Abstract
Sporotrichosis is one of the most frequent subcutaneous fungal infections in humans and animals caused by members of the plant-associated, dimorphic genus Sporothrix. Three of the four medically important Sporothrix species found in Brazil have been considered asexual as no sexual stage has ever been reported in Sporothrix schenckii, Sporothrix brasiliensis, or Sporothrix globosa. We have identified the mating type (MAT) loci in the S. schenckii (strain 1099-18/ATCC MYA-4821) and S. brasiliensis (strain 5110/ATCC MYA-4823) genomes by using comparative genomic approaches to determine the mating type ratio in these pathogen populations. Our analysis revealed the presence of a MAT1-1 locus in S. schenckii while a MAT1-2 locus was found in S. brasiliensis representing genomic synteny to other Sordariomycetes. Furthermore, the components of the mitogen-activated protein kinase (MAPK)-pheromone pathway, pheromone processing enzymes, and meiotic regulators have also been identified in the two pathogens, suggesting the potential for sexual reproduction. The ratio of MAT1-1 to MAT1-2 was not significantly different from 1:1 for all three Sporothrix species, but the population of S. brasiliensis in the outbreaks originated from a single mating type. We also explored the population genetic structure of these pathogens using sequence data of two loci to improve our knowledge of the pattern of geographic distribution, genetic variation, and virulence phenotypes. Population genetics data showed significant population differentiation and clonality with a low level of haplotype diversity in S. brasiliensis isolates from different regions of sporotrichosis outbreaks in Brazil. In contrast, S. schenckii isolates demonstrated a high degree of genetic variability without significant geographic differentiation, indicating the presence of recombination. This study demonstrated that two species causing the same disease have contrasting reproductive strategies and genetic variability patterns.
INTRODUCTION
Fungi exhibit a wide diversity of reproductive modes, including sexual, asexual, and parasexual strategies. Sexual reproduction generates genetic variation by meiotic recombination, which may alter virulence, increase fitness in new ecological niches, and purge deleterious mutations from the genome (1, 2). A strictly clonal mode of reproduction may be advantageous where genotypes are adapted to specific hosts and habitats (3). Pathogenic species with unknown sexual cycles usually have phylogenetically close environmental, sexually active counterparts, suggesting that asexual propagation greatly outpaces any outcrossing that might be coupled with the emergence of pathogenic status (4–9).
Members of Sporothrix species are fungal pathogens associated with sporotrichosis, a subcutaneous disease of humans and animals, especially felines (10–12). The infection is mediated by the traumatic inoculation of fungal elements into the cutaneous/subcutaneous tissues or, occasionally, by inhalation followed by pulmonary lesions. The fungi are thermo-dimorphic, living as saprophytes in association with plant debris and decaying organic matter in soil, and exhibiting an invasive yeast-like form in the warm-blooded host (11). Multiple gene genealogies of chitin synthase, β-tubulin, and calmodulin have shown that several Sporothrix species are involved (13), of which S. schenckii sensu stricto, S. brasiliensis, S. globosa, and S. luriei form a monophyletic clade (14). With the exception of rare infections caused by S. mexicana and S. pallida, sporotrichosis is almost exclusively associated with species in this group (15). Infection experiments in mouse models have shown that clinical isolates of S. brasiliensis, S. schenckii, and S. globosa express different levels of virulence (16, 17). Sporotrichosis is commonly observed in felines (Felis catus), and zoonotic outbreaks have been reported. A 1998-2010 outbreak of 3,244 cases of sporotrichosis in cats, 120 cases in dogs, and 2,200 cases in humans occurred in Brazil, suggesting a sharp increase in human cases correlated with one in domestic cats (12, 18, 19). In the outbreak region of Rio de Janeiro (RJ), S. brasiliensis proved to be overwhelmingly prevalent among cats and humans (20); however, the population structure, epidemiology, and emergence of these pathogens have not been elucidated.
Sporothrix belongs to the order Ophiostomatales (Sordariomycetes, Ascomycota) and is closely related to Ophiostoma, a global genus of plant pathogens distributed by arthropods (15, 21, 22). Sporothrix reproduces asexually, whereas in many Ophiostoma species elaborate sexual fruiting bodies are observed in addition to a Sporothrix-type of sporulation. For many years S. schenckii has been considered to be the asexual stage of Ophiostoma stenoceras (23, 24); however, phylogenetic analysis revealed them to be two distinct species (21, 22, 25). Sexual structures (long-necked perithecia) in Ophiostoma can be easily obtained by in vitro mating on special medium (26). Ophiostoma harbors homothallic species (e.g., Ophiostoma stenoceras and Ophiostoma nigrocarpum) in which isolates are self-fertile, as well as heterothallic species (e.g., Ophiostoma quercus and Ophiostoma novo-ulmi), where two self-sterile individuals are necessary to induce mating (27–30).
Genomic comparisons of various fungi have increased our knowledge of the evolution of reproduction in both homothallic and heterothallic species (31–33). The mating process and sexual reproduction in Ascomycota are controlled by the mating type genes (MAT) which encode transcription factors that are essential for mating (34, 35). However, little is known at the molecular level of the sexual cycle of Ophiostomatales; MAT loci and flanking genes have been identified only in Ophiostoma ulmi, O. novo-ulmi, Ophiostoma himal-ulmi, and O. quercus (27, 36–38). Recently the MAT genes have been characterized in six Grosmannia species, as well as in Ophiostoma montium, and unequal recombination has been suggested to account for the presence of a truncated MAT1-1-1 in isolates of the opposite mating type (39). Although sexual reproduction has never been observed, the sexual state of Sporothrix species is predicted to be Ophiostoma-like based on the observation that Sporothrix is phylogenetically nested within Ophiostoma (21, 22).
The present study aimed to identify the MAT locus and sex-related genes in the genomes of S. schenckii (strain 1099-18/ATCC MYA-4821/CBS 132984) and S. brasiliensis (strain 5110/ATCC MYA-4823/CBS 132021) using comparative genomic analysis. Furthermore, this study assessed the distribution of sexual idiomorphs MAT1-1 and MAT1-2 in isolates of the S. schenckii relatives and the influence of breeding strategies in the genetic structure of the disease, especially in the recent outbreaks in Brazil. For this, we used both comparative genomic and phylogenomics approaches to identify the role of sexual recombination in the genetic structure of this epidemic. We aim to answer three specific questions: (i) Do the Sporothrix genomes demonstrate evidence of sex? (ii) What are the population structures with regard to their mating type/sex ratio? (iii) What are the genetic variations in populations of S. schenckii and S. brasiliensis? Together, the data reveal differences in population structures and breeding strategies of S. schenckii and S. brasiliensis.
MATERIALS AND METHODS
Mating type locus characterization and evolution.
The mating type loci of S. schenckii and S. brasiliensis were identified using the MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT-1-2-1 genes of Neurospora crassa and Magnaporthe grisea as queries. Sequences were searched (tBLAST) against the assembled scaffolds from the S. schenckii (strain 1099-18/ATCC MYA-4821/CBS 132984; GenBank accession number AXCR00000000) and S. brasiliensis genomes (strain 5110/ATCC MYA-4823/CBS 132021; accession number AWTV00000000) (40). The boundaries of both MAT1-1 and MAT1-2 loci within a 20-kb scaffold fragment containing both mating types were determined by reciprocal alignment using ClustalW (41) implemented in the BioEdit software (42). We also characterized the MAT1-2 idiomorph of S. globosa, another human pathogen, using long-range PCR amplification with a primer-walking sequencing approach (39).
Identification of sex-related genes in S. schenckii and S. brasiliensis genomes.
