Abstract
Membrane reshaping resides at the core of many important cellular processes, and among its mediators are the BAR (Bin, Amphiphysin, Rvs) domain-containing proteins. We have explored the diversity and function of the Rvs BAR proteins in Candida albicans and identified a novel family member, Rvs167-3 (orf19.1861). We show that Rvs167-3 specifically interacts with Rvs162 to form a stable BAR heterodimer able to bind liposomes in vitro. A second, distinct heterodimer is formed by the canonical BAR proteins Rvs161 and Rvs167. Purified Rvs161/Rvs167 complex also binds liposomes, indicating that C. albicans expresses two functional BAR heterodimers. We used live-cell imaging to localize green fluorescent protein (GFP)-tagged Rvs167-3 and Rvs167 and show that both proteins concentrate in small cortical spots. However, while Rvs167 strictly colocalizes with the endocytic marker protein Abp1, we do not observe any colocalization of Rvs167-3 with sites of endocytosis marked by Abp1. Furthermore, the rvs167-3Δ/Δ mutant is not defective in endocytosis and strains lacking Rvs167-3 or its partner Rvs162 do not display increased sensitivity to high salt concentrations or decreased cell wall integrity, phenotypes which have been observed for rvs167Δ/Δ and rvs161Δ/Δ strains and which are linked to endocytosis defects. Taken together, our results indicate different roles for the two BAR heterodimers in C. albicans: the canonical Rvs161/Rvs167 heterodimer functions in endocytosis, whereas the novel Rvs162/Rvs167-3 heterodimer seems not to be involved in this process. Nevertheless, despite their different roles, our phenotypic analysis revealed a genetic interaction between the two BAR heterodimers, suggesting that they may have related but distinct membrane-associated functions.
INTRODUCTION
Membrane reshaping is key to many cellular processes such as cell division, vesicle transport, and endocytosis. Several proteins have been implicated in membrane reshaping, among them proteins that are able to sense, promote, and stabilize local membrane curvature. One prominent protein module is the BAR (Bin/Amphiphysin/Rvs) domain (1–3). The BAR monomer consists of 3 α-helixes (see Fig. S1 in the supplemental material) that, upon dimerization, provide its characteristic “banana shape” structure. Positively charged residues in its concave face interact with negatively charged lipid head groups localized on the cytosolic side of the membrane bilayer (4–6). While BAR domains do not always display high sequence identity, the conservation of their structure is striking (7, 8).
The degrees of intrinsic curvature differ among the members of the BAR superfamily (9). Three groups can be distinguished: the classical BAR domain, the F-BAR domain that consists of an FCH domain and a coiled-coil region, and the I-BAR domain. A subgroup of the classical BAR is the N-BAR, which is characterized by an N-terminal amphipathic helix (also called helix 0 [H0]). BAR, N-BAR, and F-BAR domains can sense and promote positive membrane curvature, whereas the I-BAR domains promote negative curvature (1, 8). Pinkbar is a newly identified domain that seems to stabilize planar membranes (10).
Two major models have been proposed to explain the membrane binding and curvature-inducing properties of BAR domains: the “scaffolding” model and the “amphipathic insertion” model. According to the first model, the electrostatic interactions between the positively charged BAR amino acids and the negatively charged phospholipids result in membrane bending as the BAR domains impose the curvature of their shape on the membrane (11). The second model proposes that membrane curvature is initially generated by insertion of amphipathic helixes into the lipid bilayer (12) and is further stabilized by the BAR domain itself (13). Despite the substantial experimental evidence in support of each model, the exact mechanism of membrane bending remains elusive.
BAR domains are found as dimers, and the importance of their dimerization is illustrated by experiments where dimerization-impaired endophilin mutants are unable to bind and bend membranes (14, 15). Both homodimerization and heterodimerization have been observed. Heterodimerization often but not exclusively occurs between proteins of the same family, such as mammalian amphiphysin 1 and amphiphysin 2 (16) and the amphiphysin homologs in yeast, Rvs161 and Rvs167 (17, 18). However, amphiphysin 2 has also been found to heterodimerize with Snx4 (19), representing an additional layer of complexity to the BAR-BAR protein interaction network. The exact mechanisms that determine homodimerization or heterodimerization remain largely unknown. Specific amino acids that promote homodimerization and inhibit heterodimerization have been identified only for Snx33 (20). BAR domain-containing proteins (endophilin, amphiphysin, sortin nexins) play an essential role in clathrin-mediated endocytosis, a process that connects membrane curvature and actin organization. In this highly orchestrated process, the cell is able to internalize part of its plasma membrane, receptors, and cargo from its surface and subsequently release them in a vesicular form in the cytoplasm as a response to external (i.e., absence or presence of nutrients) or internal (i.e., polarized growth) signals (21–23). The complexity of this process is illustrated by the fact that more than 50 proteins are involved in clathrin-mediated endocytosis in Saccharomyces cerevisiae. Evidence obtained from live-cell imaging analysis places the N-BAR proteins Rvs161 and Rvs167 at the late stage of endocytosis, promoting the scission of the invaginated vesicle from the plasma membrane (24, 25).
In addition to an N-terminal BAR domain, Rvs167 proteins contain an Src-homology 3 (SH3) domain in their C terminus. SH3 domains are relatively small domains consisting of about 60 amino acids and are known to mediate transient protein-protein interactions (26).Their structure is widely studied and consists of five well-conserved antiparallel β strands, forming two β sheets that are joined together by three less-well-conserved loop sequences, the RT loop, the n-src loop, and the distal loop. They are known to bind to proline-rich sequences in their binding partners. The minimum binding consensus for SH3 binding is PxxP, in which P denotes proline and x any amino acid. The specificity of SH3 domains is determined by a conserved hydrophobic motif and sequences in the RT and the n-src loops. Many proteins involved in signaling cascades as well as in endocytosis contain SH3 interaction domains.
