Abstract
In kinetoplastid parasites, regulation of mitochondrial gene expression occurs posttranscriptionally via RNA stability and RNA editing. In addition to the 20S editosome that contains the enzymes required for RNA editing, a dynamic complex called the mitochondrial RNA binding 1 (MRB1) complex is also essential for editing. Trypanosoma brucei RGG3 (TbRGG3) was originally identified through its interaction with the guide RNA-associated proteins 1 and 2 (GAP1/2), components of the MRB1 complex. Both the arginine-glycine-rich character of TbRGG3, which suggests a function in RNA binding, and its interaction with MRB1 implicate TbRGG3 in mitochondrial gene regulation. Here, we report an in vitro and in vivo characterization of TbRGG3 function in T. brucei mitochondria. We show that in vitro TbRGG3 binds RNA with broad sequence specificity and has the capacity to modulate RNA-RNA interactions. In vivo, inducible RNA interference (RNAi) studies demonstrate that TbRGG3 is essential for proliferation of insect vector stage T. brucei. TbRGG3 ablation does not cause a defect in RNA editing but, rather, specifically affects the abundance of two preedited transcripts as well as their edited counterparts. Protein-protein interaction studies show that TbRGG3 associates with GAP1/2 apart from the remainder of the MRB1 complex, as well as with several non-MRB1 proteins that are required for mitochondrial RNA editing and/or stability. Together, these studies demonstrate that TbRGG3 is an essential mitochondrial gene regulatory factor that impacts the stabilities of specific RNAs.
INTRODUCTION
Kinetoplastid parasites, including Trypanosoma brucei, Trypanosoma cruzi, and Leishmania spp., are transmitted by insect vectors and infect 20 million people, mainly in the most impoverished regions of the world. T. brucei is the causative agent of human African trypanosomiasis, which is a health threat to millions in sub-Saharan Africa and is 100% fatal if left untreated (1). Investigations into the novel biology of kinetoplastids may provide new chemotherapeutic avenues. Indeed, these early-branching eukaryotes utilize unusual processes for gene expression regulation and metabolic organization, and a large number of predicted kinetoplastid proteins lack any homology to proteins in their mammalian hosts.
Kinetoplastids are named for their distinctive mitochondrial DNA network, known as a kinetoplast, or kDNA, localized within their single mitochondrion. In T. brucei, kDNA is a giant catenated network of circular molecules comprised of a few dozen copies of a maxicircle (∼23 to 40 kb) and approximately 10,000 minicircles (∼1 kb) representing over 100 different sequence classes (2, 3). Maxicircles encode 18 mRNAs and 2 ribosomal RNAs. The mitochondrial transcriptome is largely shaped by posttranscriptional processes regulating RNA stability and RNA editing (4–7). RNA editing in kinetoplastids is unique to this group and entails the specific addition and deletion of uridine residues to mRNAs to create translatable open reading frames. The sequence information for editing is contained in minicircles, which encode small guide RNAs (gRNAs) that specify correctly edited mRNA sequence through base pairing interactions. The enzymes that catalyze editing, including endonucleolytic cleavage, uridine (U) addition or U removal, and the final resealing by ligation, are found in a stable multiprotein structure termed the RNA editing core complex (RECC) or 20S editosome (8–11). In addition, several noneditosome proteins facilitate the editing of one or more mRNAs in vivo (12–16). More recently, a large, dynamic macromolecular complex, termed the mitochondrial RNA binding 1 (MRB1) complex, otherwise known as RESC (RNA editing substrate binding complex [17]), has emerged as essential for mitochondrial RNA editing (4). MRB1 was originally isolated through immunoaffinity purification of the gRNA-associated proteins 1 and 2 (GAP1/2, or GRBC1/2) that are required for gRNA stabilization (13, 18, 19). The current model posits two major subcomplexes comprising the MRB1 complex. The MRB1 core is essential for editing initiation and contains GAP1/2 and at least four additional proteins (4, 20). The T. brucei RGG2 (TbRGG2) subcomplex interacts with the core in an RNA-enhanced manner (20–22) and is involved in the 3′-to-5′ progression of editing and proper utilization of gRNA (21–23).
Regulation of RNA levels in trypanosome mitochondria also occurs via RNA turnover. RNA stability seems to be linked to short nonencoded nucleotide extensions, or tails, on the 3′ ends of mRNA. These tails are often comprised of both adenosine (A) and U (24–26). Depletion of the primary poly(A) polymerase [KPAP1, for kinetoplast poly(A) polymerase 1] or a terminal uridyltransferase (RET1) leads to untailed RNAs and results in changes in mRNA abundances (24, 27). However, adenylation and uridylation differently impact the stabilities of mitochondrial RNAs in a transcript-specific manner, suggesting that additional factors also affect transcript stability (28). Several proteins whose depletion affects mitochondrial RNA stabilization have been identified. Some of these exhibit a broad impact on the mitochondrial transcriptome, while others are specific for distinct subsets of RNAs. For example, RNA binding protein 16 (RBP16) and the mitochondrial RNA binding protein 1 and 2 (MRP1/2) complex are essential for stabilizing the never-edited ND4 and COI transcripts (12, 29). Additionally, simultaneous depletion of RBP16 and MRP1/2 revealed that these proteins have a redundant function in the maintenance of edited A6 (ATPase subunit 6) and cytochrome oxidase subunit III (COIII) RNAs (14). Depletion of the p22 protein resulted in a specific defect in editing of the COII RNA (15). The MERS1 (mitochondrial edited mRNA stability) Nudix hydrolase and TbRGG1 exhibit more global effects. MERS1 depletion leads to a destabilization of all edited RNAs, and the complex containing this enzyme interacts with MRB1 in an RNA-dependent manner (13, 19). Finally, the arginine-glycine-rich TbRGG1 protein is also involved in stabilizing the majority of edited RNAs or in editing efficiency (13). While TbRGG1 has been detected in association with the GAP1/2 proteins, it is not typically identified in purifications of the MRB1 complex (18–20, 30).