The presence of 108 genes involved in the mating process, mating signaling, fruiting body development, karyogamy, and meiosis was assessed in the S. schenckii and S. brasiliensis genomes. The sex-related predicted proteins previously characterized experimentally in model organisms such as Aspergillus nidulans and N. crassa were used as queries to identify putative orthologs in the predicted proteomes from two analyzed species (43). Homology was inferred via BLASTp search, and putative orthologs were considered with a minimum query/subject coverage of 60% and a minimum positive of 50% using an E value cutoff of 1e−13.
Long-range PCR and primer walking.
Primers ER (GCCACGTCGTTCAACAACTA) and ScHMGf (GCCTCCTTTACAGCTCTGTG) targeting the SLA and the MAT1-2-1 genes of the MAT locus were designed for long-range PCR in Sporothrix globosa. Long-range PCR amplifications of DNA were carried out in 50 μl using an Applied BioSystems 2720 thermal cycler (Life Technologies Corp., Carlsbad, CA, USA). Reaction mixtures contained 100 ng of DNA, 1× PCR buffer, 200 μM each deoxynucleoside triphosphate (dNTP), 1 μM each primer (Eurofins MWG Operon, Huntsville, AL, USA), 3% dimethyl sulfoxide (DMSO), and 2 U of Phusion DNA polymerase (Finnzymes, BioLabs, New England, USA). The PCR amplifications were performed for 30 s at 98°C, followed by 35 cycles of 10 s at 98°C, 30 s at 60°C, and 4 min at 72°C, with a final extension at 72°C for 10 min. Sequencing reactions with primer walking were performed at the Centre de recherche du CHUQ (CHUL), Québec, Canada. Primers for sequencing are listed in Table S1 in the supplemental material. Sequence reads after primer walking were assembled using the Staden package (44) and Geneious Pro (Biomatters, Auckland, New Zealand), and sequences were compared with genes present in GenBank using BLASTx and BLASTn. The assembled sequences were submitted to FGENESH+ in Softberry (Softberry, Inc., Mount Kisco, NY) for gene prediction and to determine the location of the coding/noncoding regions. The nucleotide sequences of the MAT idiomorphs were compared using dot plot (matrix) analyses implemented in Geneious, version 5. The sequences of the MAT loci were also compared across different species using BLASTn in the Artemis comparison tool (http://www.webact.org/WebACT/home). The amino acid sequences encoded by the MAT1-1 α-box and the MAT1-2 high-mobility group (HMG) domain in Sporothrix species were aligned to sequences from other ascomycetes using ClustalW (41) implemented in Geneious. Sequences from Verticillium dahliae and Neurospora tetresperma (Sordariomycetes) were selected as outgroups. Phylogenetic analysis was carried out with the neighbor-joining (NJ) method in MEGA, version 5 (45), and the maximum-likelihood (ML) method in PhyML (46) (available at http://www.phylogeny.fr/version2_cgi/index.cgi).
Quantitative expression of MAT genes in yeast and mycelial forms of dimorphic S. schenckii and S. brasiliensis.
We used reverse-transcription (RT) real-time quantitative PCR (qPCR) to assess the levels of expression of MAT1-1-1, MAT1-1-2, MAT1-1-3, MAT-1-2-1, and truncated MAT1-1-1 genes. S. schenckii 1099-18 and S. brasiliensis 5110 isolates were cultured in both yeast and mycelium forms. Mycelia were cultured in Sabouraud broth for 10 days at 25°C under agitation. Yeast cells were obtained after transfer of 106 conidia per ml in brain heart infusion (BHI) broth for 10 days at 37°C under agitation. Cells were then collected by centrifugation and plated on BHI slants to stabilize the yeast phase. Yeast cells were then grown again in BHI broth for 10 days at 37°C under agitation. Cells were collected by centrifugation (5,000 × g for 10 min) and washed three times with 1× phosphate-buffered saline (PBS), and RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. DNA was removed using RNase-free DNase I (Promega), followed by ethanol precipitation. RNA samples were then used for real-time RT-qPCR. Two micrograms of RNA was reverse transcribed (Superscript III; Invitrogen) using an oligo(dT)12–18 primer and then subjected to real-time RT-PCR. Amplification assays were performed in a 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA), using a 10-μl reaction volume containing 0.2 μM (each) primers (see Table S1 in the supplemental material), 5 μl of 2× SYBR green PCR master mix, 0.2 μl of cDNA template, and H2O. After initial denaturation at 95°C for 20 s, amplifications were performed for 40 cycles at 95°C for 3 s and 60°C for 20 s. All reactions were carried out in triplicate. In order to confirm the amplification specificity, PCR products were subjected to a melting-curve analysis. The comparative crossing threshold (CT) method, employing the constitutive S. schenckii and S. brasiliensis L34 ribosomal gene for normalization, was used to evaluate the expression value for each gene of interest. Primers targeting the five genes at the opposite MAT idiomorphs were designed using Primer, version 3 (http://primer3.wi.mit.edu/).
DNA extraction and MAT locus distribution in the Sporothrix complex.
Genomic DNA was extracted and purified from mycelial colonies using a Fast DNA kit protocol (MP Biomedicals, Vista, CA, USA). Mycelial cells were homogenized three times with a Precellys 24 instrument (Bertin, Montigny le Bretonneux, France). DNA integrity was confirmed by 0.8% agarose gel electrophoresis with ethidium bromide (0.5 μl/ml) staining and quantified with a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA). For PCR amplification, 20 to 50 ng of genomic DNA from each isolate (see Table S2 in the supplemental material) was used for PCR amplifications. Primers were designed to target regions inside the MAT1-1-1 (Mat1_1_1F, AAGCACGCCAGTTTCATTCT; Mat1_1_1R, CACCAACGAGCATCTCATGT) and MAT-1-2-1 (Mat1_2F, GATCTCAACGGCCATCTTGT; Mat1_2R, GCTACATACTTCCGCCCTGA) open reading frames (ORFs). PCR amplifications were done in a Mastercycler Pro thermocycler (Eppendorf) and using 2× PCR master mix (Promega) as recommended by the manufacturers. The annealing temperature for both regions was 60°C. PCRs were performed separately for each idiomorph, and amplicons were loaded onto a 0.8% agarose gel and analyzed by electrophoresis. Amplified bands corresponding to each sexual idiomorph of each isolate were assigned. Mating type distributions were tested for deviation from the expected ratio of 1:1 using a chi-square test in Microsoft Excel.
Population genetic studies between S. schenckii and S. brasiliensis.
Population studies were done based on the large data sets (n = 133) of calmodulin and EF1α sequences from S. schenckii and S. brasiliensis species (see Table S2 in the supplemental material). Sequences from both loci were aligned using ClustalW (41) and manually adjusted. Nucleotide substitution models were selected using lower Bayesian information criteria (BIC) scores based on ML values computed automatically in a given tree topology implemented in MEGA, version 5, software (45). The evolutionary distances used for phylogenetic inferences were determined using the Kimura two-parameter method (47), and the rate variation among sites was modeled with a gamma distribution (shape parameter, 1). Phylogenetic analyses were carried out using maximum-likelihood and Bayesian inferences implemented in the MEGA, version 5 (45), and MrBayes, version 3.1, software programs (48), respectively. For maximum-likelihood analysis, 1,000 bootstrap replicates were used to estimate confidence values for individual clades. For Bayesian analysis using the Markov chain Monte Carlo (MCMC) method, two independent analyses of four chains each as a default were initiated from a random tree and processed for 1 million generations; sample trees were retrieved every 100 generations. Log-likelihood scores were plotted against the generation number in order to evaluate convergence; samples collected prior to burn-in (25%) were ignored. The remaining samples were used to determine the distribution of posterior probability values (49). Trees were visualized using FigTree software, version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Polymorphism comparisons between and within S. schenckii and S. brasiliensis populations were measured using DnaSP, version 5 (50), and the number of variable sites (S), total number of mutations (Eta; H), nucleotide diversity per site (π), number of haplotypes (h), haplotype diversity (hd), average number of nucleotide differences (k), mean evolutionary diversity for the entire population (Med), and substitutions per site (Sps) were estimated for cal and ef1 loci. In order to evaluate the reproductive modes of both S. schenckii and S. brasiliensis, we performed recombination analysis using different approaches. Recombination events were inferred using the split decomposition method via the LogDet algorithm and the pairwise homoplasy index (PHI) test for phylogenetic heterogeneity, both implemented in the SplitsTree, version 4.1 (51).
