4+ T cells in vitro under Th0 or Th1 conditions in the presence or absence of IL-27. We found that NFIL3 deficiency resulted in reduced Tim-3 and IL-10 expression in all conditions, suggesting that NFIL3 is critical for the expression of both Tim-3 and IL-10 (Fig. 3a). Importantly, when compared with IL-27-treated WT Th0 cells, IL-27-induced Tim-3 and IL-10 expression was largely abrogated in NFIL3−/− Th0 cells. Tim-3 expression was similarly, attenuated in NFIL3−/− CD8+ T cells (Supplementary Fig. 4). Together these data suggested that NFIL3 is a critical transcription factor for IL-27-mediated Tim-3 expression.
Figure 3.
IL-27-mediated Tim-3 expression is dependent on the functional cooperation between T-bet and NFIL3. (a) Naïve CD4+ T cells from wild type (WT) C56BL/6 mice and NFIL3−/− mice were activated with anti-CD3 and anti-CD28 under Th0 or Th1 (IL-12 treatment) condition with or without IL-27. Five days after TcR activation, cells were restimulated with anti-CD3 and anti-CD28 for 24 hours and subjected to detection of the expression of Tim-3 and IL-10 by flow cytometry. (b) Naïve CD4+ T cells were transduced with retrovirus carrying NFIL3 cDNA (NFIL3), T-bet cDNA (T-bet), or both NFIL3 and T-bet. Retrovirus empty vector-transduced T cells were used as controls (GFP for NFIL3, Thy1.1 for T-bet). Four days after TcR activation, the expression of Tim-3, IL-10 and PD-1 was examined by flow cytometry. The percentage represents the expression of Tim-3, IL-10, or PD-1 in NFIL3- or T-bet-transduced, or NFIL3/T-bet-cotransduced cells relative to GFP- or Thy1.1-expressing empty vector-transduced, or GFP/Thy1.1 empty vector-cotransduced cells respectively. (c) Naïve CD4+ T cells were activated by anti-CD3 and anti-CD28 under Th0 and Th1 condition (IL-12 treatment) with or without IL-27. Three days after TcR activation, the expression of NFIL3 expression was detected by Western blot (left) and its relative expression to β-actin was quantified by LI-COR Odyssey Imaging System (right). Data are representative of at least 3 independent experiments with similar results.
To further examine the role of NFIL3 in the regulation of Tim-3 expression, we generated a retroviral expression construct for NFIL3 (NFIL3) and ectopically expressed NFIL3 in naïve CD4+ T cells. Compared to cells transduced with empty retroviral vector (GFP), overexpression of NFIL3 dramatically increased Tim-3 expression (Fig. 3b). This effect was highly specific for Tim-3 since NFIL3 only slightly increased the expression of other inhibitory receptors such as PD-1, LAG3, and CD160 (Fig. 3b and Supplementary Fig. 5). We also analyzed cytokine production and found that NFIL3-transduced cells exhibit significantly increased IL-10 expression (Fig. 3b).
While our data clearly show that NFIL3 induces Tim-3, the expression of Tim-3 expression is much higher on NFIL3 transduced cells cultured under Th1 conditions (Supplementary Fig. 5), indicating a potential functional cooperation between NFIL3 and Th1 associated transcription factor(s). We therefore examined the transcriptional cooperation between NFIL3 and T-bet in the regulation of Tim-3 and IL-10 expression. We found that NFIL3 was much more potent in inducing both Tim-3 and IL-10 expression than T-bet. However, overexpression of both NFIL3 and T-bet synergistically enhanced both Tim-3 and IL-10 expression (Fig. 3b). The fact that overexpression of T-bet in Th0 cells only slightly enhances Tim-3 expression once again indicates that T-bet alone is not sufficient for optimal Tim-3 expression without significant induction of other transcription factors. Interestingly, neither NFIL3 nor T-bet significantly induced PD-1 expression (Fig. 3b). Thus, our data indicate a fundamental difference in the transcriptional regulation of PD-1 and Tim-3 expression.
To determine whether IL-27-mediated induction of Tim-3 and IL-10 also is linked to its induction of NFIL3, we examined the amounts of NFIL3 protein in Th0 and Th1 cells in the presence or absence of IL-27 stimulation. A low level of NFIL3 protein was detected in Th0 cells, and no significant increase was found in Th1 cells. However, NFIL3 was strongly induced by IL-27 and this was further enhanced in IL-27-treated Th1 cells (Fig. 3c, Supplementary Fig. 12a). Thus, IL-27 promotes NFIL3 expression, which in turn activates Tim-3 and IL-10 expression.
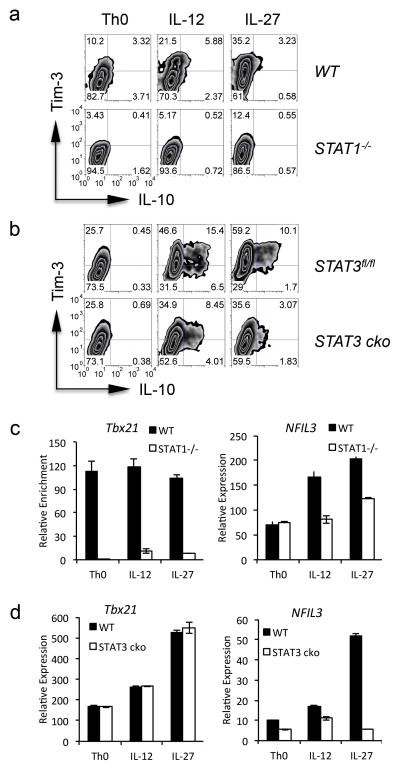
IL-27 induces Tim-3 and IL-10 expression via STAT1 and STAT3
STAT1 and STAT3 are two major transducers in the IL-27 signaling pathway. To determine whether IL-27-driven Tim-3 and IL-10 expression is dependent on either STAT1 or STAT3, we activated naïve CD4+ T cells from STAT1−/− and STAT3fl/fl x CD4-Cre (STAT3 cko) mice in Th0 conditions or in the presence of either IL-12 or IL-27. We found that STAT1−/− CD4+ T cells exhibited significant defects in both Tim-3 and IL-10 expression when compared to WT CD4+ T cells. This defect became more pronounced upon stimulation by either IL-27 or IL-12 (Fig. 4a). Interestingly, STAT3 deficiency did not affect Tim-3 or IL-10 expression in cells cultured under Th0 condition. However, IL-27-treated STAT3 cko CD4+ T cells clearly displayed reduced Tim-3 and IL-10 expression relative to WT cells. Surprisingly, this defect in Tim-3 and IL-10 expression was also observed in IL-12 treated cells, indicating a role for STAT3 in both IL-12- and IL-27-mediated Tim-3 and IL-10 expression (Fig. 4b).