In this study, we present the functional characterization of the TbRGG3 protein, which was previously annotated as MRB1820 (20). TbRGG3 was originally identified through its association with the GAP1/2 proteins and TbRGG1 (13) although later studies failed to reveal a stable association with the MRB1 complex, prompting us to rename the protein here. Due to these findings, and in keeping with nomenclature of mitochondrial proteins with the sequence character of arginine-glycine-rich stretches, we renamed this protein TbRGG3. TbRGG3 is implicated in mitochondrial RNA biology not only by association with GAP1/2 and TbRGG1 but also through its identification in pulldown of the large ribosomal subunit (25) and the REH2 (RNA editing helicase 2) complex (31). Moreover, the arginine-glycine-rich character of TbRGG3 suggests a function in RNA binding (32, 33). Here, we characterize the role of TbRGG3 in mitochondrial gene expression regulation using both in vivo and in vitro approaches. We show that, in vitro, TbRGG3 binds both gRNA and mRNA with broad sequence specificity. In addition, TbRGG3 modulates RNA-RNA interactions in vitro. In vivo, TbRGG3 is essential for growth in procyclic form (PF) T. brucei and for the stability of both preedited and edited versions of two pan-edited transcripts. Interestingly, analysis of protein-protein interactions demonstrates that TbRGG3 interacts with GAP1/2 in an RNA-independent manner, and this association is largely independent of the remainder of the MRB1 complex. TbRGG3 also associates with several proteins that are required for mitochondrial RNA editing and/or stability. Thus, TbRGG3 is an essential mitochondrial gene regulatory factor that impacts the stabilities of specific RNAs.
MATERIALS AND METHODS
Cells.
Strain 29-13 procyclic form (PF) T. brucei, which expresses the T7 RNA polymerase under the control of a tetracycline (Tet)-inducible promoter, as well as derivatives described below, was grown under standard conditions (12, 34). To construct an RNA interference (RNAi) vector for TbRGG3, 454 nucleotides downstream of the stop codon were PCR amplified from oligo(dT)-primed cDNA with the addition of 5′ BamHI and 3′ XhoI restriction sites using the following primers: TbRGG33′UTR5′BamHI (5′-GAGGATCCGATATGTAGAAGCGGAAAGCAAGC-3′; UTR is untranslated region) and TbRGG33′UTR3′XhoI (5′-GACTCGAGCATGCAGTAGCGAGAACCGTAG-3′). The PCR product was cloned into pJET1.2/blunt cloning vector (Fermentas), and the resultant vector was digested with BamHI and XhoI to yield the TbRGG3 3′ UTR fragment, which was gel purified. The excised product was then cloned into the BamHI and XhoI sites of the p2T7-177 RNAi vector (35) to yield p2T7-177-TbRGG3 3′ UTR. For transfection, 50 μg of p2T7-177-TbRGG3 3′ UTR was electroporated into 29-13 cells, and transformants were selected with 2.5 μg/ml phleomycin. Clones were isolated by limiting dilution to produce PF monoclonal cell lines expressing inducible TbRGG3 RNAi. RNAi was induced by addition of 2.5 μg/ml Tet to the growth medium, and growth of cells in the absence or presence of Tet was monitored for 12 days.
To generate a cell line expressing tagged TbRGG3, we first created a pLew100 derivative that expresses proteins followed by a tobacco etch virus (TEV) protease cleavage site and two copies of a Myc tag (2×Myc). To this end, a TEV 2×Myc insert was created using the following primers: 7A (5′-GATCCTCTAGAGAGAATTTGTATTTTCA-3′), 7B (5′-AATACAAATTCTCTCTAGAG-3′), 8A (5′-GGGTCATATGGAACAGAAACTGATCTCTG-3′), 8B (5′-GATCAGTTTCTGTTCCATATGACCCTGAA-3′), 9A (5′-AAGAAGACCTGAACGGGATCGAACAGAAACT-3′), and 9B (5′-TCTGTTCGATCCCGTTCAGGTCTTCTTCAGA-3′). Fifteen picomoles of each primer was phosphorylated with T4 polynucleotide kinase (PNK) and 1 mM ATP in buffer A (50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 5 mM dithiothreitol [DTT], and 0.1 mM spermidine) for 30 min at 37°C, followed by inactivation for 10 min at 75°C. Primer pairs were pooled and incubated at 94°C for 1 min, 50°C for 1 min, and 24°C for 1 min to anneal together. The primer mix was then ligated into a HindIII/BamHI-digested pLEW100 vector (34) to generate pLEW100-TEV-2×Myc. The primers 5′ TbRGG3 XhoI F (5′-GACTCGAGATGTCTGCCGCATTTGGTATCG-3′) and 3′ TbRGG3 XbaI R (5′-GATCTAGAGCGGCGGTATTGCCCCCCAAATC-3′) were then used to amplify the TbRGG3 open reading frame, and the amplicon was cloned into the pLEW100-TEV-2×Myc vector to create the pTbRGG3-TEV-2×Myc plasmid. This plasmid was transfected into 29-13 cells as stated above, except that transformants were selected with 1 μg/ml puromycin.
Pulldown experiments and Western blotting.
Immunoaffinity purification of TbRGG3 was carried out using 5 × 1010 PF cells containing induced TbRGG3-TEV-2×Myc. Cells were lysed in IPP50 buffer (10 mM Tris-HCl, 150 mM NaCl, and 0.1% NP-40) with 1% Triton X-100 and split in half. One-half was incubated with 50 U of RNase inhibitor (Applied Biosystems) and DNase 1 (0.002 U/μl) (Fermentas). The other half was treated with a nuclease cocktail containing RNase A (0.1 U/μl), RNase T1 (0.1 U/μl), RNase H (0.01 U/μl), RNase 1 (0.1 U/μl), RNase V1 (0.002 U/μl), DNase 1 (0.002 U/μl), and micrococcal nuclease (0.25 U/μl) (Fermentas) for 60 min on ice. Both nuclease-inhibited and nuclease-treated lysates were then incubated with an agarose rabbit anti-c-Myc column (ICL) for 2 h at 4°C in the presence of Complete protease inhibitor cocktail (Roche). TbRGG3-Myc was eluted using 100 mM glycine (pH 2.5) and neutralized in 1 M Tris-HCl (pH 8.7). Fractions were analyzed by Western blotting, and TbRGG3 was detected using anti-c-Myc antibodies (ICL). Additional proteins were probed with polyclonal antibodies against GAP1 (36), TbRGG2 (21), MRB3010 (20), TbRGG1 (13), MRP2 (14), and RBP16 (37).
Quantitative RT-PCR.