Haplotypic networks were built to visualize differences and diversity among S. schenckii and S. brasiliensis populations in Brazil. The distribution and diversity of haplotypes for cal plus ef1 were estimated using the software DnaSP, version 5, and used for input in Network, version 4.610, software (Fluxus Technology, Clare, Suffolk, England). Gaps and missing data were excluded in the analysis. Median-joining networks (52) for the concatenated data set were obtained and visualized using the software Network, version 4.610.
Genetic differentiation among local populations of S. brasiliensis (four) and S. schenckii (seven) was calculated using the analysis of molecular variance (AMOVA) with Arlequin, version 3.11 (53). ΦST is analogous to the fixation index, FST, and incorporates sequence divergence among haplotypes in addition to changes in haplotype frequency to assess genetic differentiation among populations (54). The statistical significance of the PHI statistic was tested based on 1,000 permutations (default settings).
RESULTS
Genomic characterization of mating type locus.
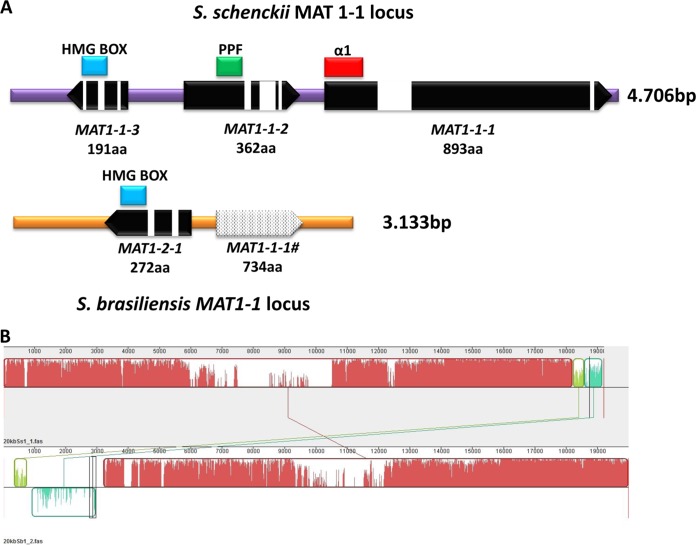
The whole-genome sequences of S. schenckii and S. brasiliensis isolates obtained, respectively, from human and cat infections were generated using Roche's 454 pyrosequencing (40). Genome searches using the MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2-1 genes of N. crassa and Magnaporthe grisea as queries revealed the presence of a single MAT1-1 locus in the S. schenckii genome (GenBank accession number JX105435) and a MAT1-2 locus in the S. brasiliensis genome(GenBank accession number JX101596), indicating that the two Sporothrix species are heterothallic (Fig. 1A). The MAT1-2 locus of S. globosa isolate KMU 3314 was characterized using a primer-walking approach (GenBank accession number KF021931). The gene order and orientation flanking the Sporothrix MAT loci were syntenic in all species investigated (see Fig. S1 in the supplemental material). The MAT1-1 locus of S. schenckii contains a region of 4,706 bp that is absent in the S. brasiliensis genome. In contrast, the MAT1-2 locus of S. brasiliensis is composed of 3,133 bp corresponding to a nonhomologous region in the S. schenckii scaffold (Fig. 1B).
FIG 1.
Genomic representation of mating type (MAT) locus of S. schenckii and S. brasiliensis. (A) Genomic analysis indicated that S. schenckii and S. brasiliensis harbor the MAT1-1 (MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2 (MAT1-2-1) locus, as well its corresponding domains: α1, α-box domain; PPF, proline-proline-phenylalanine domain; HMG, high mobility group domain. A truncated MAT1-1-1 was detected in the S. brasiliensis genome (MAT 1-1-1#) showing the lack of the α1 domain. The protein product size and conserved domains as well intron distribution along the genes are shown. (B) Genomic alignment of a 20-kb MAT-containing scaffold between S. schenckii and S. brasiliensis showing the nonhomologous region corresponding to the MAT locus.
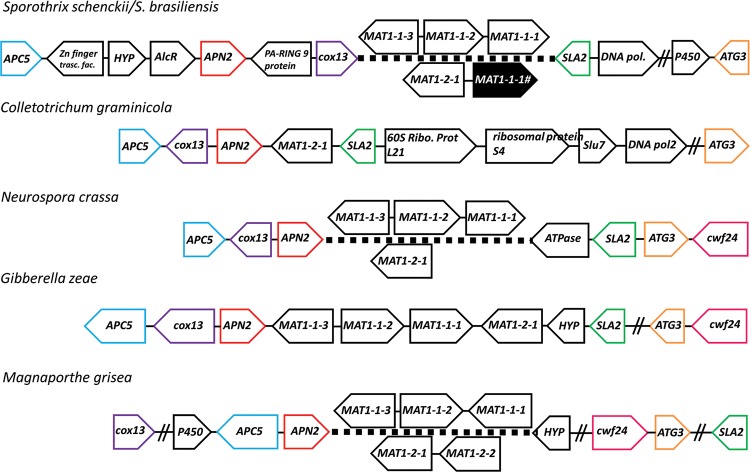
Similar to other representatives in Sordariomycetes, the S. schenckii and S. brasiliensis MAT locus is flanked by SLA2, which encodes the cytoskeleton assembly control protein (ORFs SPSK05860 and SPBR04592), cytochrome c oxidase subunit COX13 (ORFs SPBR05223 and SPSK06524), and DNA lyase APN2 (ORFs SPSK05918 and SPBR04646) (55) (Fig. 2). The deduced amino acid sequences of SLA2 and APN2 shared, respectively, 89% and 62% identity with those of Grosmannia clavigera, a tree pathogen in the Ophiostomatales (39). Three open reading frames (ORFs) were predicted in the MAT1-1 idiomorph of the S. schenckii genome: MAT1-1-1, MAT1-1-2, and MAT1-1-3, encoding proteins of 893, 362, and 191 amino acids (aa), respectively (Fig. 1A). The first protein had the α-box domain and was homologous (50% similarity) to the MAT1-1-1 of Ophiostoma novo-ulmi subsp. novo-ulmi (GenBank accession number ACZ53927.1). The second protein had an invariant PPF domain and was 35% similar to MAT1-1-2 of O. montium (GenBank accession number AGH03255.1). The third ORF encoded an HMG box domain protein sharing 65% similarity to the MAT1-1-3 of O. montium (GenBank accession number AGH03256.1). Two ORFs were predicted in the MAT1-2 idiomorphs of S. brasiliensis and S. globosa. The translated protein bearing the HMG domain (272 aa from S. brasiliensis) shared 90% similarity to that of S. globosa and was homologous to the MAT1-2-1 genes of other Sordariomycetes (see Fig. S1 in the supplemental material). tBLAST searches using the α-box domain containing genes from S. schenckii as queries did not return any significant matches in the S. brasiliensis genome, confirming the absence of the MAT1-1-1 gene.
FIG 2.