Figure 4.
STAT1/T-bet and STAT3/NFIL3 mediate IL-27-induced Tim-3, and IL-10, expression. (a) and (b) Naïve CD4+ T cells purified from STAT1−/− mice (STAT1−/−) (a) or STAT3fl/fl x CD4-Cre mice (STAT3 cko) (b) and their wild type littermates (WT for STAT1−/− and STAT3fl/fl for STAT3 cko) were activated with anti-CD3 and anti-CD28 antibodies under neutral (Th0) condition, or under the stimulation of IL-12 or IL-27. Five days after activation, the cells were restimulated by anti-CD3 and anti-CD28 for 24 hours and were subjected to the detection of Tim-3 and IL-10 by flow cytometry. (c) and (d) Naïve CD4+ T cells as described in (a) and (b) were stimulated with anti-CD3 and anti-CD28 antibodies for 72 hours and were subjected to qPCR analysis for the transcription of T-bet (Tbx21) and NFIL3. The expression was normalized to the β-actin. Data are representative of at least 2 independent experiments with similar results (a, b: mean ± s.d.).
We next examined T-bet and NFIL3 transcription in STAT1−/− or STAT3 cko CD4+ T cells. The absence of STAT1, but not STAT3, resulted in a profound reduction of T-bet transcription in all conditions (Fig. 4c and 4d). This is consistent with previous data indicating that STAT1, but not STAT3, is essential for T-bet expression 22. In contrast, STAT1 deficiency only resulted in a modest reduction in NFIL3 expression in IL-12- and IL-27-stimulated cells but not cells cultured in Th0 condition (Fig. 4c), indicating that STAT1 partially affects NFIL3 expression. Further, we found that STAT3 deficient T cells exhibited partially reduced NFIL3 transcription under Th0 condition and IL-12 treatment. Importantly, these STAT3 deficient T cells showed a near complete loss of IL-27-induced NFIL3 expression (Fig. 4d), indicating that STAT3 is essential for IL-27-induced NFIL3 transcription. Thus, IL-27 triggers two distinct and non-redundant pathways: a STAT1/T-bet and a STAT3/NFIL3 pathway, both of which are critical for the induction of Tim-3 and IL-10 expression in T cells.
IL-27 directly induces Tim-3 expression via NFIL3 and T-bet
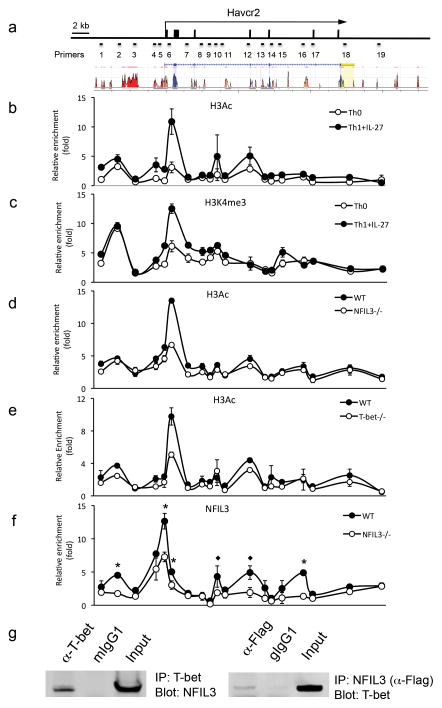
To elucidate the molecular mechanism of IL-27-mediated activation of Tim-3 transcription, we performed chromatin immunoprecipitation (ChIP) assays to examine histone modification of the Tim-3 locus. The ChIP products were subjected to quantitative PCR (qPCR) using primer sets that cover the whole Tim-3 locus (Fig. 5a and Supplementary Table 1). We examined histone H3 acetylation, a marker for permissive chromatin modification favoring gene expression, in chromatin samples prepared from Th0 cells and IL-27-treated Th1 cells. We found that IL-27-treated Th1 cells exhibited significantly higher enrichment of histone H3 acetylated at N-terminal lysine residues (H3Ac) in the proximal promoter region and in intron 1 of the Tim-3 locus (Fig. 5b). In addition, the same regions in the Tim-3 locus were also highly enriched with histone H3 trimethylation at lysine 4 (H3K4me3), another permissive chromatin modification marker (Fig. 5c). Together these data indicate that IL-27 signaling promotes chromatin modifications in the Tim-3 locus that favor the transcription of Tim-3 mRNA in Th1 cells.
Figure 5.
IL-27-induced permissive chromatin modification in the Tim-3 locus is both NFIL3 and T-bet dependent. (a) Mouse Tim-3 locus and the location of PCR primers for ChIP-qPCR analysis. The histogram represent CNSs which stand for 70% or greater identity over at least 100 bp stretches of the genomic DNA sequences between human and mouse Tim-3 locus (Blue: coding exons, red: intergenic regions; Orange: intronic regions; yellow: UTRs). ChIP PCR primers were marked according to their relative location in the Tim-3 locus (b) and (c) Naïve CD4+ T cells from C57BL/6 mice were activated by anti-CD3 and anti-CD28 antibodies under Th0 or IL-27-treated Th1 condition. The cells were restimulated on day 5 for 24 hours and were subjected to ChIP-qPCR for detection of H3Ac (b) and H3K4me3 (c) enrichment in the Tim-3 locus. (d) and (e) In a similar experimental setting, H3Ac enrichment in the Tim-3 locus was examined in NFIL3−/− CD4+ T cells and T-bet−/− CD4+ T cells. (f) NFIL3−/− and WT CD4+ T cells were activated by anti-CD3 and anti-CD28 antibodies under Th1 culture condition in the presence of IL-27. Four days after T cell activation, the cells were subjected to ChIP-qPCR to analyze NFIL3 enrichment in the Tim-3 locus. NFIL3 enriched areas marked with * represent the potential NFIL3 binding sites in both human and mouse homologous sequences, whereas those marked with ◆ represent the potential NFIL3 binding sites only in mouse sequences. (g) Expression plasmids for NFIL3 and T-bet were transiently transfected into 293T cells. Whole cell lysates were harvested 48 hours after transfection for co-IP to detect the interaction between NFIL3 and T-bet. Data are representative of at least 2 independent experiments with similar results (a, b, c, d, and e: mean ± s.d.).