Total RNA was extracted from uninduced and induced TbRGG3 RNAi cells using TRIzol reagent (Invitrogen) at 3 and 4 days postinduction. RNA was phenol-chloroform extracted and ethanol precipitated before final resuspension of the pellet in diethyl pyrocarbonate (DEPC)-treated water. Ten micrograms of RNA was DNase treated with a DNA-free DNase kit (Ambion). RNA was reverse transcribed to cDNA using random hexamer primers and a Taq-Man reverse transcription kit (Applied Biosciences). Quantitative reverse transcription-PCR (qRT-PCR) reactions were then performed using established primers specific to the preedited and edited mRNAs and preprocessed mitochondrial transcripts from T. brucei (21, 38). TbRGG3 mRNA was amplified using the following primers: RGG3fwdq1 (5′-GTGAGGAGCGTGGATTTGG-3′) and RGG3revq1 (5′-ACCCACCGTAAACGGATAAC-3′). These primers amplify 35 nucleotides upstream and 64 nucleotides downstream of the stop codon. The qRT-PCRs (n = 6 to 15) were performed in a final volume of 25 μl, and the cDNA was amplified using an iQ5 real-time PCR detection system (Bio-Rad). Results were analyzed using iQ5 software, and RNA levels were normalized to levels of steady-state 18S rRNA using the standard-curve method.
Recombinant protein expression.
Glutathione S-transferase (GST)-tagged TbRGG2 was purified as described previously (21, 22). To generate GST-tagged TbRGG3, the TbRGG3 open reading frame was PCR amplified from oligo(dT)-primed cDNA from PF 29-13 T. brucei using the primers TbRGG3-5 EcoRI (5′-GAGAATTCATGCGAACCCAAGGATC-3′) and TbRGG3-3 XhoI (5′-GACTCGAGCTAGCGGCGGTATTGCC-3′), and the product was cloned into pGEX4T-1 (Sigma). The resultant pGEX4T-1-TbRGG3 was then transformed into Escherichia coli Rosetta strain cells (Novagen) for expression. Cells were grown to an optical density (OD) of ∼0.6, and protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C overnight. Recombinant protein was purified using a standard GST purification method using glutathione-agarose (Invitrogen).
UV cross-linking assays.
[α-32P]UTP (800 Ci/mmol) was purchased from PerkinElmer Life Sciences, and homoribopolymer RNAs [poly(A), poly(G), poly(U), and poly(C)] were purchased from Sigma. A6U5 shorter pre-mRNA, gA6[14]NX gRNA, 5′ CYbU pre-mRNA, and gCYb[558] gRNA (39–42) were all in vitro transcribed and body labeled with [α-32P]UTP using a T7 Maxiscript kit (Ambion). All UV cross-linking experiments contained 5 fmol of radiolabeled RNA and the amount of protein indicated in the figure legends. RNA and protein were incubated in a reaction mixture containing 6 mM HEPES (pH 7.5), 2.1 mM MgCl2, 0.5 M DTT, 1.5 mM ATP, 5 mM creatine phosphate, 0.1 mM EDTA, 10 μg/ml torula (Candida utilis) yeast RNA, 6% glycerol, and 20 μg/ml bovine serum albumin (BSA) for 20 min at room temperature. Reaction mixtures were then UV cross-linked using a Stratalinker 2400 (Stratagene) for 10 min on ice, followed by treatment with RNase A for 15 min at 37°C. Reactions were stopped by the addition of SDS-PAGE loading buffer and analyzed by 10% SDS-PAGE, followed by phosphorimaging. For competitor analyses, 7.5 pmol of unlabeled competitor homoribopolymer was incubated with 7.5 pmol of recombinant protein immediately prior to the addition of 5 fmol of radiolabeled RNA.
RNA annealing assays.
Annealing reactions were performed in an initial volume of 20 μl containing 6 mM HEPES-KOH (pH 7.5), 40 mM KCl, 1.9 mM MgCl2, 0.09 mM EDTA, 0.45 mM DTT, 750 nM BSA, 10 nM 5′ radiolabeled A6U5 41-nt pre-mRNA, and 10 nM unlabeled gA6[14]NX cognate gRNA. RNA substrates were allowed to anneal for 2 min at 70°C and cooled to room temperature for 15 min. The reaction mixtures were incubated with the indicated amount of protein for 20 min at room temperature. Reactions were stopped by the addition of stop buffer containing 40 μg of proteinase K, 0.1% SDS, and 2.5 mM EDTA followed by a 30-min incubation at room temperature. Reaction products were diluted to a final volume of 40 μl containing 5% glycerol and 20 μl of each reaction product and analyzed by 8% native PAGE followed by phosphorimaging (22, 23, 42).
Immunofluorescence.
PF cells harboring the TbRGG3-TEV-2×Myc construct were either left uninduced or induced with 2.5 μg/ml of Tet. Mitochondria were labeled by treating cells with 250 nm MitoTracker Red CMXRos (Invitrogen) for 15 min. Cells were then harvested by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended at 1.5 × 107 cells/ml. Cells were fixed with 4% formaldehyde in PBS for 30 min before permeabilization using 0.5% NP-40. Permeable cells were blocked using 10% normal rabbit serum and then incubated for 1 h with monoclonal mouse anti-Myc antibody [c-Myc(9E10); Santa Cruz] diluted 1:50 in PBS-BSA. A secondary Alexa Fluor 488 goat anti-mouse antibody (a kind gift from Jay Bangs) was diluted at 1:200 and incubated with cells for 30 min at room temperature. Cells were mounted using ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen). Images were taken with a Zeiss Axio Imager.M2 microscope using Volocity software.
RESULTS
TbRGG3 predicted amino acid sequence.
TbRGG3 (Tb927.3.1820) is annotated in TriTrypDB as a 245-amino-acid protein. However, three genome-wide studies identified multiple trans-splice sites on TbRGG3 mRNA, all of which predict utilization of an alternate upstream start codon (43–45). Using 5′ rapid amplification of cDNA ends (RACE) with a spliced leader primer, we confirmed one of the major trans-splice sites identified by the genome-wide studies at position −547 relative to the more upstream ATG codon (data not shown). Based on this analysis, the predicted TbRGG3 preprotein is 289 amino acids, with a predicted molecular mass of 30.2 kDa and pI of 11.6 (Fig. 1). The MitoProt II program (46) predicts a mitochondrial import sequence that is cleaved following amino acid 54 (underlined in Fig. 1), which would result in a mature protein of 24.3 kDa with a very basic pI of 11.5. The predicted mature protein is characterized by two arginine-glycine-rich stretches, a short one of 25 amino acids at the N terminus and a longer one of almost 100 amino acids at the C terminus. Figure 1 shows the alignment of preprocessed T. brucei TbRGG3 with the homologous proteins from Trypanosoma congolense, Trypanosoma vivax, and T. cruzi. The central portion of the protein, spanning the region between residues 94 and 208, is highly conserved, exhibiting 66% identity and 88% similarity among the four species. No conserved domains or motifs are apparent in this region of the protein. In striking contrast, the sequences of the arginine-glycine-rich N and C termini are much less conserved. Interestingly, however, the sequence character of these regions is quite similar, with all four proteins enriched in arginine and glycine. This suggests that the relatively basic and likely unstructured nature of the N- and C-terminal regions is important for TbRGG3 function, while the specific amino acid sequence is less so. Finally, we recently reported the identification of 14 methylarginine residues in TbRGG3 (green in Fig. 1) (47), 2 in the N terminus and 12 in the C terminus. This observation marks TbRGG3 as one of the most heavily methylated proteins identified to date in T. brucei (47, 48) and suggests that posttranslational modifications will impact the biological functions of this protein.