Besides well-defined roles in endocytosis, other cellular functions have been associated with the Rvs proteins. Rvs167 has been implicated in polarized exocytosis (27), whereas Rvs161 seems to play an active role in cell fusion through its interaction with Fus2 (28) and in polarizing actin filaments (29). In addition, there is compelling genetic evidence to suggest a connection between Rvs161 and Rvs167 and sphingolipid biosynthesis (30, 31).
Here we focus on the Rvs family in the human fungal pathogen Candida albicans. To date, four proteins belonging to the Rvs family have been identified in C. albicans: two proteins with a domain composition similar to that of S. cerevisiae Rvs167 (ScRvs167), named Candida albicans Rvs167 (CaRvs167) and CaRvs167-2, and two proteins with a domain structure similar to that of ScRvs161, denoted CaRvs161 and CaRvs162 (32, 33). Genetic and biochemical experiments have shown an important role for CaRvs161 and CaRvs167 in endocytosis, hyphal formation, and virulence, but the functions of CaRvs167-2 and CaRvs162 have remained enigmatic (32, 33). We have identified a novel fifth member of the C. albicans Rvs family, which we have termed Rvs167-3 (orf19.1861), and provide strong in vitro and in vivo evidence for the existence of two specific BAR heterodimers: Rvs161/Rvs167 and Rvs162/Rvs167-3. Both BAR heterodimers are able to bind liposomes in vitro, an observation that is in line with the reported membrane binding properties of BAR modules. We report for the first time the localization of Rvs167 and Rvs167-3 in C. albicans and show that both proteins can be found in small cortical puncta. However, Rvs167, but not Rvs167-3, colocalizes with sites of endocytosis marked by Abp1. Consistent with this, we show that the rvs167-3Δ/Δ mutant is not deficient in endocytosis. Finally, a phenotypic screen performed with single- and double-deletion strains revealed a genetic interaction between the two BAR heterodimers, suggesting that, although they do not colocalize to the same membrane structures, they may have partially overlapping functions.
MATERIALS AND METHODS
Media and culture conditions.
C. albicans strains were grown at 28°C, unless otherwise stated. Cells were grown in rich yeast extract-peptone-dextrose (YPD) medium [1% (wt/vol) Bacto yeast extract, 2% (wt/vol) Bacto peptone, 2% (wt/vol) d(+)-glucose] supplemented with uridine (80 mg/liter) or on minimal medium containing 2% glucose, 0.67% yeast nitrogen base (YNB) without amino acids (Difco), and amino acids as required.
The S. cerevisiae yeast-2-hybrid (Y2H) Gold strain (Clontech) was grown in rich YPD medium supplemented with adenine (40 mg/liter) for use in transformation performed with bait and prey plasmids. Transformants were selected on minimal medium (2% glucose, 0.67% YNB) lacking tryptophan and leucine and supplemented with uracil (20 mg/liter), histidine (20 mg/liter), lysine (30 mg/liter), adenine (20 mg/liter), and methionine (20 mg/liter).
Escherichia coli strain DH5α was grown in Luria-Bertani (LB) medium (1% [wt/vol] tryptone, 0.5% [wt/vol] Bacto yeast extract, 1% [wt/vol] NaCl), liquid or solid, with the addition of the appropriate antibiotics. E. coli strain BL21-CodonPlus(DE3)-RIPL (Stratagene) was grown in LB medium enriched with 2% glucose, 50 mM Tris (pH 7.8), and the appropriate antibiotics.
Strains and plasmids.
E. coli strain DH5α [genotype F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1] was used for cloning purposes and BL21-CodonPlus(DE3)-RIPL {genotype F− ompT hsdS(rB− mB−) dcm+ Tetr galλ(DE3) endA Hte [argU proL Camr] [argU ileY leuW Strep/Specr]} for protein expression.
For yeast two-hybrid experiments, a S. cerevisiae yeast-2-hybrid Gold strain (MATa trp1-901 leu2-3 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-Gal1TATA-His3 GAL2UAS-Gal2TATA-Ade2 URA3::MEL1UAS-MelTATA AUR1-C MEL1; Clontech) was used. C. albicans strains used in this study are listed in Table 1 and are derivatives of SN148 or SN76 (34). Primers and plasmids used in this study are listed in Table S1 and Table S2 in the supplemental material, respectively.
TABLE 1.