To test whether NFIL3 is required for this permissive chromatin modification in the Tim-3 locus, we compared H3Ac enrichment in IL-27-treated Th1 cells derived from WT and NFIL3−/− mice. The absence of NFIL3 resulted in profound attenuation of H3Ac enrichment in the proximal promoter region and intron1 (Fig. 5d). Interestingly, a similar reduction of H3Ac enrichment in the same region of the Tim-3 locus was also found in T-bet deficient cells (Fig. 5e). Taken together, our data suggest T-bet and NFIL3 are both essential for IL-27-mediated permissive chromatin modification in the Tim-3 locus.
Our previous ChIP-qPCR data demonstrated that T-bet binds to the Tim-3 proximal promoter region 14, thus providing evidence supporting the direct involvement of T-bet in the chromatin remodeling of the Tim-3 locus. To identify whether NFIL3 directly binds to the Tim-3 locus, we conducted ChIP-qPCR using chromatin samples prepared from WT and IL-27-treated NFIL3−/− Th1 cells and identified multiple NFIL3 binding regions scattered in the Tim-3 distal and proximal promoter regions and introns 3, 4, and 5 (Fig. 5f).
We further assembled a high-resolution map of NFIL3 binding in the Tim-3 locus by performing NFIL3 ChIP-Seq and observed that NFIL3 binding in the Tim-3 locus is in line with our ChIP-qPCR data (Supplementary Fig. 6a). In addition, we analyzed published T-bet ChIP-Seq in Th1 cells 23(GSE33802). Interestingly, the alignment of T-bet and NFIL3 binding revealed a high degree of co-localization of these two transcription factors in the Tim-3 locus, particularly in the introns of Tim-3, indicating that they occupy common cis-regulatory regions with very close proximity (Supplementary Fig. 6a). Importantly, we were able to confirm the ChIP-Seq-identified T-bet binding sites to the Tim-3 locus by ChIP-qPCR further supporting the potential functional interaction between T-bet and NFIL3 (Supplementary Fig. 6b). Further, a similar co-localization of T-bet and NFIL3 is also found in the IL-10 locus (Supplementary Fig. 6a). Thus, the ChIP-Seq data provided preliminary evidence for T-bet-NFIL3 interaction and their functional cooperation in both Tim-3 and IL-10 expression. Interestingly, both T-bet and NFIL3 ChIP-Seq and ChIP-qPCR results indicate stronger binding in the intragenic regions than in the proximal promoter region of the Tim-3 locus (Fig. 5f, Supplementary Figs. 6a and 6b), suggesting that T-bet and NFIL3 may function to optimize Tim-3 transcription through permissive chromatin modification of the Tim-3 locus. Indeed, we did not observe strong transcriptional activity from either transcription factors in driving Tim-3 promoter-luciferase reporter activity (Supplementary Fig. 6d). Given that NFIL3 and T-bet binding sites are co-localized in multiple regions in the Tim-3 locus, we hypothesized that NFIL3 and T-bet may physically interact with each other to coordinate Tim-3 transcription. To test this hypothesis, we performed a co-immunoprecipitation assay and confirmed the physical interaction between NFIL3 and T-bet (Fig. 5g, Supplementary Fig. 12b). In addition, we performed a competition ChIP-qPCR experiment by preclearing the chromatin sample with antibodies to NFIL3, or control IgG prior to T-bet ChIP, we found that NFIL3-preclearance attenuates T-bet ChIP at most of the identified T-bet binding sites in the Tim-3 locus (Supplementary Fig. 6c), suggesting the physical interaction between T-bet and NFIL3 that we identified in co-IP represents true binding of NFIL3 and T-bet to the Tim-3 locus. Taken together, our data indicate that IL-27-enhanced Tim-3 expression in Th1 cells is dependent on the combinatorial action of NFIL3 and T-bet in remodeling the Tim-3 locus.
We further performed computational analysis of approximately 40 kb of genomic DNA sequence in the mouse Tim-3 locus aligned with the human homologous sequence. We identified conserved non-coding sequences (CNSs) located in the distal and proximal promoter regions (CNSs 6, 12, 13) as well as introns 1 and 5 (CNSs 15, 28) that contain putative NFIL3 binding sites. In ChIP-qPCR analysis, these regions exhibited significantly higher enrichment of NFIL3 binding in WT cells than in NFIL3−/− cells (Fig. 5f and Supplementary Fig. 7), suggesting that the cis-regulatory function of NFIL3 may also be critical for human Tim-3 transcription.