FIG 1.
Sequence analysis of TbRGG3. ClustalW alignment of TbRGG3 from T. brucei (Tb927.3.1820) with homologues from T. congolense (TcIL3000.0.25010), T. vivax (TvY486_0301090), and T. cruzi (Tc00.1047053510091.110). The underlined segment at the N terminus indicates the predicted mitochondrial import sequence identified using the MitoProt II program. Highlighted in red is the previously misannotated start codon within the mitochondrial import sequence. Green arginine (R) residues represent the 14 methlyarginine residues recently reported (47). An asterisk indicates identical amino acids, a colon indicates similar amino acids with strongly similar properties, and a period indicates similar amino acids with weakly similar properties.
Subcellular localization.
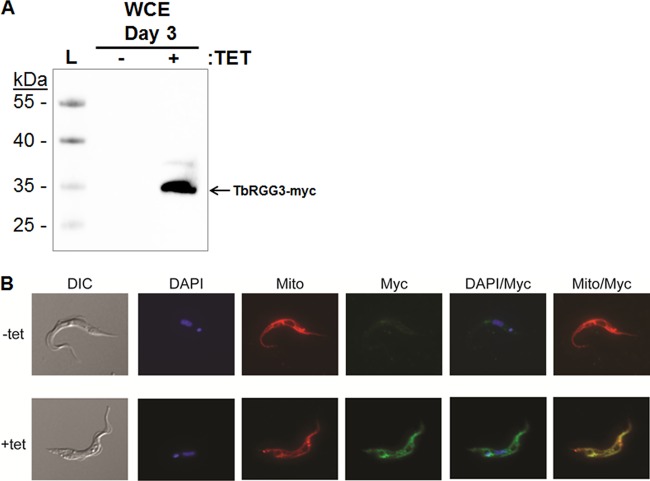
To begin to understand the functions of TbRGG3, we first wanted to determine its localization within the cell. In addition to harboring a predicted mitochondrial import sequence, TbRGG3 was designated “mitochondria likely” by the mitochondrial proteome study of Panigrahi et al. (49) (www.trypsproteome.org). Moreover, TbRGG3 was isolated by tandem affinity purification in association with known mitochondrial proteins GAP1, GAP2, and TbRGG1 (13), RNA editing helicase 2 (REH2) (31), and mitochondrial RPL3 (25). To directly determine the subcellular localization of TbRGG3, we generated PF T. brucei cells expressing TbRGG3 with a 2×Myc tag at its C terminus in a Tet-regulatable manner (Fig. 2A). Overexpression of 2×Myc-tagged TbRGG3 following Tet addition had no effect on cell growth. Uninduced and induced TbRGG3-2×Myc cells were incubated with MitoTracker Red to identify the reticulated mitochondrion, fixed, and probed with an anti-Myc monoclonal antibody. Analysis of these cells by immunofluorescence revealed that TbRGG3 colocalizes with the mitochondrial marker throughout the mitochondrion (Fig. 2B). These data confirm that TbRGG3 is mitochondrially localized, in accordance with bioinformatic and proteomic predictions.
FIG 2.
TbRGG3 overexpression and subcellular localization. (A) PF 29-13 cells harboring a Tet-inducible TbRGG3-Myc construct were grown in the absence or presence of Tet for 3 days. Proteins from 1 × 106 total cells were resolved by 10% SDS-PAGE, and TbRGG3-Myc was detected using anti-c-Myc antibodies. L, molecular mass ladder. WCE, whole-cell extract. (B) The subcellular localization of TbRGG3-Myc was determined by indirect immunofluorescence (Myc; green). Mitochondria were detected using MitoTracker Red CMXRos (Mito; red). Nuclei and kinetoplasts were stained blue with DAPI. The signals are shown merged on the right (DAPI/Myc and Mito/Myc). DIC, differential interference contrast.
RNA binding and annealing activity of TbRGG3.