Strains used in this study
| Strain | Description | Genotype | Source |
|---|---|---|---|
| SN76 | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | 34 | |
| SN76-P | arg4Δ/ARG4 his1Δ/HIS1 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 | 35 | |
| SN148 | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | 34 | |
| SN148-P | arg4Δ/ARG4 leu2Δ/LEU2 his1Δ/HIS1 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 iro1Δ::imm434/iro1Δ::imm434::IRO1 | This study | |
| CJB142b | RVS167-HA | arg4Δ/arg4Δ leu2Δ/leu2Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-HA::CdHIS1 | This study |
| CJB129b | RVS167-2-HA | leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-2-HA::CdARG4 | This study |
| CJB132b | RVS167-3-HA | leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-3-HA::CdARG4 | This study |
| CJB126b | RVS161-HA | leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS161-HA::CdARG4 | This study |
| CJB128b | RVS162-MYC | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS162-MYC::CmLEU2 | This study |
| CJB141b | RVS167-MYC | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-MYC::CmLEU2 | This study |
| CJB127b | RVS167-MYC/RVS161-HA | his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS161-HA::CdARG4 RVS167-MYC::CmLEU2 | This study |
| CJB133b | RVS162-MYC/RVS167-3-HA | his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-3-HA::CdARG4 RVS162-MYC::CmLEU2 | This study |
| CJB160b | rvs161ΔΔ | his1Δ/HIS1 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs161Δ::CmLEU2/rvs161Δ::CdARG4 | This study |
| CJB175b | rvs167ΔΔ | arg4Δ/ARG4 leu2Δ/LEU2 iro::imm434/iro::imm434::IRO1 rvs167Δ::CdHIS1/rvs167Δ::URA3 | This study |
| CJB163b | rvs162ΔΔ | leu2Δ/LEU2 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs162Δ::CdARG4/rvs162Δ::CdHIS1 | This study |
| CJB122b | rvs167-3ΔΔ | leu2Δ/leu2Δ ura3Δ::imm434/ura3Δ::imm434:: ura3Δ iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 | This study |
| CJB123b | rvs167-3ΔΔ | leu2Δ/LEU2 ura3Δ::imm434/ura3Δ::imm434::URA3 iro1Δ::imm434/iro1Δ::imm434::IRO1 Rvs167-3Δ::CdARG4/Rvs167-3Δ::CdHIS1 | This study |
| CJB168b | rvs161ΔΔ/rvs167ΔΔ | iro::imm434/iro::imm434::IRO1 rvs167Δ::CdHIS1/rvs167Δ::URA3 rvs161Δ::LEU2/rvs161Δ::ARG4 | This study |
| CJB166b | rvs161ΔΔ/rvs162ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs161Δ::CdARG4/rvs161Δ::CmLEU2 rvs162Δ::CdHIS1/rvs162Δ:URA3 | This study |
| CJB167b | rvs167-3ΔΔ/rvs167ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167Δ::CdHIS1/rvs167Δ::URA3 rvs167-3Δ::LEU2/rvs167-3Δ::ARG4 | This study |
| CJB165b | rvs167-3ΔΔ/rvs162ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 rvs162Δ::LEU2/rvs162Δ:URA3 | This study |
| CJB184b | rvs167-3ΔΔ/rvs161ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 rvs161Δ::LEU2/rvs161Δ:URA3 | This study |
| CSB4a | Abp1-RFP | his1Δ/his1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 ABP1-RFP:: ARG4 | This study |
| CSB6a | RVS167-GFPγ | arg4Δ/arg4Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 Rvs167-GFPγ::CdHIS1 | This study |
| CSB9a | RVS167-3-GFPγ | arg4Δ/arg4Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 Rvs167-3-GFPγ::CdHIS1 | This study |
| CJB158a | RVS167-3-GFPγ/ABP1-RFP | ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 ABP1-RFP:: ARG4 RVS167-3-GFPγ::CdHIS1 | This study |
| CJB215a | RVS167- GFPγ/ABP1-RFP | ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-GFPγ:: CdHIS1 ABP1-RFP:: CdARG4 | This study |
| CJB151b | RVS167-HA/rvs161ΔΔ | ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS167-HA::CdHIS1 rvs161Δ::CdARG4/rvs161Δ::CmLEU2 | This study |
| CJB136b | RVS161-HA/rvs167ΔΔ | leu2Δ/leu2Δ iro1Δ::imm434/iro1Δ::imm434 RVS161-HA::CdARG4 rvs167Δ::CdHIS1/rvs167Δ::URA3 | This study |
| CJB135c | RVS167-3-HA/rvs162ΔΔ | iro1Δ::imm434/iro1Δ::imm434 RVS167-3-HA::URA3 rvs162Δ::CdHIS1/rvs162Δ::CdARG4 | This study |
| CJB140b | RVS162-MYC/rvs167-3ΔΔ | ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 RVS162-MYC::CmLEU2 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 | This study |
| CJB138b | RVS162-MYC/RVS167-2 ΔΔ | arg4Δ/arg4Δ iro1Δ::imm434/iro1Δ::imm434::IRO1 Rvs167Δ::CaURA3 Rvs1672Δ::CdHIS1 RVS162-MYC::CmLEU2 | This study |
| CJB180b | RVS162-HA/RVS167-3-HA | his1Δ/his1Δ, ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434::IRO1 RVS167-3-HA::CdARG4 RVS162-HA::CmLEU2 | This study |
| CJB209b | RVS167-2-HA/rvs167-3ΔΔ | ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 RVS167-2-HA::CmLEU2 | This study |
| CJB212b | RVS167-2-HA/rvs167-3ΔΔ | leu2Δ/leu2Δ iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-3Δ::CdARG4/rvs167-3Δ::CdHIS1 RVS167-2-HA::URA3 | This study |
| CJB221b | RVS162-MYC/rvs167-3ΔΔ/rvs167-2ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-2Δ::CaURA3/rvs167-2Δ::CdHIS1 RVS162-MYC::CmLEU2 rvs167-3Δ::CdARG4/rvs167-3Δ::SAT1 | This study |
| CJB222b | RVS162-MYC/rvs167-3ΔΔ/rvs167-2ΔΔ | iro1Δ::imm434/iro1Δ::imm434::IRO1 rvs167-2Δ::CaURA3/rvs167-2Δ::CdHIS1 RVS162-MYC::CmLEU2 rvs167-3Δ::CdARG4/rvs167-3Δ::SAT1 | This study |
Derived from strain SN76.
Derived from strain SN148.
Derived from strain BWP17.
C. albicans deletion strains were constructed by sequential deletion of both copies of the targeted gene using a PCR-based procedure as described previously (35). Proper integration of deletion cassettes was confirmed by PCR using a combination of primers flanking the site of cassette integration and internal primers. The rvs deletion strains, used in the phenotypic screen, were transformed with linearized plasmids containing the appropriate marker genes to make them prototrophic.
C. albicans RVS alleles were C-terminally tagged by the insertion of the 3HA (hemagglutinin) epitope, the 6MYC (c-Myc) epitope, the gamma green fluorescent protein (GFPγ) gene (36), or the yEmRFP gene (37) using a PCR-based procedure and homologous recombination (38). Construction of the pFA-3HA-URA3 C-terminal tagging plasmid was described before (38). To create pFA-3HA-HIS1 and pFA-3HA-ARG4, the HIS1 gene and ARG4 gene were isolated from pFA-GFP-HIS1 and pFA-GFP-ARG4 by the use of AscI and PmeI (39), respectively, and then inserted into pFA-3HA-URA3 cut with AscI and PmeI.