Role of NFIL3 in gut inflammation
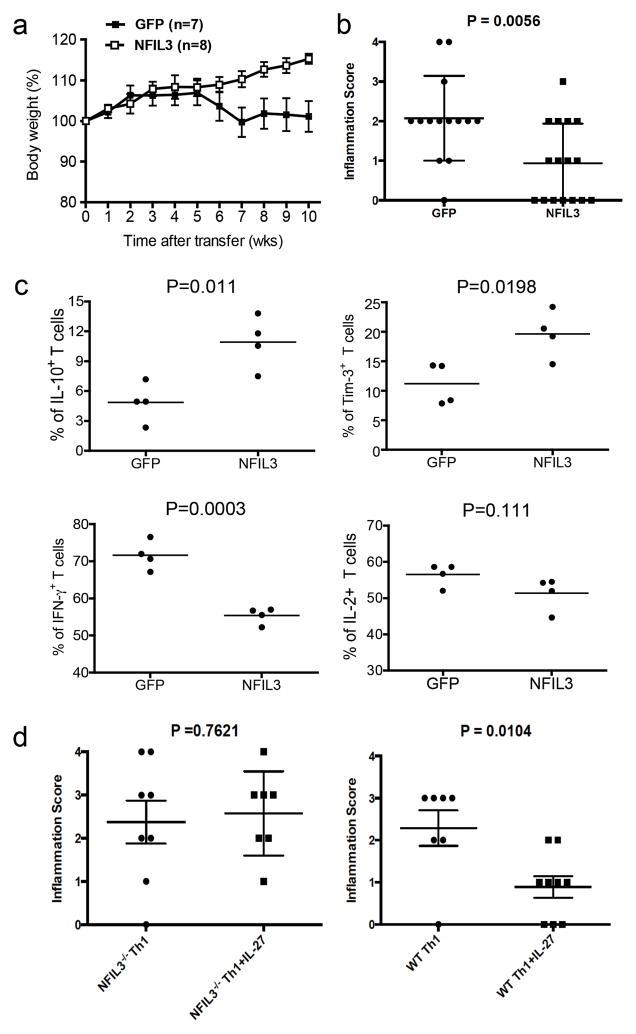
Our data indicate that IL-27-induced NFIL3 plays a key role in the induction of Tim-3 and IL-10 in effector T cells and increases dysfunctional phenotype in effector CD4+ T cells. We therefore hypothesized that overexpression of NFIL3 could dampen effector T cell function. Thus, we transduced naïve CD4+ T cells with NFIL3-expressing retrovirus (NFIL3) and transferred these cells into Rag1−/− 13 recipient mice and observed these mice for the development of gut inflammation. Recipient mice that received NFIL3-transduced CD4+ T cells failed to develop wasting disease 10 weeks after transfer (Fig. 6a), when compared with the control mice that received empty virus-transduced (GFP) CD4+ T cells. Histological analysis further revealed that empty virus-transduced T cells induced significantly higher mucosal inflammation than NFIL3 transduced T cells. After grading the crypt damage and cell infiltration (as described in Supplementary Fig. 8a), we observed that almost all of the recipient mice that received empty virus-transduced CD4+ T cells developed clear histological signs of both enteritis and colitis. In contrast, about half of the mice transferred with NFIL3-transduced T cells did not have any signs of gut inflammation and the rest of the mice had a milder inflammation when compared to the mice transferred with control empty virus-transduced T cells (Fig. 6b). We also found that NFIL3-transduced T cells were localized in Peyer’s patches, suggesting that failure to induce inflammation in the gut was not due to a defect of cell migration (Supplementary Fig. 8b). In addition, when we harvested empty virus- and NFIL3-transduced cells from the mesenteric lymph nodes of recipient mice 6 weeks post transfer, we found that NFIL3-transduced cells exhibited high Tim-3 expression and IL-10 production, but low IFN-γ production. Although not statistically significant, a clear trend towards low IL-2 production was also observed (Fig. 6c). Collectively, these data indicate that the effector function of NFIL3-transduced CD4+ T cells was suppressed in vivo, thereby attenuating the induction of gut inflammation.
Figure 6.
Ectopic expression of NFIL3 in CD4+ T cells attenuates gut pathology in adoptively transferred enteritis/colitis. Naïve CD4+ T cells from C57BL/6 mice were transduced with NFIL3-expressing retrovirus (NFIL3) or control empty retrovirus (GFP). Cells were injected i.p. into Rag1−/− recipient mice to induce gut inflammation (0.7 × 107 cells/mouse). (a) Wasting disease was monitored for 10 weeks after transfer. Statistical analysis was performed on the combination of total animals from two independent experiments. Data are shown as mean ± SEM. Mann Whitney test two-tailed P=0.0064. (b) Immunopathology of gut inflammation. Two randomly selected tissue sections from each animal were graded for crypt damage and T cell infiltration. Two scores were given to each animal to represent the overall gut immunopathology. Data are shown as mean ± SEM. Unpaired t test two-tailed P=0.0056. (c) The recipient mice were sacrificed 6 weeks after T cell transfer for ex vivo analysis of cytokine production (IL-2, IL-10, and IFN-γ) and Tim-3 expression by flow cytometry. (d) Naïve CD4+ T cells from WT FoxP3-GFP KI and NFIL3−/− x FoxP3-GFP KI mice (CD4+CD62L+GFP−) were differentiated into Th1 cells with or without the presence of IL-27. On day 6, GFP+ (FoxP3+) cells were isolated by cell sorting and were injected i.p. into Rag1−/− recipients to induce colitis. Colon pathology was analyzed 8 weeks after transfer. Each score represents the pathology from one recipient mouse (NFIL3−/− Th1 n=8; NFIL3−/− Th1+IL-27 n=7; WT Th1 n=7; WT Th1+IL-27 n=9. Data are shown as mean ± SEM, t test).
We further addressed the role of IL-27-driven NFIL3 in regulating Th1 driven gut inflammation. For this, we crossed NFIL3−/− mice to FoxP3-GFP KI mice. We then differentiated the CD4+CD62L+FoxP3− (GFP−) cells from NFIL3−/− x FoxP3 KI and WT FoxP3 KI mice into Th1 effector cells with or without IL-27 and transferred the cells into Rag1−/− mice to induce colitis. Mice that received WT Th1 cells showed clear signs of T cell infiltration and tissue damage, but IL-27-treated WT Th1 cells were much less colitogenic. By contrast, both NFIL3−/− Th1 cells and IL-27-treated NFIL3−/− Th1 cells induced similar degrees of colonic inflammation. IL-27 treatment failed to suppress the pathogenicity of these cells in the absence of NFIL3 (Fig. 6d, Supplementary Fig. 9a). Such IL-27-mediated regulatory effect seems to be associated with elevated expression of Tim-3 in IL-27-treated WT Th1 cells, which is abrogated in the absence of NFIL3 (Supplementary Fig 9b). Although, we did not find significantly elevated IL-10 production from IL-27-treated WT Th1 cells (Supplementary Fig. 9b). A clear trend of IL-10 induction by IL-27 in WT Th1 cells was detected. Taken together, our data indicate that IL-27 can directly suppress Th1 cell-mediated colitis, and NFIL3 is essential for such regulatory effect.