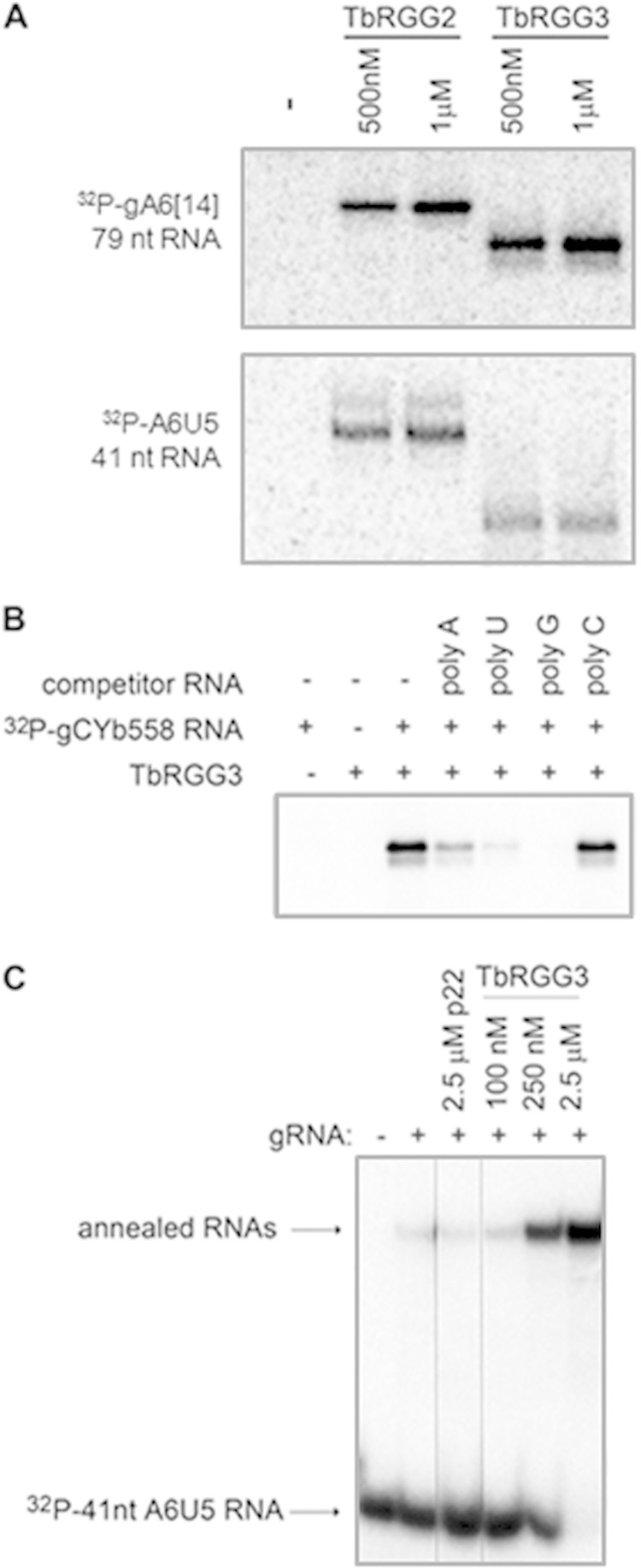
Having identified TbRGG3 as a highly basic mitochondrial protein, we next wanted to characterize its biochemical activities. Arginine-glycine-rich amino acid stretches frequently possess the capacity to interact with RNA (21, 50). To determine whether TbRGG3 is an RNA binding protein, we expressed recombinant GST-TbRGG3 in E. coli and purified the protein by affinity chromatography on glutathione agarose. We incubated either 500 nM or 1 μM TbRGG3 with 5 fmol of in vitro-transcribed, body-labeled gA6[14] guide RNA (gRNA) harboring a 17-nt oligo(U) tail and exposed the reaction mixtures to UV irradiation at 254 nm to induce label transfer in RNA-protein complexes. Reactions containing the RNA binding protein TbRGG2 (23) were performed in parallel as a positive control, and reactions lacking protein (other than the BSA added as a nonspecific protein in all reaction mixtures) were performed as a negative control. Figure 3A (top) clearly demonstrates label transfer from radiolabeled gRNA to TbRGG3, indicating that the protein binds gRNA. To examine the specificity of TbRGG3 RNA binding activity, we also performed UV cross-linking assays with a 41-nucleotide RNA corresponding to a portion of unedited ATPase subunit 6 (A6) mRNA and demonstrated that TbRGG3 binds this RNA as well (Fig. 3A, bottom). Finally, we examined the affinity of TbRGG3 for ribohomopolymers of differing sequences by performing competition assays. Unlabeled poly(A), poly(U), poly(G), or poly(C) (7.5 pmol) was incubated with TbRGG3 prior to addition of radiolabeled gCYb[558] gRNA, and UV cross-linking experiments were performed as described above. TbRGG3 displayed the highest affinity for poly(G) and moderate affinity for poly(U) and poly(A) and did not interact with poly(C) (Fig. 3B). Preferential binding of TbRGG3 to poly(G) and poly(U) RNAs could indicate a preferential interaction for mitochondrial preedited mRNA, which is rich in G and U content. From these data, we conclude that TbRGG3 is an RNA binding protein with broad sequence specificity.
FIG 3.
RNA binding and annealing activity of TbRGG3. (A) UV cross-linking assays were performed using 5 fmol of radiolabeled guide RNA (gA6[14]) or a fragment of preedited A6 mRNA (A6U5) with either 500 nM or 1 μM purified GST-TbRGG3. Reaction mixtures were subjected to UV cross-linking and treated with RNase A, and proteins were resolved by 10% SDS-PAGE. Proteins that cross-linked to radiolabeled RNA were visualized by PhosphorImager analysis. TbRGG2 binding was performed in parallel as a positive control, while reactions using mixtures containing no protein (−) were performed as a negative control. (B) Competition experiments were performed by incubating 7.5 pmol of unlabeled homoribopolymer RNA with GST-TbRGG3 prior to the addition of radiolabeled gRNA (gCYb[558]). Reaction mixtures were UV cross-linked and visualized as stated above. (C) RNA annealing assays were performed with 10 nM radiolabeled A6U5 41-nt pre-mRNA and 10 nM unlabeled gA6[14] gRNA at the indicated protein concentrations. Negative-control reactions were performed in the absence of gRNA (−) or with both RNAs in the absence of protein (+). The reaction mixture containing p22 served as a negative control. Reaction mixtures were incubated for 20 min, treated with proteinase K, and analyzed by 8% native PAGE.
One way in which RNA binding proteins can affect the fate of associated RNAs is through alteration of RNA structure. Having shown that TbRGG3 has the capacity to bind RNA, we next asked whether it is able to modulate RNA-RNA interactions. To this end, we employed our established RNA annealing assay, which utilizes a radiolabeled 41-nucleotide fragment of unedited A6 mRNA and cognate gRNA (39, 42). Single-stranded and double-stranded (annealed) RNAs were resolved on 8% native polyacrylamide gels following protease treatment. As expected, no annealed RNA is evident in the absence of gRNA (Fig. 3C). When cognate unlabeled gRNA is added to the labeled mRNA fragment, a small amount of annealed RNA is generated. The degree of annealing is unaffected by addition of a 2.5 μM concentration of the negative-control protein, p22 (51). We next titrated increasing concentrations of recombinant TbRGG3 into the annealing reaction mixture. Addition of TbRGG3 resulted in a very strong, concentration-dependent stimulation of annealed product. Thus, TbRGG3 possesses the capacity to modulate RNA-RNA interactions.
Effect of TbRGG3 silencing on growth and mitochondrial RNA levels.