Construction of both pFA-3HA-C. maltosa LEU2 (CmLEU2) and pFA-6MYC-CmLEU2 involved the AscI-PmeI isolation of the CmLEU2 from plasmid pFA-GFP-CmLEU2 (number 697; see reference 40), which was subsequently ligated into AscI-PmeI-digested pFA-3HA-C. dubliniensis HIS1 (CdHIS1). The 6MYC tag was obtained by PCR amplification using pFA6a-13MYC-KanMX6 (41) with primers containing 5′ PstI and 3′ AscI sites and used to exchange the 6MYC tag for 3HA in pFA-3HA-CmLEU2. Correct transformants were identified by both PCR and immunoblotting using antibodies raised against the tags.
BAR domain fragments were amplified from C. albicans or S. cerevisiae genomic DNA using a combination of forward and reverse primers as given in Table S1 in the supplemental material, thereby introducing restriction sites at the 5′ and 3′ ends of the gene fragment. All PCR fragments were cloned into the pGEM-T Easy vector using the A-tailing procedure (Promega). The presence of the correct insertion was confirmed by restriction endonuclease digestion analysis and sequencing. Next, pGEM-T Easy-BAR plasmids were digested using NcoI and NotI or PciI and NotI (Sc167BAR) and the resulting fragments were ligated into NcoI-NotI-digested pPC97mMYC, pPC86mHA, or pET-21d(+), respectively. pPC97mMYC and pPC86mHA are derivatives of the yeast two-hybrid (Y2H) pPC97 and pPC86 vectors (42), respectively, harboring an epitope tag (myc or HA) upstream of the multiple cloning site.
For coexpression in E. coli, the BAR fragments were cloned in pETM41m, a derivative of pETM30 and pETM41 (both generously provided by G. Stier, EMBL Heidelberg). pETM41m was constructed as follows: an annealed double-stranded (ds) oligonucleotide was inserted between the XhoI and NotI sites of pETM30, introducing four additional restriction sites (AatII, AvrII, NheI, and PmeI). Next, the 1,242-bp NcoI-XbaI fragment of pETM41 encompassing 6His-MBP (maltose binding protein) to TEV (Tobacco etch virus cleavage site) was inserted into the modified pETM30 between the XbaI and NcoI sites, thereby replacing glutathione S-transferase (GST) with MBP. To construct polycistronic vector 12, the NcoI-NotI fragment harboring CaRVS167-BAR(−H0) was cloned into NcoI-NotI-digested pETM41m. The resulting plasmid was cut using DraIII (in the f1 origin of replication) and NheI and the 6,800-bp fragment isolated and ligated to the 1,198-bp XbaI-DraIII fragment encompassing CaRVS161(−H0) preceded by a ribosome binding site (rbs) and half of the f1 origin of replication, both derived from the pET21d(+) plasmid. The resulting polycistronic vector coexpresses 6His-MBP-CaRVS167-BAR(−H0) and CaRVS161-(−H0) from a single T7 promoter. Polycistronic vector 20 was constructed similarly. First, CaRVS167-3-BAR(−H0) was cloned by the use of NcoI-NotI into pETM41m. The resulting plasmid was digested with NheI and XhoI and the vector isolated and ligated to a 1,058-bp fragment encompassing CaRVS162_FL (full length) preceded by the rbs derived from pET-21d(+). The resulting plasmid coexpresses 6His-MBP-CaRVS167-3BAR(−H0) and CaRVS162-FL.
The yEmRFP was amplified from its original vector using primers AG169 and AG170, and the resulting PCR product was digested by the use of PstI and AscI and ligated to a PstI-AscI-digested pFA-3HA-CdARG4 vector, replacing 3HA with yEmRFP.
Transformation.
C. albicans and S. cerevisiae transformations were done as described previously using lithium acetate (43, 44).
Yeast two-hybrid interactions.
The constructed Y2H plasmids (see Table S2 in the supplemental material) were transformed to the S. cerevisiae Y2H Gold strain, and the strength of interaction was assessed by spotting serial dilutions of logarithmically grown cells onto minimal plates without histidine (His−), without histidine and containing 20 mM 3-amino-1,2,4-triazole (3AT) (Sigma-Aldrich), or without adenine (Ade−). Growth on plates without adenine indicated a stronger interaction, as the ADE2 reporter gene has a weaker GAL4 promoter sequence than the HIS3 reporter gene whereas 3AT increases the stringency of the histidine selection.
Purification of Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimers.
Polycistronic vector plasmids 12 and 20 were transformed to E. coli BL21-CodonPlus(DE3)-RIPL (Stratagene) cells, grown at 37°C until an optical density at 600 nm (OD600) of 0.7 was reached, and induced overnight at 16°C with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTGl Invitrogen Life Technologies). Pelleted cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) with protease inhibitor cocktail (Roche) by sonication (Sanyo Soniprep). The CaRvs161/6His-MBP-CaRvs167 and CaRvs162/6His-MBP-CaRvs167-3 heterodimers were purified over an amylose column (New England BioLabs) followed by removal of the 6His-MBP tag from the heterodimers by TEV protease treatment. After the TEV treatment step, an ultracentrifugation step (30 min at 100,000 × g) was included to remove insoluble protein. The soluble fraction was poured over a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen) to remove the 6His-MBP tag, and the flowthrough fraction containing the free heterodimers was buffer exchanged (PD10 desalting column; GE Healthcare) into 20 mM HEPES (pH 7.4)–100 mM NaCl.
Coimmunoprecipitation.