Syntenic organization of mating type (MAT) loci among Sordariomycetes. MAT genes are indicated by black arrows showing gene positions and orientation along the scaffolds. Overlapping genes presented in MAT1-1 and MAT1-2 loci are shown for heterothallic species, while both sets of MAT genes are shown in homothallic species. The surrounding genes of the MAT locus in Sordariomycetes are conserved. The MAT1-1 or MAT1-2 locus is flanked by APC5, cox13, and APN2 on one side and by SLA2 and ATG3 on the other side. Ribo. Prot., ribosomal protein; pol, polymerase; trasc. fac., transcription factor. Double slashes indicate a span of over 50 kb.
The 734-amino-acid hypothetical proteins upstream from the MAT1-2-1 genes in both S. globosa and S. brasiliensis had no significant similarity to any genes in the NCBI sequence database. Surprisingly this putative protein in S. brasiliensis (GenBank accession number JX101596) and S. globosa (GenBank accession number KF021931) was partially homologous (>60% similarity in amino acids) to the MAT1-1-1 gene in the MAT1-1 of S. schenckii but shorter in length and with no introns (see Fig. S2 in the supplemental material). Sequence comparison also revealed an α-box domain truncation/deletion at the N terminus of the putative protein. The start codon, however, was present in the putative protein coding sequence (called truncated MAT1-1-1), followed by fragments that are not homologous to the MAT1-1-1 in the MAT1-1 idiomorph (see Fig. S2 in the supplemental material). This 179-bp N-terminal sequence was used to search the genome, as well as the NCBI database, but no significant match was returned. Phylogenetic analysis of a 225-amino-acid stretch from the HMG domain corresponding to the MAT1-2-1 gene grouped S. brasiliensis and S. globosa in a strongly supported monophyletic clade, and they are divergent from representatives of Grosmannia and Ophiostoma (see Fig. S3). Representatives of ophiostomatoid fungi clustered with members of Neurospora and other Sordariales, with strong likelihood support (data not shown). This suggested that the human/animal pathogens might have evolved from the plant and forest pathogens within the Ophiostomatales.
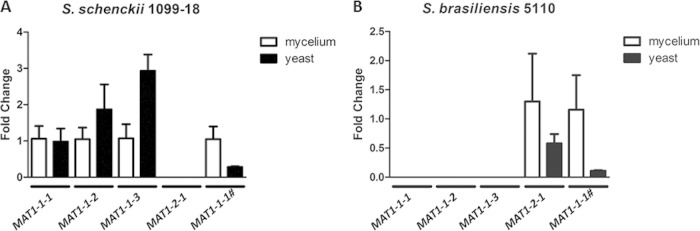
The mRNA levels of the MAT1-1-1, MAT1-1-2, MAT1-1-3, MAT-1-2-1, and the truncated MAT1-1-1 genes were quantified by qPCR analysis of S. schenckii and S. brasiliensis in both mycelial and yeast stages. The genes MAT1-1-1, MAT1-1-2, and MAT1-1-3 were expressed in S. schenckii 1099-18 while MAT-1-2-1 was expressed in S. brasiliensis 5110 (Fig. 3). Surprisingly, mRNA from the truncated MAT1-1-1 gene of S. brasiliensis was detected, suggesting that this ORF is transcribed despite the absence of the α-box domain.
FIG 3.
Quantitative expression of the MAT1-1-1, MAT1-1-2, MAT1-1-3, MAT-1-2-1, and truncated MAT1-1-1 (MAT1-1-1#) genes in Sporothrix species. Gene expression was measured between mycelium and yeast forms from S. schenckii (1099-18) and S. brasiliensis (5110).
Identification of sex-related genes in Sporothrix genomes.
We evaluated the presence or absence of 108 genes involved in the mating process, mating signaling, fruiting body development, karyogamy, and meiosis known from a wide range of fungi in the S. schenckii and S. brasiliensis genomes. Reciprocal best BLAST hits revealed the presence of 106 putative orthologs involved in sexual development in the Sporothrix genomes (see Table S3 in the supplemental material). In addition to the mating type idiomorphs, pheromone precursor and pheromone receptor genes, pheromone degrading enzymes, and meiotic genes were detected in the two Sporothrix genomes. The genes involved in the classical mating mitogen-activated protein kinase (MAPK) signaling cascade were also found highly conserved in these two pathogens. The presence of the MAP kinases, pheromone G-coupled protein subunits, and the transcriptional activator containing the homeodomain DNA-binding factor ste12 could also be recognized. Additionally, the majority of genes involved in fruiting body development were also found in both Sporothrix genomes. The only two genes not identified in the Sporothrix genomes related to fruiting body development were rhamnogalacuronase B and mutanase. The Sporothrix species harbor the majority of genes involved in pheromone activation, karyogamy, and meiosis in their genomes, thus indicating that both S. schenckii and S. brasiliensis possess the genomic apparatus to complete a sexual cycle.
Mating type distribution.
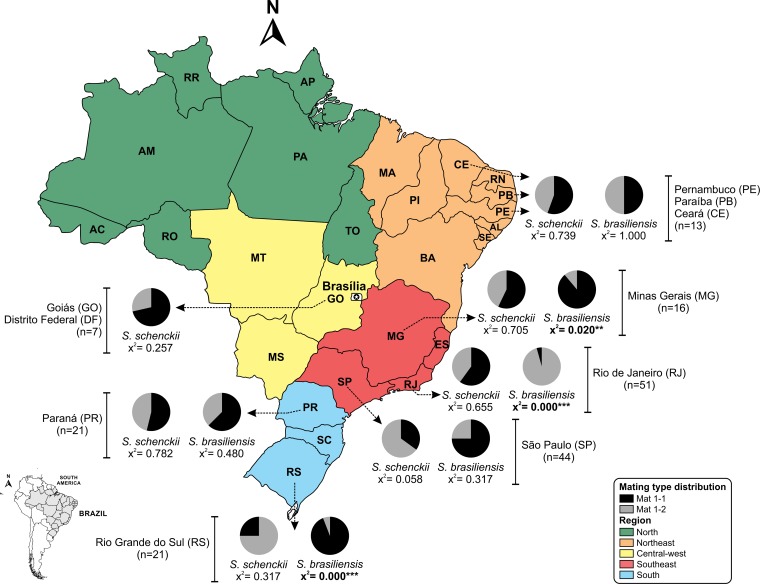
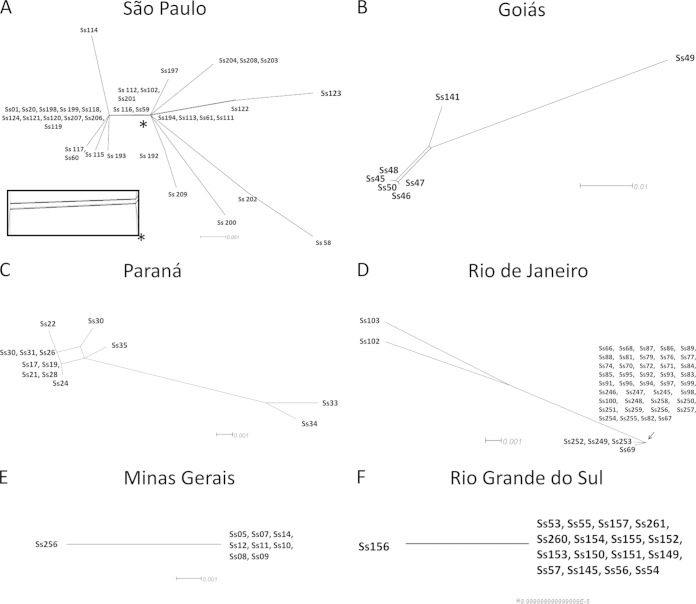
Primers targeting MAT1-1 and MAT1-2 loci were designed to investigate the mating type distribution in the Sporothrix populations in Brazil. A total of 193 isolates comprising S. schenckii (n = 99), S. brasiliensis (n = 90), and S. globosa (n = 4) were examined, revealing the presence of a single copy of the MAT1-1-1 or MAT1-2-1 locus in their genomes (single amplified band), suggesting that they are all heterothallic (see Fig. S4 and Table S2 in the supplemental material). All three Sporothrix species have approximately equal ratios of the MAT1-1 and the MAT1-2 idiomorphs, suggesting the presence of random mating within each species (chi-square test, P values of ≫0.05) (Table 1). Analysis of the mating type frequencies in populations of S. schenckii and S. brasiliensis also indicated a 1:1 ratio of both mating types in most states of Brazil (Fig. 4). However, in Rio Grande do Sul (RS) and Minas Gerais (MG), the frequency of MAT1-1 isolates was significantly higher in S. brasiliensis (P < 0.001) (Fig. 4), while MAT1-2 isolates were dominant in Rio de Janeiro (RJ) (P < 0.001). In contrast, S. schenckii appeared to have a 1:1 ratio of both mating types in the states of São Paulo (SP), Paraná (PR), and Goiás (GO) (P > 0.05) (Fig. 4).