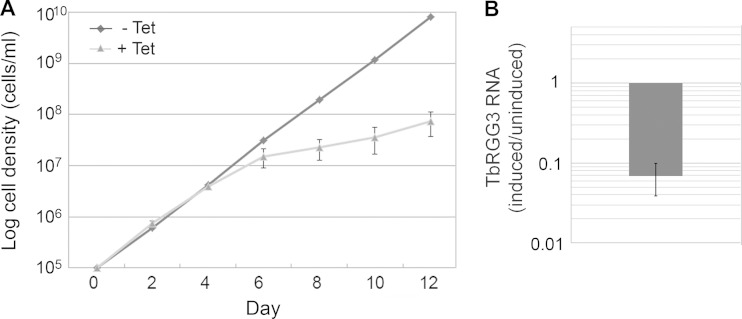
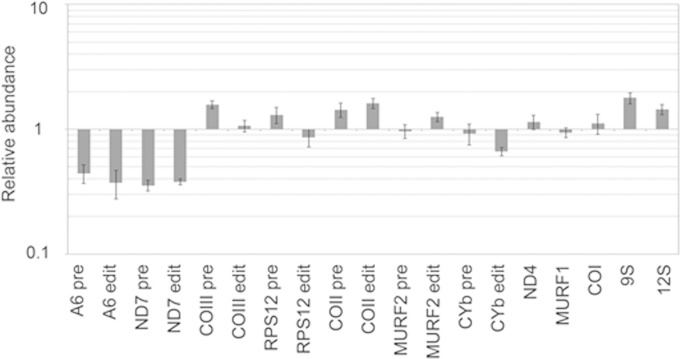
The ability of TbRGG3 to bind RNA and modulate RNA structure implicates TbRGG3 in shaping the mitochondrial transcriptome. Additionally, mass spectrometry studies from several laboratories identified TbRGG3 in association with proteins involved in mitochondrial RNA stability, editing, and translation (13, 25, 31), pointing to a function for TbRGG3 in mitochondrial RNA metabolism. To examine the role of TbRGG3 in mitochondrial gene expression, we generated PF T. brucei cells in which RNAi silenced TbRGG3 expression in a Tet-regulated manner. qRT-PCR analysis showed that TbRGG3 mRNA levels were decreased to less than 10% of wild-type levels by day 3 postinduction (Fig. 4B). Silencing of TbRGG3 led to a substantial growth defect, evident by day 6 and persisting through day 12 postinduction (Fig. 4A). We next analyzed the levels of several classes of mitochondrial RNAs by qRT-PCR in TbRGG3 RNAi cells either uninduced or induced with Tet for 3 or 4 days (Fig. 5). TbRGG3 silencing had little or no effect on the abundance of the never-edited mRNAs, ND4, MURF1, and COI, or 12S rRNA. A modest increase, less than 2-fold, in 9S rRNAs was reproducibly detected. We next turned to analysis of RNAs that undergo editing, quantifying both preedited and edited versions of these RNAs using established primer pairs (21, 38). Neither the minimally edited RNAs, COII and MURF2, nor the pan-edited RPS12 and COIII RNAs were substantially affected by TbRGG3 silencing. However, upon TbRGG3 ablation, we reproducibly observed a 55 to 65% decrease in the levels of both preedited and edited A6 and ND7 mRNAs. We also analyzed prepreprocessed RNAs spanning the 9S rRNA-ND8, CYb-A6, and RPS12-ND5 boundaries to ask whether TbRGG3 affects processing of primary transcripts, but levels of these preprocessed RNAs were essentially unchanged upon TbRGG3 silencing (data not shown). Because TbRGG3 silencing equally impacts the preedited and edited versions of A6 and ND7 RNAs, these data strongly suggest that TbRGG3 functions in the stabilization of specific preedited RNAs. Observed decreases in corresponding edited RNAs likely result from the decrease in substrate for the editing reaction.
FIG 4.
Effect of TbRGG3 silencing on growth. (A) The RNAi vector p2T7-177-TbRGG3 was transfected into the PF strain 29-13 of T. brucei. RNAi was induced by the addition of 2.5 μg/ml tetracycline to the cell medium. Cell growth was monitored in triplicate cultures of uninduced and induced cells for 12 days. (B) qRT-PCR analysis of TbRGG3 mRNA levels (induced/uninduced) on day 3 postinduction. RNA levels were standardized to 18S rRNA, and the value represents the mean and standard error of 6 determinations.
FIG 5.
Effect of TbRGG3 knockdown on mitochondrial RNA levels. RNA was isolated from PF T. brucei on day 3 or 4 postinduction. RNAs were quantified by qRT-PCR using primer sets specific for selected never-edited, pan-edited, and minimally edited RNAs. Relative RNA abundance indicates RNA levels in Tet-induced cells compared to those in uninduced cells. RNA levels were standardized to 18S rRNA levels, and values represent the means and standard errors of 6 to 15 determinations.
Association of TbRGG3 with other mitochondrial proteins.
Having shown that TbRGG3 affects the abundance of specific mitochondrial RNAs, we next wanted to determine whether it interacts with known modulators of the mitochondrial transcriptome. TbRGG3 was originally identified in pulldown of GAP1, GAP2, and TbRGG1 during an early investigation of the MRB1 complex (13). We subsequently defined the architecture of the MRB1 complex to include a core complex comprising GAP1, GAP2, and at least four other proteins, including MRB11870, MRB3010, MRB5390, and MRB8620 (20). In that study, TbRGG3 exhibited a strong yeast two-hybrid interaction with GAP1 in both directions but only weak and/or unidirectional interactions with MRB3010 and MRB8620. Moreover, TbRGG3 was not identified in pulldown of MRB3010, MRB5390, or MRB11870 (20, 36). To clarify the interaction of TbRGG3 with the MRB1 complex, we probed TbRGG3-2×Myc pulldown from extracts that were either RNase inhibited or RNase treated with anti-GAP1 or anti-MRB3010 antibodies. Analysis of these RNA populations by electrophoresis verified the efficiency of RNase treatment (data not shown). As shown in Fig. 6, we confirmed that TbRGG3 stably interacts with GAP1 in an RNA-independent manner. In contrast, we observed almost no interaction of TbRGG3 with MRB3010. The MRB1 core associates in an RNA-enhanced manner with a subcomplex containing TbRGG2 and additional components. Strikingly, we also detected no interaction of TbRGG3 with TbRGG2. Because TbRGG3 appears to mediate RNA stabilization (Fig. 5), we next asked whether TbRGG3 associates with other trypanosome mitochondrial proteins that have been implicated in RNA stabilization, including the TbRGG1 protein with which TbRGG3 was originally identified. Indeed, TbRGG3 clearly displays RNA-independent interactions with TbRGG1, MRP2, and RBP16. Collectively, these data demonstrate that TbRGG3 interacts with the GAP1/2 heterotetramer (19) independently of the remainder of the MRB1 complex. It also participates in numerous additional interactions with proteins that impact the mitochondrial transcriptome.
FIG 6.