YPD-grown cells (20 OD600 units) of singly (CJB141, CJB128, CJB126, and CJB132) and doubly (CJB127, and CJB133) tagged C. albicans strains were lysed by vortex mixing with glass beads for 20 min at 4°C in lysis buffer (1× phosphate-buffered saline [PBS], 0.5% NP-40, 1 mM PMSF, 0.5 mM dithiothreitol [DTT], yeast protease inhibitor cocktail [Roche]). After centrifugation (13,000 rpm for 10 min at 4°C), 1/10 of the sample was set aside (total lysate), and the remainder of the sample was precleared with 25 μl (10% [wt/vol]) protein G-Sepharose beads (Amersham Biosciences) and then incubated with α-HA (or α-MYC) antibody linked to protein G-Sepharose beads for 2 h at 4°C. Precipitates were washed two times with lysis buffer and two times with PBS, and elution was carried out by heating the beads in 1× Laemmli sample buffer (0.2 M Tris-HCl [pH 6.8], 1.5% sodium dodecyl sulfate [SDS], 10% glycerol, 1 mM EDTA, 0.004% bromophenol blue) prior to SDS-polyacrylamide gel electrophoresis (PAGE). Total lysates and immunoprecipitates were analyzed by Western blotting using the appropriate antibodies.
Immunoblotting.
Protein extracts were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Whatman Protran; GE Healthcare) using a semidry transfer system (Bio-Rad). The membranes were incubated in blocking solution consisting of 2% powdered milk–Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 8.0], 150 mM NaCl) and then overnight at 4°C with primary antibody solution, followed by incubation with the appropriate anti-horseradish peroxidase-conjugated secondary antibody, when required. Chemiluminescence detection of the cross-reacting bands was performed using a SuperSignal West Dura kit (Pierce Biotechnology, Inc.) and an ImageQuant LAS 4000 system (GE Healthcare Life Sciences). The primary antibodies used included α-HA rat monoclonal peroxidase-conjugated antibody (3F10; Roche), α-MYC mouse monoclonal antibody (9B11; Cell Signaling Technology), and α-HXK (hexokinase).
Lipid cosedimentation assay.
Liposomes were prepared as previously described (6). Prior to use, liposomes were sequentially passed through a 0.2-μm-pore-size Nuclepore polycarbonate filter (Whatman) by syringe extrusion. Two different lipid cocktails were used for liposome binding experiments. Liposome cocktail 1 consisted of 40% (vol/vol) Sigma brain extract from bovine brain, type I, containing Folch fraction I (Sigma; B1502), 40% (vol/vol) Avanti total brain extract (Avanti 131101P), 15% brain PS (l-α-phosphatidylserine; Avanti), and 5% brain PI(4,5)P2 (Avanti l-α-phosphatidylinositol-4,5-bisphosphate). Cocktail 2 contained 42.5% Folch fraction I (Sigma), 42.5% Avanti total brain extract (Avanti 131101P), and 15% brain PS (Avanti l-α-phosphatidylserine). Heterodimers Rvs161 and Rvs167 (1.7 μM) and heterodimers Rvs162 and Rvs167-3 (5.3 μM) were incubated with liposomes in a 1:3 ratio for 30 min at room temperature and then centrifuged in a benchtop ultracentrifuge (Beckman Coulter Optima MAX-XP Ultracentrifuge) for 20 min at 100,000 × g. The pellet and the supernatant fractions were taken up in equal volumes of sample buffer and loaded on a 4% to 12% Tris-SDS polyacrylamide gel (Invitrogen). After separation, protein bands were visualized with Coomassie brilliant blue (Thermo Scientific PageBlue protein staining solution).
Fluorescence microscopy.
The endocytic capacity of strains was determined as described previously (32). Briefly, logarithmically growing cells were incubated for 15 min on ice with 20 μM SynapseRed C2 (equivalent to FM4-64; PromoCell, Germany), washed, diluted in fresh YPD medium, and incubated for different time periods. Dye internalization was evaluated by fluorescence microscopy. For live-cell imaging, GFP-tagged and red fluorescent protein (RFP)-tagged strains were grown for 16 h at 28°C with shaking and were embedded in low-melting-point agarose before imaging. All images were collected in the Core Facility Cellular Imaging/LCAM-AMC using a DM RA HC microscope (Leica Microsystems, Wetzlar, Germany) equipped with a cooled charge-coupled-device (CCD) camera (Apogee Instruments, CA, USA) and a 100× 1.4-numerical-aperture (NA) Plan-Apochromat objective.
Phenotypic screen.
Wild-type (SN148-P) and rvs knockout strains were grown in either rich (YPD) or minimal (YNB–0.3% glucose) medium to an OD600 of ∼1 to ∼2, washed twice, resuspended to an OD600 of 0.2, and serially diluted (10-fold) in water. A 4-μl volume of each dilution was spotted on agar plates supplemented with various (stress-inducing) compounds (see Table S3 in the supplemental material). Plates were incubated for 2 to 5 days at the indicated temperatures before pictures were taken and growth was scored.
RESULTS
The C. albicans Rvs protein family consists of five members.
Four Rvs proteins have been reported so far in C. albicans, namely, the canonical Rvs161 and Rvs167 proteins and their paralogs, Rvs162 and Rvs167-2, respectively (32, 33). BLAST analyses performed with the C. albicans genome revealed a putative new paralog of Rvs167, encoded by the uncharacterized orf19.1861 gene, which we named Rvs167-3. Rvs167-3 has the same domain architecture as Rvs167 and Rvs167-2, containing an N-terminal BAR domain and a C-terminal SH3 domain (Fig. 1). Secondary-structure prediction identified three long α-helices (residues 32 to 330) and an N-terminal sequence of ∼22 residues that appears to fold into an amphipathic α-helix (denoted H0), thus defining CaRvs167-3 as an N-BAR protein (see Fig. S1 in the supplemental material). Multiple sequence alignments performed with Rvs167 proteins from different yeast species revealed that CaRvs167-3 has sequence identity of 13.9% with C. albicans Rvs167, 20.2% with S. cerevisiae Rvs167, and 13.6% with Schizosaccharomyces pombe Hob1. Orthologs of CaRvs167-3 can be found in species closely related to C. albicans (see Fig. S2).
FIG 1.