TABLE 1.
Mating type distribution of Sporothrix species
| Species | No. of isolates by mating type |
Chi-square valuea | |
|---|---|---|---|
| MAT1-1 | MAT1-2 | ||
| S. schenckii | 45 | 54 | 0.366 |
| S. brasiliensis | 37 | 53 | 0.092 |
| S. globosa | 2 | 2 | 1.000 |
| Total | 84 | 109 | 0.072 |
Chi-square test was used to test mating type distributions for deviations from equal ratios, suggesting random mating. All of the values were nonsignificant.
FIG 4.
Geographic and mating type distribution of S. schenckii and S. brasiliensis isolates along the Brazilian territory discriminated by states. Mating type distributions (MAT1-1 and MAT1-2) were tested for deviation from the expected ratios of 1:1 using a chi-square test, and P values were added to graphs in order to infer statistical support. AC, Acre; AL, Alagoas; AM, Amazonas; AP, Amapá; BA, Bahia; MA, Maranhão; MS, Mato Grosso do Sul; MT, Mato Grosso; PA, Pará; PI, Piauí; RN, Rio Grande do Norte; RO, Rondonia; RR, Roraima; SC, Santa Catarina; SE, Sergipe; TO, Tocantins.
Population genetics and recombination analysis.
All S. brasiliensis isolates formed a single, well-supported, monophyletic group based on the phylogenetic analyses of partial sequences of calmodulin (cal) and translation elongation factor 1-alpha (ef1) (see Fig. S5 in the supplemental material). S. schenckii isolates also formed a monophyletic group, but the population was divided into multiple subclades, with low to moderate statistical support (see Fig. S5). Therefore, the S. brasiliensis population is dominated by a single genotype, while the S. schenckii population is composed of multiple genetic lineages. Polymorphism analysis also revealed large variations between the genetic profiles of these two species. We evaluated the polymorphisms in the cal and ef1 loci considering both species separately. The cal locus was more polymorphic than that of ef1 in all aspects analyzed. The measurements of the number of variable sites (S), total number of mutations (Eta; H), nucleotide diversity per site (π), number of haplotypes (h), haplotype diversity (hd), average number of nucleotide differences (k), mean evolutionary diversity for the entire population (Med), and substitutions per site (Sps) clearly show that S. schenckii is highly diverse compared to S. brasiliensis, and the pattern is in agreement with the phylogenetic analysis (Table 2).
TABLE 2.
Polymorphism analyses of S. brasiliensis and S. schenckii species
| Parameterb | Value for the parameter at the indicated locusa |
|||
|---|---|---|---|---|
|
S. schenckii |
S. brasiliensis |
|||
| cal | ef1 | cal | ef1 | |
| S | 114 | 28 | 43 | 12 |
| H | 123 | 28 | 45 | 12 |
| π | 0.01177 | 0.00432 | 0.00243 | 0.00135 |
| h | 35 | 21 | 9 | 7 |
| hd | 0.800 | 0.914 | 0.250 | 0.493 |
| k | 6.72300 | 3.01982 | 1.52196 | 0.94675 |
| Med | 0.012 (000.2) | 0.008 (0.001) | 0.002 (0.000) | 0.001 (0.001) |
Polymorphisms were evaluated using cal and ef1 loci and considering each species separately.
S, number of variable sites; H, total number of mutations; π, nucleotide diversity per site; h, number of haplotypes; hd, haplotype diversity; k, average number of nucleotide differences; Med, mean evolutionary diversity for the entire population followed by its standard deviation.
In order to evaluate reproductive isolation within S. schenckii and S. brasiliensis, we performed various recombination analyses. The split decomposition method generated by the LogDet algorithm showed recombination networks linking isolates from S. schenckii in the São Paulo, Goiás, and Paraná states of Brazil (Fig. 5A to C). A PHI test also detected recombination (P = 0.018) between isolates of S. schenckii in the São Paulo state. On the other hand, we have not identified any recombination network in S. brasiliensis isolates, suggesting a clonal reproductive mode there (Fig. 5D to F), which supported the uneven distribution of different mating types along the Brazilian territory.
FIG 5.
Split decomposition analysis of cal locus in S. schenckii and S. brasiliensis geographically restricted populations. The observation of networks connecting isolates was attributable to recombination events detected in S. schenckii populations from São Paulo (A), Goiás (B), and Paraná (C) but not detected in S. brasiliensis populations from Rio de Janeiro (D), Minas Gerais (E), and Rio Grande do Sul (F), suggesting clonal complexes. The enlarged boxed graph of the São Paulo data, indicated by an asterisk, is approximate to infer the precise topology.
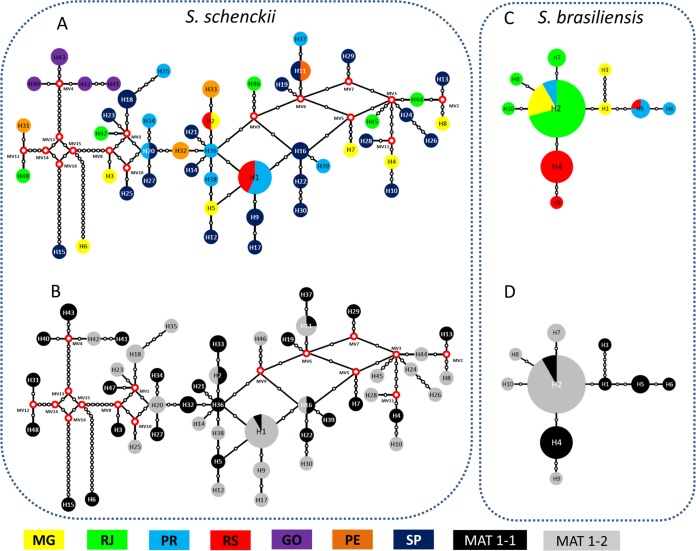
Median-joining networks were generated using the concatenated sequences of cal and ef1 loci for S. schenckii and S. brasiliensis populations independently (Fig. 6). The small number of haplotypes in S. brasiliensis suggests low genetic diversity, while in S. schenckii an increased haplotype diversity was observed. The haplotype networks show that the S. schenckii population presents high genetic variability and recombination of S. schenckii in Brazil, as deduced by the presence of several medial vectors linking different haplotypes (Fig. 6A). No strong geographic differentiation structure was observed, and mixed mating types were observed in the S. schenckii population (Fig. 6A and C). On the other hand, strong geography-based structuring was observed in the S. brasiliensis population (Fig. 6B). Four major populations were observed, each geographically restricted and with a predominant mating type, suggesting clonal expansions (Fig. 6D). The RS population composed of haplotypes H4 and H9 predominantly had the mating type MAT1-1, while the in RJ population composed of the haplotypes H2, H7, H8, and H10, the MAT1-2 idiomorph was predominant (Fig. 6D).