IL-27R deficient mice are resistant to tumor growth
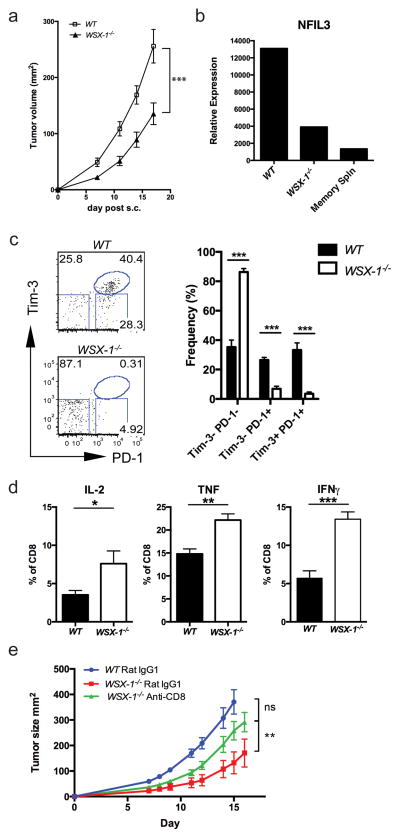
Our in vitro studies identified a potential mechanism by which IL-27 induces the expression of Tim-3 through upregulation of NFIL3 and T-bet. Tim-3 has a critical role in the development of exhausted T cells in chronic diseases such as cancer and chronic viral infection 6–8,24,25. We therefore reasoned that the IL-27 signaling pathway may also play a key role in regulating effector T cell phenotype in chronic diseases. To examine this, we implanted B16F10 melanoma into WSX-1−/− (IL-27R−/−) and wild type C57BL/6 (WT) mice, and monitored tumor growth. WSX-1−/− mice exhibited reduced tumor burden (Fig. 7a) and TILs from these mice exhibited significantly reduced NFIL3 expression (Fig. 7b). Moreover, compared to WT mice, WSX-1−/− mice failed to develop Tim-3+PD-1+ T cells, which we have previously identified as the most exhausted T cell population among TILs 6 (Fig. 7c). Indeed, the CD8+ T cells from WSX-1−/− mice did not exhibit exhausted phenotype, but rather produced more IL-2, IFN-γ and TNF than CD8+ T cells from WT recipient mice (Fig. 7d). Importantly, resistance to tumor growth and failure to develop Tim-3+PD-1+ TILs is also observed in WSX-1−/− mice bearing Lewis Lung carcinoma (LLC) (Supplementary Fig. 10).
Figure 7.
IL-27R deficient mice (WSX-1−/−) are resistant to tumor growth. (a) B16F10 melanoma cells were implanted into C57BL/6 (WT) and WSX-1−/− mice and tumor growth was monitored in two dimensions by caliper as the product of two perpendicular diameters. Statistics was based on combination of total animals from two independent experiments (WT: n=16; WSX-1−/−: n=14. Data are shown as mean ± SEM, P<0.0001, linear regression). (b) Tumor-infiltrating lymphoctyes (TILs) from these mice were isolated and were subjected to RT-qPCR analysis for NFIL3 expression. (c) The expression of Tim-3 and PD-1 on CD8+ TILs from WT (n=5) and WSX-1−/− (n=5) recipient tumor-bearing mice was determined by flow cytometry. Tim-3−PD-1−: P<0.0001; Tim-3−PD-1+: P<0.0001; Tim-3+PD-1+: P=0.0004, t test. (d) Production of IL-2, IFN-γ and TNF from peripheral CD8+ T cells from WT and WSX-1−/− mice was determined by flow cytometry. Data are representative of at least 2 independent experiments with similar results (b, c, and d: mean ± SEM. IL-2: P=0.013; TNF: P=0.0012; IFNγ: P=0.0002, t test). (e) WT and WSX-1−/− mice were implanted with 5×105 B16F10 tumor cells. On days 1, 2, 8, and 9, 100 μg of anti-CD8 antibody (53–5.8) or Rat IgG1 was injected i.p.. Tumor growth was monitored in two dimensions by caliper. (WT: n=9; WSX-1−/− Rat IgG1: n=7; WSX-1−/− Anti-CD8: n=9. P=0.0021, linear regression)
To determine whether the reduced tumor growth in WSX-1−/− mice is CD8+ T cell dependent, we depleted CD8+ T cells in mice implanted with B16F10 melanoma. Isotype rat IgG treatment in WSX-1−/− and in WT mice did not alter the phenotypic difference in tumor growth as we found in previous experiments. By contrast, CD8+ T cell depletion in WSX-1−/− mice significantly accelerated tumor growth and abrogated the protective effects of WSX-1 deficiency (Fig 7e). Together these data indicate that IL-27 signaling is critical for the induction of NFIL-3 and Tim-3 expression and mediating T cell dysfunction in cancer.
We further addressed the role of NFIL3 in tumor growth. For this, we transferred wild type or NFIL3−/− T cells into Rag1−/− mice, and subsequently inoculated with B16F10 melanoma cells. We found that tumor growth in NFIL3−/− T cell recipient mice was significantly delayed (Supplementary Figs. 11a and 11b) and that Tim-3 expression on CD4+ TILs was significantly reduced compared with WT. Interestingly, we did not see large defects in Tim-3 expression on CD8+ TILs (Supplementary Fig. 11c). These data suggest that Tim-3 expression on CD8+ TILs is relatively less NFIL3-dependent compared with CD4+ TILs. This likely reflects functional redundancy by other transcriptional factors that are induced in the tumor environment to drive Tim-3 expression in CD8+ T cells.
DISCUSSION
It has been well established that interaction between Tim-3 and its ligand galectin-9 inhibits Th1 responses 9,26,27 and induces peripheral tolerance 28,29. More importantly, Tim-3 is now recognized as having a key role in the induction of T cell exhaustion during chronic viral infections. Blockade of Tim-3 enhances proliferation and cytokine production in both HIV-1- and HCV-specific CD8+ T cells 3,30. Similarly, blockade of Tim-3 signaling also improves functional responses in tumor infiltrating lymphocytes in cancers 6–8. In contrast, a reduction in the expression of Tim-3 has been associated with the development of autoimmune diseases 13. These observations highlight the role of Tim-3 in regulating T cell effector function in chronic diseases and underscore the need for elucidating how Tim-3 expression is regulated. We report that the immunosuppressive cytokine IL-27 sets in motion the Tim-3-dependent transcriptional program of T cell inhibition. IL-27 induces expression of the transcription factor NFIL3, which co-regulates the expression of both Tim-3 and IL-10, two molecules associated with dysfunctional T cells.
T-bet is the first transcription factor directly linked to Tim-3 transcription. Although multiple studies showed that Tim-3+ exhausted T cells proportionally express high amounts of T-bet 3,31, the role of T-bet in regulating Tim-3 was contradictory. For example, while T-bet transgenic T cells exhibited enhanced Tim-3 expression 14, T cells with retrovirally transduced with T-bet exhibited slightly reduced Tim-3 expression during chronic LCMV infection 19. Taken together, these observations raised doubt as to whether T-bet alone is sufficient to drive Tim-3 expression, and pointed to the possibility that additional mechanisms may be involved in both the induction of Tim-3 and T cell dysfunction.