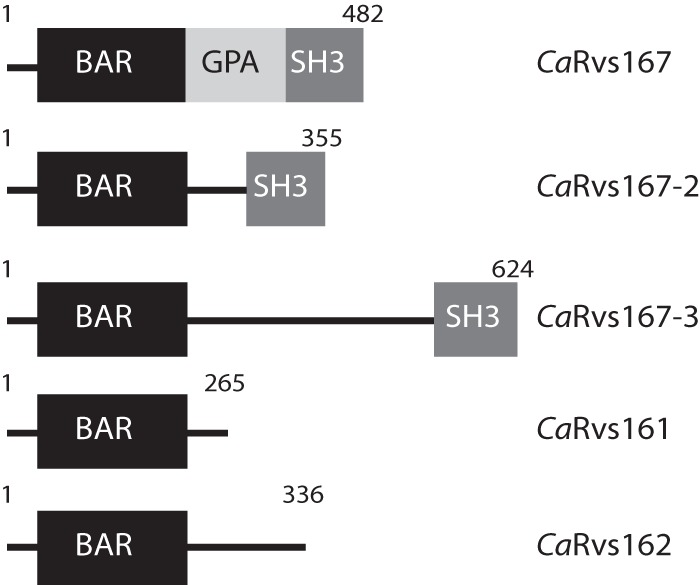
Domain architecture of the Rvs protein family members in Candida albicans. The Rvs family consists of five members, CaRvs167, CaRvs167-2, CaRvs161, CaRvs162, and a novel member, CaRvs167-3. The diagram shows the N-terminal BAR domain in each family member, the SH3 (Src-homology 3) domain in the three Rvs167 members, and the GPA (glycine-, proline-, and alanine-rich domain) in Rvs167.
Expression of Rvs family members.
To determine whether all five Rvs proteins are expressed in C. albicans yeast cells, one allele of each RVS gene was C-terminally epitope tagged either with 3-HA or with 6-MYC. Western blot analysis showed that, with the exception of Rvs167-2, the Rvs proteins were expressed in mid-exponential-growth-stage yeast cells in YPD medium (Fig. 2). As we have been unable to detect the expression of tagged Rvs167-2 under various culture conditions, including hyphal growth and H2O2-induced oxidative stress conditions (see Fig. S3 in the supplemental material), the in vivo function of this protein was not further addressed in this study. The migration of the other four Rvs proteins in SDS-PAGE gels corresponded to their calculated molecular masses (for RVS167-HA, 55.7 kDa; for RVS167-3, 77 kDa; for RVS161-HA, 34.5 kDa; and for RVS162-MYC, 49.5 kDa). We observed that Rvs167 was highly expressed compared to the other Rvs proteins; therefore, five times less total protein was loaded on the gel for analysis. In line with previous observations (32), slower-migrating Rvs167 bands were detected, suggesting that Rvs167 undergoes posttranslational modifications such as phosphorylation or possibly ubiquitination. The nature of these slower-migrating bands was not further analyzed in this study.
FIG 2.
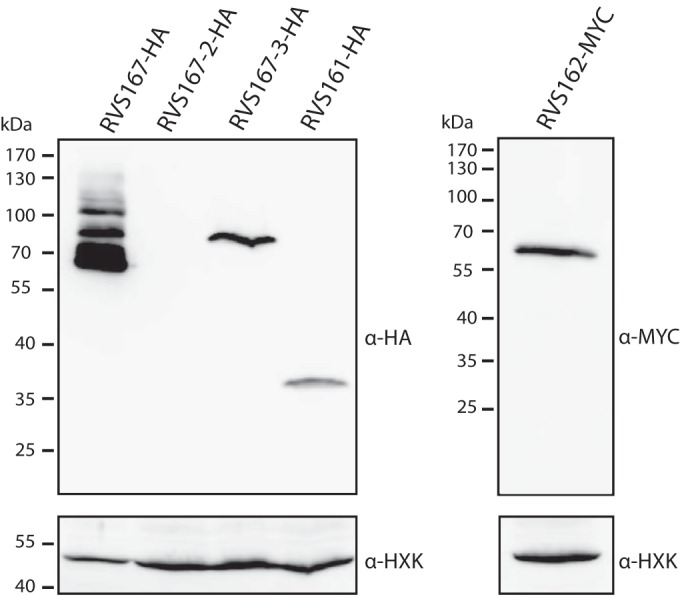
Expression of Rvs proteins in C. albicans. The results of immunoblot analysis of C. albicans strains in which the different Rvs proteins were C-terminally tagged with either 3HA (triple hemagglutinin epitope) or 6MYC (sextuple myc epitope) are shown. Cells were grown to mid-exponential phase in YPD medium, and equal amounts of total lysates were separated on a 10% acrylamide gel, except for RVS167-HA, for which five times less total protein was loaded because of the high expression level. Blots were probed with antibodies directed against HA (left) or MYC (right) and against hexokinase (HXK) as a loading control.
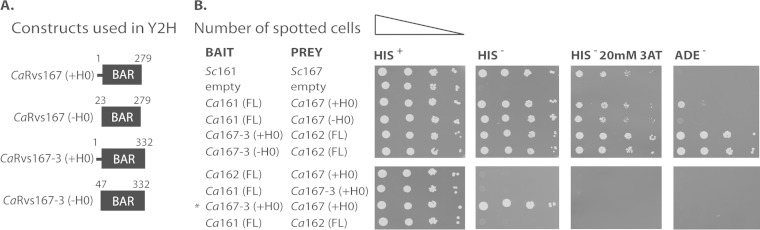
Yeast two-hybrid analysis reveals specific BAR-BAR interactions among the five Rvs family members.