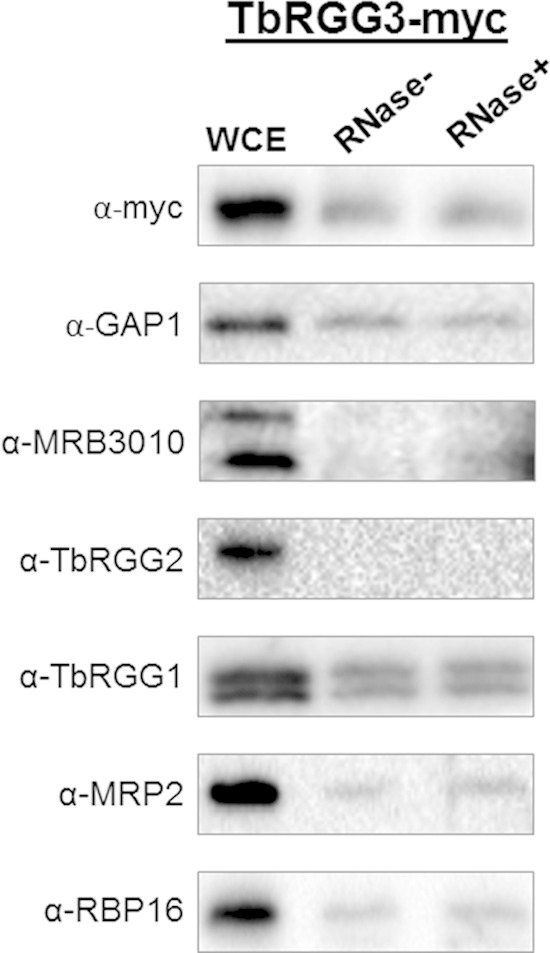
Association of TbRGG3 with other mitochondrial proteins. TbRGG3-Myc and associated proteins were affinity purified from cell extracts that were either RNase treated (RNase+) or left untreated (RNase−). Proteins were eluted from c-Myc-agarose using 100 mM glycine, pH 2.5, and electrophoresed on 10% SDS-PAGE gels, followed by Western blotting with antibodies specific to c-Myc (to detect TbRGG3) and various mitochondrial proteins. WCE, whole-cell extract. α, anti.
DISCUSSION
We hypothesized that the T. brucei TbRGG3 protein plays a role in mitochondrial RNA biology due to its reported association with the RNA binding proteins, GAP1/2, TbRGG1, and REH2, as well as the large ribosomal subunit (13, 25, 31). The arginine-glycine-rich nature of TbRGG3 further suggested that it possesses intrinsic RNA binding ability (21, 32, 33). Here, we performed in vivo and in vitro characterizations of TbRGG3 function. The N terminus of TbRGG3, which is extended compared to its current TriTrypDB entry, is predicted to encode a large mitochondrial import sequence, and we confirmed the protein's mitochondrial localization. In vitro, TbRGG3 exhibits RNA binding activity with broad sequence specificity, and it strongly promotes RNA-RNA annealing. Our in vivo studies show that TbRGG3 is essential for growth in PF T. brucei. However, despite its reported interaction with RNA editing-related proteins, TbRGG3 does not appear to affect the process of RNA editing. Rather, TbRGG3 is required for maintenance of both the preedited and edited versions of two pan-edited transcripts, A6 and ND7. Decreased abundance of edited A6 and ND7 RNAs is most likely due to the degradation of the corresponding preedited mRNAs, thus implicating TbRGG3 in the stabilization of specific mitochondrial RNA populations.
Interestingly, we show that TbRGG3 displays RNA-independent interactions with several other proteins that affect the stability of mitochondrial RNAs. TbRGG3 associates with both RBP16 and MRP1/2, which are required for the stability of never-edited ND4 and COI transcripts (12, 29) along with the maintenance of edited A6 and COIII RNAs (14). Despite extensive study, there is no evidence that RBP16 and MRP1/2 physically interact; thus, TbRGG3 likely undergoes distinct interactions with these proteins. We also confirmed by coimmunoprecipitation the previous mass spectrometry results indicating an interaction between TbRGG3 and another arginine-glycine-rich protein, TbRGG1 (52). TbRGG1 is involved in the maintenance of mitochondrial RNAs and is required for stabilizing the majority of edited RNAs (13). The interaction of TbRGG3 with numerous proteins involved in RNA stability further supports its role in the physical and functional coordination of mitochondrial mRNA levels.
The RNA-independent interaction of TbRGG3 with GAP1 is especially noteworthy. The GAP1/2 heterotetramer stabilizes the entire mitochondrial gRNA population (19, 52) and has been best characterized as a component of the MRB1 core complex that is involved in initiation of RNA editing (20, 36). We show that while TbRGG3 associates with GAP1/2, it does not stably interact with another MRB1 core component, MRB3010. It also fails to interact with the MRB1 subcomplex component, TbRGG2, even in the presence of RNA. These findings are consistent with the recent report that GAP1/2 associates with the REH2 helicase apart from the MRB1 core (53), as well as with our previous data suggesting a similar association of GAP1/2 with the mitochondrial zinc finger protein, MRB6070 (20). Given the interaction of REH2 with the RNA editing machinery and its impact on RNA editing, it is easy to envision a role for GAP1/2-bound gRNAs in association with REH2 (31, 52, 53). In contrast, our results indicate that TbRGG3 is not involved in RNA editing, and thus its function likely does not involve gRNAs. Therefore, the TbRGG3-GAP1/2 interaction may be indicative of a broader role for GAP1/2 than previously envisioned, not only outside the MRB1 complex but also separate from that of gRNA binding and stabilization. To our knowledge, the ability of GAP1/2 to directly bind mRNA has not been explored, and examination of this capacity may be informative in understanding the range of its functions, including its role in TbRGG3-mediated mRNA stabilization.
The mechanism of TbRGG3 function is still unknown. The broad sequence range of TbRGG3 RNA binding, in light of its specific impacts on the mitochondrial transcriptome, suggests that additional proteins are needed for interaction of this protein with target RNAs. With regard to the mechanism by which TbRGG3 stabilizes preedited A6 and ND7 RNAs, it is feasible that its interaction with these RNAs blocks access of the decay machinery. Alternatively, the role of TbRGG3 in modulating the mitochondrial transcriptome may be indirect. For example, it may play a role in translation of specific RNAs, which in turn impacts their abundance. In support of this model, TbRGG3 has been detected in association with the large ribosomal subunit RPL3 (25). Future in vivo cross-linking immunoprecipitation/high-throughput sequencing of RNA transcripts (RNA-Seq) experiments may provide insight into the mechanism of TbRGG3 action by revealing the positions at which TbRGG3 binds along its target RNAs. Preferential binding to the 5′ UTR would support the role of TbRGG3 in translation, while binding to the 3′ UTR would be more consistent with a direct effect on stabilization through blocking access of the RNA decay machinery. In summary, TbRGG3 is an essential mitochondrial RNA binding/annealing protein that interacts with numerous proteins involved in RNA stabilization and whose depletion destabilizes a specific subset of mitochondrial RNAs.