Here, we have identified the anti-inflammatory cytokine IL-27 as a potent signal for Tim-3 induction in T cells. In addition, we characterize a molecular mechanism by which the IL-27-triggered signal cascade directly controls the transcription of Tim-3 expression via STAT3-dependent induction of NFIL3 and also via STAT1-dependent induction of T-bet. Our findings further indicate that the STAT3-NFIL3 pathway is the key pathway in the induction of Tim-3 and largely explains the potency of IL-27 in the induction of Tim-3. Importantly, NFIL3 exhibited a direct dose-dependent effect on the induction of Tim-3 expression, and as such further cooperates with T-bet to induce Tim-3 expression. These observations support our hypothesis that T-bet is necessary but not sufficient for Tim-3 transcriptional activity and that NFIL3 is the key transcription factor that collaborates with T-bet to induce optimal Tim-3 expression.
IL-27 has been reported to act as an anti-inflammatory cytokine in multiple disease settings: autoimmune diseases 32–34, bacterial infections 35,36, and acute viral infections 8. Among the molecular mechanisms implicated in the anti-inflammatory effects of IL-27 are those that block differentiation of naïve T cells into Th2 and Th17 cells by IL-27-targeting of their master transcription factors RORγt 37 and GATA3 38. In addition to suppressing the transcriptional program for effector T cell differentiation, a recent study indicated that IL-27 directly induces expression of inhibitory molecules like PD-L1 39. Together with our data showing that IL-27 induces Tim-3, these data indicate that induction of inhibitory molecules associated with T cell dysfunction in T cells might be another mechanism by which IL-27 mediates its inhibitory effects.
While in vitro stimulation with IL-27 has been shown to promote Th1 differentiation 17 and augment CTL function 40, there are also data showing that IL-27 suppresses both effector Th1 and Tc1 responses through induction of IL-10 expression 41,42. Our data support that IL-27 controls immune responses by at least two different mechanisms: 1) by inducing IL-10 production and 2) by inducing inhibitory molecules such as Tim-3 and PD-L1 on differentiated effector Th1 and Tc1 cells, thus promoting T cell dysfunction. Indeed, we have demonstrated in the current studies that; (1) IL-27-conditioned Th1 cells induce less severe colitis and (2) endogenous IL-27 signaling results in reduced tumor burden due to enhanced effector CD8 function that is associated with increased proinflammatory cytokine production and reduced expression of Tim-3 and PD-1 in TILs. In conclusion, our data indicate a key role for IL-27 in the regulation of T cell dysfunction. Thus, targeting the IL-27 pathway could be of value as a therapeutic strategy in cancer and other chronic diseases.
NFIL3 is a key suppressor of IL-12 p40 gene expression in macrophages 43 therefore it negatively regulates innate immunity. Similarly, its anti-inflammatory role in the regulation of adaptive immune responses has been appreciated in that NFIL-3 was shown to induce IL-10 production in multiple T cell subsets 44,21. NFIL3−/− effector T cells exhibit reduced IL-10 production, and exhibit severe defect in controlling effector function thereby resulting in more severe CNS autoimmunity and colitis 21. Consistent with these data, our current study shows that forced expression of NFIL3 in T cells suppresses their effector function and significantly reduces gut pathology in RAG-deficient recipient mice. Importantly, a previous study showed that STAT3 is a major transducer for IL-27-mediated suppression of cytokine production 45. Since the induction of NFIL3 by IL-27 signaling is STAT3 dependent, our data strongly indicate that NFIL3 is an important functional mediator of the IL-27-driven anti-inflammatory effect by inducing Tim-3 and IL-10 expression and rendering T cells dysfunctional. Such NFIL3-dependent mechanism in IL-27-induced Tim-3 expression has been well validated in effector CD4+ T cells in both colitis and tumor models. Interestingly, the induction of Tim-3 is not significantly affected in NFIL3−/− CD8+ TILs, suggesting that a redundant downstream pathway in IL-27 signaling might compensate for lack of NFIL3 in CD8+ T cells. This could also reflect extreme pressure in the tumor microenvironment to drive Tim-3 expression and T cell dysfunction in CD8+ T cells. Nevertheless, our data also indicate that the IL-27-NFIL3 axis has an important role in the regulation of T helper cells in anti-tumor immunity.
In summary, we provide evidence supporting that IL-27 is a critical anti-inflammatory cytokine that induces the transcription factor NFIL3, whose function is essential for the development of dysfunctional T cell phenotype: high Tim-3 and IL-10, and low IL-2 and IFN-γ. As such, our observations place the IL-27 signaling pathway upstream of the suppression program that drives the development of T cell dysfunction in cancer. Consequently, blockade of IL-27 signaling may be a highly effective means by which T cell dysfunction could be abrogated for the treatment of cancer and other chronic diseases where T cell exhaustion is a barrier in mounting an effective immune response.
METHODS
Mice
STAT1−/− mice and 129S wild type mice were purchased from Taconic. Rag-1−/− mice, T-bet−/− mice, and C57BL/6 mice were purchased from the Jackson Laboratory. Peripheral lymphoid organs from STAT3fl/fl x CD4-Cre (STAT3 cko) mice and STAT3fl/fl control mice were provided by Dr. John O’Shea at NIH. NFIL3−/− mice were provided by Dr. Tak Mak at University of Toronto. WSX-1−/− mice were provided by Dr. Chris Saris at Amgen. WT FoxP3-GFP KI and NFIL3−/− x FoxP3-GFP KI mice are bred in house.
All mice were bred and kept in pathogenic free barriers. In vivo animal experiments were done using 7–8 weeks old female mice. Protocols for animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
Antibodies and cytokines
The following antibodies for T cells surface staining and flow cytometry were purchased from Biolegend: phycoerythrin-indotricarbocyanine (PE)-anti-CD4 (clone RM4-5), allophycocyanin (APC)-anti-CD62L (clone MEL-14), PE-anti-CD8 (clone 53-6.7), anti-IL-10 (clone JES5-16E3), anti-TNF (clone TN3-19.12), anti-IL-2 (clone JES6-5H4), anti-IFN-γ (clone XMG1.2), APC-LAG3 (clone C9B7W), PE-anti-PD-1 (RMP1-30). PE-anti-CD90.1 (clone OX-7). PE-anti-CD160 (clone eBioCNX46.3) was purchased from eBioscience. APC-anti-Tim-3 (5D12) was prepared in house. Antibodies were diluted 200 times for cell staining. All of the recombinant mouse cytokines for T cell in vitro culture and Tim-3 induction were purchased from R & D systems.