Next, we wished to determine which Rvs proteins in C. albicans form dimers. We focused on the BAR domains, as previous work showed that Rvs proteins interact through their BAR domains to form a functional BAR dimer, also described as the BAR module (4, 14). While the conditions under which Rvs167-2 is expressed are currently unknown, we have included this protein in our Y2H screen to investigate BAR domain-mediated interactions among the five Rvs family members. This analysis revealed the existence of three putative BAR heterodimers: Rvs161/Rvs167, Rvs162/Rvs167-2, and Rvs162/Rvs167-3 (Table 2). In addition, Rvs167 and Rvs167-2 showed weak self-association. The specificity and strength of the observed interactions were further assessed in a semiquantitative Y2H spot assay (Fig. 3; see also Fig. S4 in the supplemental material). Consistent with the qualitative Y2H data (Table 2), strong interactions were observed between full-length Rvs161 and Rvs167-BAR (with or without helix-0), between full-length Rvs162 and RVS167-2-BAR (without helix-0), and between full-length Rvs162 and RVS167-3-BAR (with or without helix-0), as cells grew on His− plates in the presence of 20 mM 3AT and on Ade− plates (Fig. 3B, top panels; see also Fig. S4). No interaction was observed between Rvs161 and Rvs167-3-BAR, Rvs162, and Rvs167-BAR, Rvs161 and Rvs162, or Rvs167-BAR and Rvs167-3-BAR (Fig. 3B, bottom panels). Three constructs, Rvs167-2, RVS167-3, and, to a lesser extent, RVS167, grew on His− plates when tested against an empty plasmid, suggesting weak self-activation of the constructs (see Fig. S4). Together, these results suggest a specific interaction between the BAR domains of Rvs167 and Rvs161, between Rvs167-2 and Rvs162, and between Rvs167-3 and Rvs162.
TABLE 2.
Pairwise testing of BAR interactions among Rvs family members
| Bait | Interaction with preya |
||||
|---|---|---|---|---|---|
| CaRVS161 | CaRVS167 | CaRVS167-3 | CaRVS162 | CaRVS167-2 | |
| CaRVS161 | − | +++ | − | − | − |
| CaRVS167 | +++ | + | − | − | + |
| CaRVS167-3 | − | − | + | +++ | − |
| CaRVS162 | − | − | +++ | − | +++ |
| CaRVS167-2 | − | + | − | +++ | + |
Interactions were assessed by Y2H analysis using a LacZ (β-galactosidase) reporter. −, no interaction (white); +, weak interaction (light blue staining after >8 h of incubation); +++, strong interaction (dark blue staining after ∼30 min of incubation).
FIG 3.
Specific BAR-BAR interactions as revealed by yeast two-hybrid assays. (A) Schematic representation of the RVS167 and RVS167-3 constructs used in the two-hybrid experiment. Constructs harbored the BAR domain either with or without helix 0 (+H0 or −H0). (B) Serial dilutions of strains cotransformed with the indicated bait and prey constructs were spotted on minimal plates with histidine (His+), without histidine (His−), without histidine and containing 20 mM 3-amino-1,2,4-triazole (His− 20 mM 3AT), and without adenine (Ade−). Weak interactors activated only the HIS3 reporter and showed growth on His− plates, while strong interactors activated both HIS3 and ADE2 reporters and showed growth on His−/20 mM 3AT and Ade− plates. *, Rvs167-3 showed weak self-activation and growth on His− plates in the presence of an empty prey plasmid (see also Fig. S4 in the supplemental material).
Rvs161/Rvs167 and Rvs162/Rvs167-3 form stable heterodimers in vitro.
To determine if the BAR-BAR interactions between Rvs167 and Rvs161 and between Rvs167-3 and Rvs162 are direct, in vitro binding assays were performed. To this end, polycistronic vectors expressing one of the two interacting partners as a chimeric protein with the 6His-MBP epitope followed by a TEV recognition site fused to its N terminus and the other partner as an untagged protein were constructed. Figure 4A shows that His6-MBP Rvs167-BAR without H0 (−H0) and the untagged Rvs161 (−H0) copurified in a 1:1 ratio after affinity purification on amylose beads, suggesting that the interaction between Rvs167 and Rvs161 is direct. To exclude the possibility that the 6His-MBP tag was responsible for the copurification of the untagged protein, 6His-MBP was cleaved off by incubation with TEV protease and removed using a Ni-NTA column. As can be seen in Fig. 4A, the Ni-NTA flowthrough fractions (lanes 6 and 7) contained the (untagged) Rvs167-BAR and Rvs161 proteins in a 1:1 ratio, providing strong evidence that Rvs167-BAR and Rvs161 form stable heterodimers. Using the same procedure, Rvs162(+H0)/Rvs167-3-BAR(−H0) heterodimers could be purified from E. coli cells expressing His6-MBP-Rvs167-3-BAR(−H0) and untagged Rvs162(+H0) (Fig. 4B). Of note is that we consistently found that tagged or untagged Rvs BAR constructs were mostly insoluble when expressed individually in E. coli (our unpublished data). These biochemical data are consistent with previous reports showing that BAR domains do not fold properly in the absence of their partner (45, 46) and emphasize that the functional Rvs BAR module is a (hetero)dimer.
FIG 4.
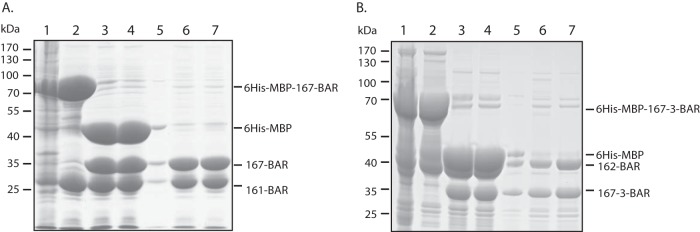
Purification of Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimers from E. coli. E. coli cells expressing 6His-MBP-RVS167BAR and untagged RVS161BAR (A) or 6His-MBP-RVS167-3BAR and untagged RVS162 (B) were lysed, and the soluble fraction (lane 1) was purified over an amylose column. The eluate (lane 2) was subjected to TEV protease treatment (lane 3) followed by an ultracentrifugation step to separate the soluble fraction (lane 4) from the insoluble fraction (lane 5). The (cleaved) 6His-MBP tag and the 6His-TEV tag were removed from the soluble fraction using a Ni-NTA column. RVS167BAR and RVS161 proteins (A) and RVS167-3BAR and RVS162 proteins (B) were recovered in a 1:1 ratio in the Ni-NTA flowthrough fractions (lanes 6 and 7), indicating copurification due to direct interaction between the BAR domains.