ACKNOWLEDGMENTS
We thank Vlad Presnyak and Kurtis Downey for technical assistance and Jay Bangs for assistance with immunofluorescence.
This work was supported by NIH grant RO1 AI061580 to L.K.R. J.N. was supported in part by NIH T32 AI007614, and K.L. was supported in part by NIH F32 AI100350.
REFERENCES
- 1.Kennedy PG. 2008. The continuing problem of human African trypanosomiasis (sleeping sickness). Ann Neurol 64:116–126. doi: 10.1002/ana.21429. [DOI] [PubMed] [Google Scholar]
- 2.Hong M, Simpson L. 2003. Genomic organization of Trypanosoma brucei kinetoplast DNA minicircles. Protist 154:265–279. doi: 10.1078/143446103322166554. [DOI] [PubMed] [Google Scholar]
- 3.Jensen RE, Englund PT. 2012. Network news: the replication of kinetoplast DNA. Annu Rev Microbiol 66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 4.Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukes J. 2013. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol 29:91–99. doi: 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aphasizhev R, Aphasizheva I. 2011. Mitochondrial RNA processing in trypanosomes. Res Microbiol 162:655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz-Reyes J, Read LK. 2013. Coordination of RNA editing with other RNA processes in kinetoplastid mitochondria, p 65–90. In Maas S. (ed), RNA editing: current research and future trends. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 7.Lukes J, Hashimi H, Zikova A. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet 48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 8.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L. 2009. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci U S A 106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. 2010. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist 161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier M, Read LK. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisk JC, Presnyak V, Ammerman ML, Read LK. 2009. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol Cell Biol 29:5214–5225. doi: 10.1128/MCB.00520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprehe M, Fisk JC, McEvoy SM, Read LK, Schumacher MA. 2010. Structure of the Trypanosoma brucei p22 protein, a cytochrome oxidase subunit II-specific RNA-editing accessory factor. J Biol Chem 285:18899–18908. doi: 10.1074/jbc.M109.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Herrera J, Zhou S, Maslov DA, Simpson L. 2011. Trypanosome REH1 is an RNA helicase involved with the 3′-5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci U S A 108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. 2014. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 34:4329–4342. doi: 10.1128/MCB.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics 7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell 32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK. 2012. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res 40:5637–5650. doi: 10.1093/nar/gks211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisk JC, Ammerman ML, Presnyak V, Read LK. 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem 283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foda BM, Downey KM, Fisk JC, Read LK. 2012. Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing. Eukaryot Cell 11:1119–1131. doi: 10.1128/EC.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. 2010. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aphasizheva I, Aphasizhev R. 2010. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol Cell Biol 30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. 2011. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Molecular Cell 42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao CY, Read LK. 2007. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol Biochem Parasitol 154:158–169. doi: 10.1016/j.molbiopara.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 2008. 3′ Adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J 27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmer SL, McEvoy SM, Menon S, Read LK. 2012. Additive and transcript-specific effects of KPAP1 and TbRND activities on 3′ non-encoded tail characteristics and mRNA stability in Trypanosoma brucei. PLoS One 7:e37639. doi: 10.1371/journal.pone.0037639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vondruskova E, van den Burg J, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem 280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- 30.Kafkova L, Ammerman ML, Faktorova D, Fisk JC, Zimmer SL, Sobotka R, Read LK, Lukes J, Hashimi H. 2012. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA 18:1846–1861. doi: 10.1261/rna.033852.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. 2010. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem 285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenan DJ, Query CC, Keene JD. 1991. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci 16:214–220. doi: 10.1016/0968-0004(91)90088-D. [DOI] [PubMed] [Google Scholar]
- 33.Bandziulis RJ, Swanson MS, Dreyfuss G. 1989. RNA-binding proteins as developmental regulators. Genes Dev 3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 34.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99:89–101. doi: 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- 35.Wickstead B, Ersfeld K, Gull K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol 125:211–216. doi: 10.1016/S0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 36.Ammerman ML, Hashimi H, Novotna L, Cicova Z, McEvoy SM, Lukes J, Read LK. 2011. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA 17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayman ML, Read LK. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J Biol Chem 274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- 38.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci U S A 102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read LK, Goringer HU, Stuart K. 1994. Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol Cell Biol 14:2629–2639. doi: 10.1128/MCB.14.4.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley GR, Corell RA, Stuart K. 1994. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J Biol Chem 269:6101–6108. [PubMed] [Google Scholar]
- 41.Koslowsky DJ, Kutas SM, Stuart K. 1996. Distinct differences in the requirements for ribonucleoprotein complex formation on differentially regulated pre-edited mRNAs in Trypanosoma brucei. Mol Biochem Parasitol 80:1–14. doi: 10.1016/0166-6851(96)02646-1. [DOI] [PubMed] [Google Scholar]
- 42.Ammerman ML, Fisk JC, Read LK. 2008. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA 14:1069–1080. doi: 10.1261/rna.982908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. 2010. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog 6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog 6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 47.Fisk JC, Li J, Wang H, Aletta JM, Qu J, Read LK. 2013. Proteomic analysis reveals diverse classes of arginine methylproteins in mitochondria of trypanosomes. Mol Cell Proteomics 12:302–311. doi: 10.1074/mcp.M112.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lott K, Li J, Fisk JC, Wang H, Aletta JM, Qu J, Read LK. 2013. Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular processes impacted by arginine methylation. J Proteomics 91:210–225. doi: 10.1016/j.jprot.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaili N, Goringer U, Benne R, Pays E. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J Biol Chem 273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 51.Hayman ML, Miller MM, Chandler DM, Goulah CC, Read LK. 2001. The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res 29:5216–5225. doi: 10.1093/nar/29.24.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. 2009. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madina BR, Kumar V, Metz R, Mooers BH, Bundschuh R, Cruz-Reyes J. 2014. Native mitochondrial RNA-binding complexes in kinetoplastid RNA editing differ in guide RNA composition. RNA 20:1142–1152. doi: 10.1261/rna.044495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]