Cell isolation and culture
Total CD4+ T cells from different lines of mice were first enriched by positive selection using CD4+ T cell isolation reagent from Miltenyi Biotec. Naïve CD4+ (CD4+CD62L+) T cells were stained by PE-anti-CD4 and APC-anti-62L antibodies and were subsequently sorted by BD FACSAria (BD Biosciences). The sorted naïve T cells were then activated with plate-bound anti-CD3 (1 μg ml−1; 145-2C11) and anti-CD28 (1 μg ml−1; PV-1) (both were made in house) for 2 days and were rested afterwards in the presence of 10 ng ml−1 of IL-2. Th1 cells were cultured under the presence of IL-12 (10 ng ml−1). In some conditions, 25 ng ml−1 of IL-27 were added.
Gene expression profiling
Naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 antibodies with or without the presence of IL-27 for 60 hours. Total RNA was isolated with an RNeasy Plus kit (Qiagen) and then was amplified with an Ovation Biotin RNA Amplification and Labeling System (NuGEN). The cDNA (10 μg) was fragmented and labeled and then was hybridized to a GeneChip Mouse Genome 430 2.0 array (Affymetrix). Comparative gene expression between Th0 and IL-27-treated CD4+ T cells was analyzed by GenePattern software. The data have been deposited in NCBI Gene Expression Omnibus (GEO) under the accession code GSE63628.
Retroviral transduction
cDNAs encoding mouse NFIL3 and T-bet were subcloned into modified pMSCV vector that bicistronically expresses GFP (for NFIL3), and Thy1.1 (for Thy1.1) respectively. Retroviruses were packed in 293 T cells and were used to transduce mouse naïve CD4+ T cells activated by plate-bound anti-CD3 and anti-CD28 antibodies.
Intracellular cytokine staining
Naïve CD4+ T cells were activated by plate-bound anti-CD3 and anti-CD28 antibodies for 2 days. Cells were then rested for 3 days in the presence of 10 ng ml−1 of IL-2, and restimulated with 0.1 μg ml−1 of plate-bound anti-CD3 and anti-CD28 for 24 hours before they were subjected to PMA and ionomycin stimulation in the presence of Golgi Stop™ (BD Biosciences) for intracellular cytokine detection. All data were collected on LSR II (BD Biosciences) or Calibor (BD Biosciences) and analyzed by FlowJo software (Tree Star, Inc).
ChIP assays
Naïve CD4+ T cells from C57BL/6 mice were purified by naïve CD4+ T cell negative selection kit (Miltenyi Biotec), and were activated by plate-bound anti-CD3 and anti-CD28 (2 μg ml−1 each) under either neutral condition (Th0) or Th1 differentiating condition in the presence of IL-27 (Th1+IL-27) for 2 days. Cells were rested for additional 3 days in the presence of 10 ng ml−1 of IL-2 and were restimulated with 0.1 μg ml−1 of plate-bound anti-CD3 and anti-CD28 for 24 hours before they were subjected to chromatin preparation for the ChIP analysis. Chromatin fraction preparation and chromatin IP were performed using SimpleChIP™ Enzymatic Chromatin IP Kit (Cell Signaling Technology). Antibody against NFIL3 (C-18) was purchased from Santa Cruz Biotechnology; anti-acetylated Histone 3 antibody (Cat# 06-599) was purchased from Millipore; and an anti-trimethylated Histone Lysine 4 antibody (Cat# ab8580) was purchased from Abcam.
Real-time PCR analysis
RNA was extracted with RNeasy Plus kits (Qiagen) and cDNA was made by Iscript (BioRad). All of the Real-time PCR probes were purchased from Applied Biosystems. Quantitative PCR were performed by the GeneAmp7500 Sequence Detection System and ViiA™ 7 Real-Time PCR System (Applied Biosystems).
Tumor challenge and phenotypic/functional studies
B16F10 melanoma (CRL-6475) cell line was purchased from ATCC. 5×105 cells were injected subcutaneously at the flanks of the mice. Tumors were measured in two dimensions by caliper as the product of two perpendicular diameters. TILs were isolated on day 14 to day 20 post tumor implantation as they were reaching 200mm2 in size 6. Single cells suspensions were stained for CD8, CD4, Tim-3, and PD-1. For functional assay, intracellular cytokine staining was conducted as described before 6. For gene expression analysis, CD8+ 7AAD− TILs of B16 melanoma on WT C57BL/c mice or WSX-1−/− mice were sorted by BD FACSAria after magnetic separation by Dynabeads FlowComp Mouse CD8 (Invitrogen). CD44hi CD62Llow memory CD8+ splenocytes from B6 non-tumor bearing mice were also sorted as a control. RNA from sorted CD8+ cells were then extracted and reverse transcribed to cDNA. Gene expressions were quantified by qPCR.
Gut pathology
Naïve CD4+ T cells from C57BL/6 mice were subjected to TcR activation by anti-CD3 and anti-CD28 antibodies. Cells were subsequently transduced with NFIL3-expressing retrovirus or GFP-expressing empty retrovirus in the next day. On day 5 after activation, GFP positive T cells were sorted by BD FACSAria (BD Biosciences) and were transferred intraperitoneally into Rag-1−/− recipient mice. To analyze Tim-3 expression and cytokine production, NFIL3 or GFP transduced cells were i.p. injected to Rag1−/− mice. Mice were sacrificed 6 weeks after injection. Cells from spleen and mesenteric lymph nodes were restimulated with PMA/ionomycin in the presence of Golgi Stop™ (BD Biosciences) for detection of cytokine production and Tim-3 expression by flow cytometry.
Body weight and symptoms of disease were monitored up to 10 weeks. At the end point of the experiment, mice were sacrificed. Intestines were fixed with 10% Formalin and sections were stained with hematoxylin and eosin. The slides were evaluated independently by Dr. Vijay Yajnik at Massachusetts General Hospital and Dr. Roderick Bronson at The Rodent Histopathology Core, Harvard Medical School.
Additional transfer colitis was performed using naïve CD4+ T cells from WT FoxP3-GFP KI and NFIL3−/− x FoxP3-GFP KI mice (CD4+CD62L+GFP−) that were activated by plate-bound anti-CD3/CD28 in the presence of 10 ng ml−1 of IL-12, 10 μg ml−1 of anit-TGFβ1-2-3 antibody, 10 μg ml−1 of anti-IL-4 antibody. The cells were simultaneously treated with or without 25 ng ml−1 of IL-27. On day 6, cells were sorted again to deplete GFP+ (FoxP3+) cells, and were peritoneally injected into Rag1−/− recipients (0.6×106 cells per mouse) to induce colitis.