FIG 6.
Median-joining haplotype network of Sporothrix schenckii (A and C) and Sporothrix brasiliensis (B and D) populations using concatenated nucleotide sequences of the calmodulin-encoding gene (cal) and elongation factor-1 alpha (ef1). Median vectors are displayed by red dots shown in the S. schenckii population (A and C). The mating type distribution was plotted in each haplotype of S. schenckii (C) and S. brasiliensis (D) populations. The circumference size is proportional to the haplotype frequency. MG, Minas Gerais; RJ, Rio de Janeiro; PR, Paraná; RS, Rio Grande do Sul; GO, Goiás; PE, Pernambuco; SP, São Paulo.
AMOVA was used to estimate population differentiation in each of two Sporothrix species. Only 27.56% of the genetic variation in S. schenckii was attributed to differences between populations; in contrast, between-population variation in S. brasiliensis was more than double that observed in S. schenckii (66.4%), taking into account that the samples were collected on a smaller geographic scale (see Table S4 in the supplemental material).
DISCUSSION
Sexual reproduction in fungi promotes recombination and is thought to be more efficient in removing deleterious mutations (1). However, many pathogenic fungi could maintain the ability to carry out the sexual cycle and recombination, yet they drive clonal expansion after adaption to newly acquired environmental and host conditions. As a result, it is not uncommon that the core genes required for sexual development are found in the genomes of limiting-sex fungi (32, 55). In asexual pathogenic fungi such as Paracoccidioides brasiliensis, Paracoccidioides lutzii, and A. fumigatus, the sex-related genes were identified in their genomes prior to identification of sexual morphology (56, 57). In our investigation, different pathways of mating processes present in the genomic architecture of sexual fungi, such as the mating signaling cascade, fruiting body development, karyogamy, and meiosis, were detected in S. schenckii and S. brasiliensis genomes, providing molecular evidence to support the potential to perform mating/sexual recombination in these species (see Table S3 in the supplemental material). The identification of a single mating type locus bearing either an HMG or an α-box indicates that these species are heterothallic (Fig. 1). The presumed heterothallism in these species is also confirmed by the amplification of a single mating type in all 196 isolates tested. The general SLA-MAT-APN pattern, as well as the synteny and orientation of the MAT1-1-1, MAT1-1-2, and MAT1-1-3 genes on the MAT1-1 idiomorph, is consistent with other representatives of Grosmannia and Ophiostoma in the Ophiostomatales (39, 58). This pattern suggests a shared common heterothallic ancestor among members of the Ophiostomatales. However, apart from heterothallic species (e.g., O. quercus and O. novo-ulmi), Ophiostoma also harbors homothallic species (e.g., O. stenoceras and O. nigrocarpum), indicating different evolutionary strategies for breeding (28, 29). Also we detected by quantitative RT-PCR the expression of the mating type genes in a sex-type-specific manner, indicating heterothallism and functional activation of the MAT locus (Fig. 3).
The occurrence of truncated MAT1-1-1 genes in the MAT1-2 idiomorphs in Sporothrix provides strong evidence for unequal recombination at the MAT locus in an ancestor of the ophiostomatoid fungi. Truncated MAT1-1-1 genes were already reported in various representatives of Grosmannia and Ophiostoma (39), as well as other ascomycetes (27, 59, 60). The loss of a functional domain (in this case an α-box) with mutations and the high level of divergence between the truncated MAT1-1-1 and the full-length MAT1-1-1 suggest that these truncated MAT genes might have drifted under relaxed selective pressure (39) or might have been inactivated due to signal interference if both the HMG and α-box domains were present. Our data also demonstrated the variation in the rate of truncated MAT1-1-1 degeneration among Grosmannia, Ophiostoma, and Sporothrix. We detected expression of the truncated MAT1-1-1 in S. brasiliensis, and hence this gene may have gained a new function apart from mating in Sporothrix. Our investigation presented challenging questions on the biology of Sporothrix. The S. schenckii species complex has been considered asexual because of a lack of morphological evidence on sexual development. However, genomic evidence indicated that S. schenckii, S. brasiliensis, and S. globosa are heterothallic, and they may retain the ability to reproduce sexually, probably in the same fashion as other sexual fungi in Ophiostomatales (61, 62).
Sexual recombination may drive the formation of a novel, successful genotype that can replicate in mass numbers through asexual propagation (1, 63). Sexual reproduction increases the rate of adaptation to new environments by increasing the mutation rates in a fungal offspring. The reduction of sexual status and of recombination rates between species is correlated with a limited potential to disperse within different environments, but once a species is well adapted to an environment, asexual reproduction may preserve a successful genotype (3, 6). The extremely skewed ratios observed between mating types in Rio Grande do Sul, Minas Gerais, and, especially, Rio de Janeiro suggest the presence of a local clonal lineage or a recent introduction of the opposite mating type of S. brasiliensis during the outbreaks. Local differences may exist in virulence-associated genes that may show chromosomal linkage to the MAT locus. Due to the presence of feline sporotrichosis outbreaks in the states of Rio de Janeiro and Rio Grande do Sul, we hypothesize that this deviation is related to the form of transmission, cat to cat and cat to human, which favors the emergence of a clonal genotype in particular. The uneven mating type ratio in these states also indicates clonal expansion of S. brasiliensis.
We hypothesize that the origin of cat/human sporotrichosis was the result of a recent spread of S. brasiliensis, with increased virulence, into new areas of Brazil where the fungus encountered susceptible cat hosts. Infection assays using mouse models revealed a higher virulence of S. brasiliensis than of S. schenckii, supporting the spread of virulent clone complexes in sporotrichosis epidemics (17, 64). According to the phylogenetic trees (see Fig. S5 in the supplemental material), a monophyletic branch of a single mating type was observed in the ef1 phylogeny that was composed of isolates from Rio Grande do Sul and, thus, suggested the dispersal of a new genetic variant of S. brasiliensis. A pronounced geography-based population structure was observed in S. brasiliensis (Fig. 6B), and four major geographically restricted populations were detected. Each S. brasiliensis population harbored a single mating type, suggesting clonal expansions of specific genotypes, even though both mating types can be found in the same clinical and natural environments, making sexual reproduction possible (65).
Two species of Sporothrix differ in their major modes of reproduction, as indicated from the incongruent population structure patterns and contrasting genetic signatures. S. schenckii was a highly diverse population compared to S. brasiliensis based on polymorphism data of cal and ef1 loci, in agreement with the phylogenies and haplotype networks. Recombination analysis by split decomposition shows networks linking isolates of S. schenckii in the São Paulo, Goiás, and Paraná states of Brazil (Fig. 5A to C), which is in agreement with sexual recombination. These findings strongly suggest other forms of acquisition of the disease, such as those based on environmental contamination with soil and decaying wood. In contrast, the lack of recombination networks and the uneven distribution of opposite mating types among various states suggest a clonal reproductive mode in S. brasiliensis isolates (Fig. 5D to F). Recently, the pulsed-field gel electrophoresis (PFGE) profile of S. brasiliensis isolates showed less genome variability than S. schenckii isolates, indicating lower genomic rearrangements in S. brasiliensis than in S. schenckii (66).