Supplementary Material
Acknowledgments
We thank Deneen Kozoriz for technical assistance; Dr. Vijay Yajnik at Massachusetts General Hospital and Dr. Roderick Bronson at The Rodent Histopathology Core, Harvard Medical School for histology analyses. This work was supported by grants from the National Institutes of Health (VKK: P01 AI073748, R01NS045937, NS030843, P01NS038037, P01AI056299; SX: K01DK090105), Ragon Institute of MGH, MIT and Harvard Innovation Award (VKK and CZ), and the American Cancer Society (ACA: RSG-11-057-01-LIB). EMD Serono Canada/endMS Network Transitional Career Development Award from the Multiple Sclerosis Society of Canada and the Multiple Sclerosis Scientific Research Foundation (MR).
Footnotes
Conflict of Interest Statement
VKK and ACA are paid consultants for Novartis Pharmaceuticals, which has interests in cancer immunotherapy. All other authors declare no competing financial interests.
Author Contributions
C.Z. and V.K.K. designed the experiments, C.Z. A.C.A., and K.S. analyzed the data and wrote the manuscript; C.Z., K.S., S.X., S.Z., G.G., C.W., D.J., C.W., M.R., T.P., H-T.J. did the experiments. Z.S. analyzed the ChIP-Seq data. A.C.A. directed tumor experiments carried out by K.S. and D.J.. R.A. provided reagents and intellectual input. Correspondence and requests for materials should be addressed to V.K.K. and A.C.A..
References
- 1.Monney L, et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536–541. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 2.Zhu C, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nature immunology. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 3.Jones RB, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. The Journal of experimental medicine. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahan RH, et al. Tim-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J Clin Invest. 2010;120:4546–4557. doi: 10.1172/JCI43127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin HT, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakuishi K, et al. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. The Journal of experimental medicine. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, et al. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117:4501–4510. doi: 10.1182/blood-2010-10-310425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourcade J, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. The Journal of experimental medicine. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 10.Petrovas C, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. The Journal of experimental medicine. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trautmann L, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 12.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 13.Zhu C, Anderson AC, Kuchroo VK. TIM-3 and its regulatory role in immune responses. Curr Top Microbiol Immunol. 2010;350:1–15. doi: 10.1007/82_2010_84. [DOI] [PubMed] [Google Scholar]
- 14.Anderson AC, et al. T-bet, a Th1 transcription factor regulates the expression of Tim-3. European journal of immunology. 2010;40:859–866. doi: 10.1002/eji.200939842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awasthi A, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nature immunology. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 16.Batten M, et al. Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. Journal of immunology. 2008;180:2752–2756. doi: 10.4049/jimmunol.180.5.2752. [DOI] [PubMed] [Google Scholar]
- 17.Takeda A, et al. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. Journal of immunology. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald DC, et al. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nature immunology. 2007;8:1372–1379. doi: 10.1038/ni1540. [DOI] [PubMed] [Google Scholar]
- 19.Kao C, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nature immunology. 2011;12:663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motomura Y, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nature immunology. 2011;12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature immunology. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 23.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejrnaes M, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. The Journal of experimental medicine. 2006;203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klibi J, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood. 2009;113:1957–1966. doi: 10.1182/blood-2008-02-142596. [DOI] [PubMed] [Google Scholar]
- 27.Sehrawat S, Suryawanshi A, Hirashima M, Rouse BT. Role of Tim-3/galectin-9 inhibitory interaction in viral-induced immunopathology: shifting the balance toward regulators. Journal of immunology. 2009;182:3191–3201. doi: 10.4049/jimmunol.0803673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Fueyo A, et al. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nature immunology. 2003;4:1093–1101. doi: 10.1038/ni987. [DOI] [PubMed] [Google Scholar]
- 29.Sabatos CA, et al. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nature immunology. 2003;4:1102–1110. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 30.Golden-Mason L, et al. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol. 2009;83:9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang ZZ, et al. IL-12 upregulates TIM-3 expression and induces T cell exhaustion in patients with follicular B cell non-Hodgkin lymphoma. J Clin Invest. 2012;122:1271–1282. doi: 10.1172/JCI59806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batten M, et al. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nature immunology. 2006;7:929–936. doi: 10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- 33.Niedbala W, et al. Interleukin 27 attenuates collagen-induced arthritis. Ann Rheum Dis. 2008;67:1474–1479. doi: 10.1136/ard.2007.083360. [DOI] [PubMed] [Google Scholar]
- 34.Sugiyama N, et al. Amelioration of human lupus-like phenotypes in MRL/lpr mice by overexpression of interleukin 27 receptor alpha (WSX-1) Ann Rheum Dis. 2008;67:1461–1467. doi: 10.1136/ard.2007.077537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarino A, et al. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity. 2003;19:645–655. doi: 10.1016/s1074-7613(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 36.Hamano S, et al. WSX-1 is required for resistance to Trypanosoma cruzi infection by regulation of proinflammatory cytokine production. Immunity. 2003;19:657–667. doi: 10.1016/s1074-7613(03)00298-x. [DOI] [PubMed] [Google Scholar]
- 37.Diveu C, et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. Journal of immunology. 2009;182:5748–5756. doi: 10.4049/jimmunol.0801162. [DOI] [PubMed] [Google Scholar]
- 38.Lucas S, Ghilardi N, Li J, de Sauvage FJ. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15047–15052. doi: 10.1073/pnas.2536517100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirahara K, et al. Interleukin-27 Priming of T Cells Controls IL-17 Production In trans via Induction of the Ligand PD-L1. Immunity. 2012;36:1017–1030. doi: 10.1016/j.immuni.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morishima N, et al. Augmentation of effector CD8+ T cell generation with enhanced granzyme B expression by IL-27. Journal of immunology. 2005;175:1686–1693. doi: 10.4049/jimmunol.175.3.1686. [DOI] [PubMed] [Google Scholar]
- 41.Axtell RC, et al. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat Med. 2010;16:406–412. doi: 10.1038/nm.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nature immunology. 2011;12:327–334. doi: 10.1038/ni.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi T, et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. Journal of immunology. 2011;186:4649–4655. doi: 10.4049/jimmunol.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashiwada M, Cassel SL, Colgan JD, Rothman PB. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. The EMBO journal. 2011;30:2071–2082. doi: 10.1038/emboj.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshimura T, et al. Two-sided roles of IL-27: induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. Journal of immunology. 2006;177:5377–5385. doi: 10.4049/jimmunol.177.8.5377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.