Sporotrichosis is an important subcutaneous mycosis affecting humans and animals and mainly occurs in the form of epidemics, which may be very large. Areas of endemicity with outbreaks of thousands of cases have been recognized in South Africa, China, and Brazil (Y. Zhang, F. Hagen, B. Stielow. A. M. Rodrigues, K. Samerpitak, Z. Xun, P. Feng, L. Yang, M. Chen, S. Deng, W. Liao, R. Y. Li, F. Li, J. F. Meis, J. Guarro, and G. S. de Hoog, unpublished data). Our study suggests that the high levels of effective clonality and endemicity found in S. brasiliensis may have more to do with MAT expansion. The fungus may have specific host interactions while S. schenckii may generate population genetic diversity through sexual recombination. Interestingly, these two fungi cause the same disease while presenting different population structures. Sexual reproduction enhances the rate of adaptation to new environments by increasing mutation rates in fungal offspring. Limiting sex and recombination between species are correlated with a reduced potential to disperse within different environments. For example, restricted dispersal and adaptation to particular niches were associated with asexuality in the human and animal pathogen Talaromyces (Penicillium) marneffei (7, 67). Similarly, clonally restricted Australian and Papua New Guinean populations of the basidiomycetous yeast Cryptococcus gattii type VGI were detected in regions of cryptococcosis endemicity (68). In addition, a clonal population structure of the thermo-dimorphic fungus Paracoccidioides brasiliensis PS3 was suggested (69) and supported by recent dispersion from the Brazilian shield to Colombia (70). The same hypothesis might apply in virulent clonal lineages of S. brasiliensis in cats in geographically restricted parts of Brazil, as deeply explored in this work.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to FAP-DF, FAPESP, CNPq, and Capes for the financial support and fellowships of the projects Pronex (grant 193000569/2009), Genoprot (grant 559572/2009-3), and FAPESP (Proc. 2009/54024-2). A.M.R. is a fellow and acknowledges the financial support of the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2011/07350-1) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (BEX 2325/11-0). G.F.F. is a fellow of FAPESP (2011/01628-8). C.K.M.T and R.C.H. acknowledge the financial assistance of Genome Canada and Genome British Columbia in support to the Tria I and Tria II Projects.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00153-14.
REFERENCES
- 1.Heitman J. 2006. Sexual reproduction and the evolution of microbial pathogens. Curr Biol 16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 2.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu Rev Genet 45:405–430. doi: 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibayrenc M, Ayala FJ. 2012. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci U S A 109:E3305-3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman BH, White TJ, Taylor JW. 1996. Human pathogeneic fungi and their close nonpathogenic relatives. Mol Phylogenet Evol 6:89–96. doi: 10.1006/mpev.1996.0061. [DOI] [PubMed] [Google Scholar]
- 5.Rydholm C, Szakacs G, Lutzoni F. 2006. Low genetic variation and no detectable population structure in aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot Cell 5:650–657. doi: 10.1128/EC.5.4.650-657.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heitman J. 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henk DA, Shahar-Golan R, Devi KR, Boyce KJ, Zhan N, Fedorova ND, Nierman WC, Hsueh PR, Yuen KY, Sieu TP, Kinh NV, Wertheim H, Baker SG, Day JN, Vanittanakom N, Bignell EM, Andrianopoulos A, Fisher MC. 2012. Clonality despite sex: the evolution of host-associated sexual neighborhoods in the pathogenic fungus Penicillium marneffei. PLoS Pathog 8:e1002851. doi: 10.1371/journal.ppat.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graser Y, De Hoog S, Summerbell RC. 2006. Dermatophytes: recognizing species of clonal fungi. Med Mycol 44:199–209. doi: 10.1080/13693780600606810. [DOI] [PubMed] [Google Scholar]
- 9.Ene IV, Bennett RJ. 2014. The cryptic sexual strategies of human fungal pathogens. Nat Rev Microbiol 12:239–251. doi: 10.1038/nrmicro3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Romero E, Reyes-Montes Mdel R, Perez-Torres A, Ruiz-Baca E, Villagomez-Castro JC, Mora-Montes HM, Flores-Carreon A, Toriello C. 2011. Sporothrix schenckii complex and sporotrichosis, an emerging health problem. Future Microbiol 6:85–102. doi: 10.2217/fmb.10.157. [DOI] [PubMed] [Google Scholar]
- 11.Lopes-Bezerra LM, Schubach A, Costa RO. 2006. Sporothrix schenckii and sporotrichosis. Anais Acad Bras Cienc 78:293–308. doi: 10.1590/S0001-37652006000200009. [DOI] [PubMed] [Google Scholar]
- 12.Barros MB, de Almeida Paes R, Schubach AO. 2011. Sporothrix schenckii and sporotrichosis. Clin Microbiol Rev 24:633–654. doi: 10.1128/CMR.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marimon R, Cano J, Gene J, Sutton DA, Kawasaki M, Guarro J. 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol 45:3198–3206. doi: 10.1128/JCM.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Rodrigues AM, Feng P, de Hoog GS. 2013. Global ITS diversity in the Sporothrix schenckii complex. Fungal Divers 66:1–13. doi: 10.1007/s13225-013-0220-2. [DOI] [Google Scholar]
- 15.Rodrigues AM, de Hoog S, de Camargo ZP. 2013. Emergence of pathogenicity in the Sporothrix schenckii complex. Med Mycol 51:405–412. doi: 10.3109/13693786.2012.719648. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes GF, dos Santos PO, Rodrigues AM, Sasaki AA, Burger E, de Camargo ZP. 2013. Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence 4:241–249. doi: 10.4161/viru.23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Marine M, Gene J, Cano J, Guarro J. 2009. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect 15:651–655. doi: 10.1111/j.1469-0691.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- 18.de Lima Barros MB, de Oliveira Schubach A, Galhardo MC, Schubach TM, dos Reis RS, Conceicao MJ, do Valle AC. 2003. Sporotrichosis with widespread cutaneous lesions: report of 24 cases related to transmission by domestic cats in Rio de Janeiro, Brazil. Int J Dermatol 42:677–681. doi: 10.1046/j.1365-4362.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- 19.de Lima Barros MB, Schubach TM, Galhardo MC, de Oliviera Schubach A, Monteiro PC, Reis RS, Zancope-Oliveira RM, dos Santos Lazera M, Cuzzi-Maya T, Blanco TC, Marzochi KB, Wanke B, do Valle AC. 2001. Sporotrichosis: an emergent zoonosis in Rio de Janeiro. Mem Inst Oswaldo Cruz 96:777–779. doi: 10.1590/S0074-02762001000600006. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues AM, de Melo Teixeira M, de Hoog GS, Schubach TM, Pereira SA, Fernandes GF, Bezerra LM, Felipe MS, de Camargo ZP. 2013. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Negl Trop Dis 7:e2281. doi: 10.1371/journal.pntd.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, Vismer HF, Wingfield MJ. 2008. Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 100:647–661. doi: 10.3852/07-157R. [DOI] [PubMed] [Google Scholar]
- 22.Berbee ML, Taylor JW. 1992. 18S ribosomal RNA gene sequence characters place the human pathogen Sporothrix schenckii in the genus Ophiostoma. Exp Mycol 16:87–91. doi: 10.1016/0147-5975(92)90044-R. [DOI] [Google Scholar]
- 23.Mariat F, Escudié A, Gaxotte P. 1968. Solement de souches de Ceratocystis sp. à forme conidienne Sporotrichum, se cuirs chevelus humains et de poils de rats. Comparaison avec l'espèce pathogène Sporotrichum schenckii. C R Acad Sci Hebd Seances Acad Sci D 267:974–976. [PubMed] [Google Scholar]
- 24.Nicot J, Mariat F. 1973. Caractères morphologiques et position systématique de Sporothrix schenckii, agent de la sporotrichose humaine. Mycopathol Mycol Appl 49:53–65. doi: 10.1007/BF02057447. [DOI] [PubMed] [Google Scholar]
- 25.de Beer ZW, Harrington TC, Vismer HF, Wingfield BD, Wingfield MJ. 2003. Phylogeny of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 95:434–441. doi: 10.2307/3761885. [DOI] [PubMed] [Google Scholar]
- 26.Aghayeva DN, Wingfield MJ, de Beer ZW, Kirisits T. 2004. Two new Ophiostoma species with Sporothrix anamorphs from Austria and Azerbaijan. Mycologia 96:866–878. doi: 10.2307/3762119. [DOI] [PubMed] [Google Scholar]
- 27.Wilken PM, Steenkamp ET, Hall TA, De Beer ZW, Wingfield MJ, Wingfield BD. 2012. Both mating types in the heterothallic fungus Ophiostoma quercus contain MAT1-1 and MAT1-2 genes. Fungal Biol 116:427–437. doi: 10.1016/j.funbio.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Brasier CM, Kirk SA. 1993. Sibling species within Ophiostoma piceae. Mycol Res 97:811–816. doi: 10.1016/S0953-7562(09)81156-8. [DOI] [Google Scholar]
- 29.Harrington TC, McNew D, Steimel J, Hofstra D, Farrell R. 2001. Phylogeny and taxonomy of the Ophiostoma piceae complex and the Dutch elm disease fungi. Mycologia 93:111–136. doi: 10.2307/3761610. [DOI] [Google Scholar]
- 30.Solla A, Dacasa MC, Nasmith C, Hubbes M, Gil L. 2008. Analysis of Spanish populations of Ophiostoma ulmi and O. novo-ulmi using phenotypic characteristics and RAPD markers. Plant Pathol 57:33–44. doi: 10.1111/j.1365-3059.2007.01692.x. [DOI] [Google Scholar]
- 31.Fraser JA, Heitman J. 2004. Evolution of fungal sex chromosomes. Mol Microbiol 51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Metin B, White TC, Heitman J. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot Cell 9:46–58. doi: 10.1128/EC.00259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin SH, Wingfield BD, Wingfield MJ, Steenkamp ET. 2011. Causes and consequences of variability in peptide mating pheromones of ascomycete fungi. Mol Biol Evol 28:1987–2003. doi: 10.1093/molbev/msr022. [DOI] [PubMed] [Google Scholar]
- 34.Yun SH, Berbee ML, Yoder OC, Turgeon BG. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A 96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turgeon BG. 1998. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol 36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 36.Paoletti M, Buck KW, Brasier CM. 2005. Cloning and sequence analysis of the MAT-B (MAT-2) genes from the three Dutch elm disease pathogens, Ophiostoma ulmi, O. novo-ulmi, and O. himal-ulmi. Mycol Res 109:983–991. doi: 10.1017/S0953756205003308. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti M, Buck KW, Brasier CM. 2006. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol Ecol 15:249–262. doi: 10.1111/j.1365-294X.2005.02728.x. [DOI] [PubMed] [Google Scholar]
- 38.Jacobi V, Dufour J, Bouvet GF, Aoun M, Bernier L. 2010. Identification of transcripts up-regulated in asexual and sexual fruiting bodies of the Dutch elm disease pathogen Ophiostoma novo-ulmi. Can J Microbiol 56:697–705. doi: 10.1139/W10-053. [DOI] [PubMed] [Google Scholar]
- 39.Tsui CK, DiGuistini S, Wang Y, Feau N, Dhillon B, Bohlmann J, Hamelin RC. 2013. Unequal recombination and evolution of the mating-type (MAT) loci in the pathogenic fungus Grosmannia clavigera and relatives. G3 3:465–480. doi: 10.1534/g3.112.004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira MM, Almeida LGP, Kubitschek-Barreira PH, Alves FL, Kioshima ES, Abadio AKR, Fernandes L, Derengowski LS, Ferreira KS, Souza RC, Jeronimo CR, Andrade NC, Mora-Montes HM, Almeida SR, Stajich JE, Lopes-Bezerra LM, Vasconcelos ATR, Felipe MS. 2014. Comparative genomics of the major fungal agents of human and animal sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis. BMC Genomics 15:943. doi: 10.1186/1471-2164-15-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 43.Zheng P, Xia Y, Xiao G, Xiong C, Hu X, Zhang S, Zheng H, Huang Y, Zhou Y, Wang S, Zhao GP, Liu X, St Leger RJ, Wang C. 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol 12:R116. doi: 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staden R. 1996. The Staden sequence analysis package. Mol Biotechnol 5:233–241. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 48.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 49.Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 50.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 51.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 52.Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 53.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 54.Cockerham CC, Weir BS. 1984. Covariances of relatives stemming from a population undergoing mixed self and random mating. Biometrics 40:157–164. doi: 10.2307/2530754. [DOI] [PubMed] [Google Scholar]
- 55.Debuchy R, Turgeon BG. 2006. Mating-type structure, evolution, and function in euascomycetes, p 293–323. In Kues U, Fischer R (ed), The mycota. I. Growth, differentiation, and sexuality Springer-Verlag, Berlin, Germany. [Google Scholar]
- 56.Poggeler S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr Genet 42:153–160. doi: 10.1007/s00294-002-0338-3. [DOI] [PubMed] [Google Scholar]
- 57.Teixeira Mde M, Theodoro RC, Derengowski Lda S, Nicola AM, Bagagli E, Felipe MS. 2013. Molecular and morphological data support the existence of a sexual cycle in species of the genus Paracoccidioides. Eukaryot Cell 12:380–389. doi: 10.1128/EC.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haridas S, Wang Y, Lim L, Massoumi Alamouti S, Jackman S, Docking R, Robertson G, Birol I, Bohlmann J, Breuil C. 2013. The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle-associated pine pathogen Grosmannia clavigera. BMC Genomics 14:373. doi: 10.1186/1471-2164-14-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quevillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collemare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Guldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuveglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Segurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun MH, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gioti A, Mushegian AA, Strandberg R, Stajich JE, Johannesson H. 2012. Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol Biol Evol 29:3215–3226. doi: 10.1093/molbev/mss132. [DOI] [PubMed] [Google Scholar]
- 61.Hunt J. 1956. Taxonomy of the genus Ceratocystis. Lloydia 19:1–59. [Google Scholar]
- 62.Wingfield MJ, Seifert KA, Webber JF (ed). 1993. Ceratocystis and Ophiostoma: taxonomy, ecology, and pathogenicity. American Phytopathological Society, St. Paul, MN. [Google Scholar]
- 63.Sun S, Heitman J. 2011. Is sex necessary? BMC Biol 9:56. doi: 10.1186/1741-7007-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castro RA, Kubitschek-Barreira PH, Teixeira PA, Sanches GF, Teixeira MM, Quintella LP, Almeida SR, Costa RO, Camargo ZP, Felipe MS, de Souza W, Lopes-Bezerra LM. 2013. Differences in cell morphometry, cell wall topography and Gp70 expression correlate with the virulence of Sporothrix brasiliensis clinical isolates. PLoS One 8:e75656. doi: 10.1371/journal.pone.0075656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nielsen K, Heitman J. 2007. Sex and virulence of human pathogenic fungi. Adv Genet 57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- 66.Sasaki AA, Fernandes GF, Rodrigues AM, Lima FM, Marini MM, Feitosa LS, Teixeira MM, Felipe MS, Silveira JF, Camargo ZP. 2014. Chromosomal polymorphism in the Sporothrix schenckii complex. PLoS One 9:e86819. doi: 10.1371/journal.pone.0086819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher MC, Hanage WP, de Hoog S, Johnson E, Smith MD, White NJ, Vanittanakom N. 2005. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathog 1:e20. doi: 10.1371/journal.ppat.0010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot Cell 4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, Bagagli E, Rauscher JT, Restrepo A, Morais F, Nino-Vega G, Taylor JW. 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol 23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- 70.Theodoro RC, Teixeira Mde M, Felipe MS, Paduan Kdos S, Ribolla PM, San-Blas G, Bagagli E. 2012. Genus Paracoccidioides: species recognition and biogeographic aspects. PLoS One 7:e37694. doi: 10.1371/journal.pone.0037694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.