In vivo evidence for Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimer formation.
To test whether the Rvs proteins interact in vivo, we performed coimmunoprecipitation experiments on C. albicans strains expressing C-terminally tagged (HA or MYC) Rvs proteins from their endogenous promoter. We constructed strains in which both proteins of the presumed heterodimer were individually tagged and control strains in which only one of the partner proteins was epitope tagged. Western blot analysis of the immunoprecipitates showed coimmunoprecipitation in both directions for Rvs161-HA with Rvs167-MYC (Fig. 5A) and Rvs167-3-HA with 162-MYC (Fig. 5B), suggesting that these proteins interact in vivo.
FIG 5.
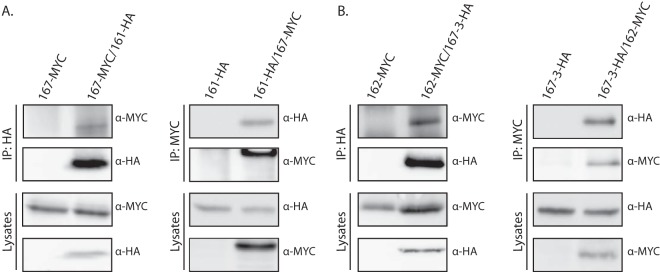
In vivo evidence for Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimer formation. Results of coimmunoprecipitation (IP) of RVS167-MYC and RVS161-HA (A) and RVS167-3 HA and RVS162-Myc (B). Strains expressing the indicated HA- or Myc-tagged Rvs proteins were lysed and subjected to immunoprecipitation with anti-HA or anti-myc antibody. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with anti-HA and anti-Myc antibodies.
Previous work in S. cerevisiae has shown reduced stability of Rvs161p in the absence of its partner Rvs167 and vice versa (17), indicating that the interaction between Rvs161 and Rvs167 is required for stability in vivo. To provide further evidence for the existence of the two Rvs heterodimers in C. albicans, we assessed protein levels of the four Rvs proteins in the presence and absence of their partners. Wild-type strains expressing a tagged Rvs protein were compared with strains in which the partner had been deleted. In concordance with previous results in S. cerevisiae, deletion of RVS161 resulted in a strong reduction of Rvs167 protein levels and vice versa (Fig. 6A and B). Also, the levels of Rvs167-3 were markedly reduced in the rvs162 deletion mutant (Fig. 6C). Remarkably, the levels seen with Rvs162 did not change significantly in the absence of its partner Rvs167-3. While our in vitro binding experiments clearly show that Rvs162 and Rvs167-3 heterodimerize, these stability experiments suggest that Rvs162 may interact with another partner in the absence of Rvs167-3 in vivo, resulting in its stabilization. Deletion of the RVS167-2 gene either in a wild-type strain or in the rvs167-3Δ/Δ mutant did not affect Rvs162 levels (see Fig. S5 in the supplemental material), indicating that Rvs167-2 is not required for Rvs162 stability in vivo. Whether another protein plays a role in Rvs162 stabilization remains to be investigated. Together, these data strongly suggest that two Rvs heterodimers exist in C. albicans: Rvs161/Rvs167 and Rvs162/Rvs167-3.
FIG 6.
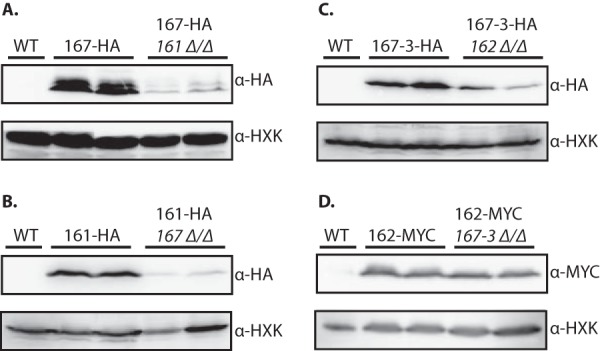
Reduced Rvs protein levels in the absence of its binding partner. Results of immunoblot analysis of protein extracts of wild-type (WT) and rvs deletion strains expressing 3HA- or 6MYC-tagged Rvs proteins are shown. The indicated strains were grown to mid-exponential phase in YPD medium and lysed, and protein extracts were analyzed by SDS-PAGE and immunoblotting. Blots were probed with antibodies directed against HA (A, B, and C) or MYC (D) and against hexokinase (HXK) as a loading control (A to D).
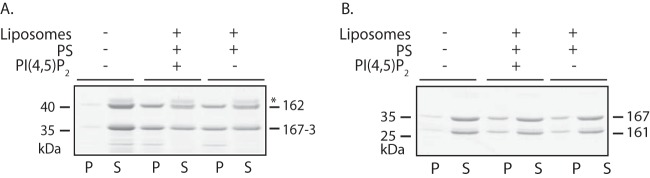
Rvs161/Rvs167 and Rvs162/Rvs167-3 bind to lipid membranes in vitro.
One of the hallmarks of the BAR domain family of proteins is their ability to bind membranes (1, 3, 4). To address the membrane binding capacity of both BAR heterodimers in C. albicans, we performed lipid cosedimentation experiments. The BAR heterodimers were purified as described for Fig. 5. Both the Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimers showed binding to liposomes (Fig. 7A and 7B, respectively; see also Fig. S6 in the supplemental material), although binding of the latter seemed more efficient, as up to 50% of the Rvs162/Rvs167-3 heterodimer was recovered in the pellet fraction. Of note is that the individual subunits of the Rvs heterodimers were consistently recovered in a 1:1 ratio in the pellet fraction, suggesting that they bind to lipids as heterodimers. The addition of PI(4,5)P2 (phosphatidylinositol-4,5-bisphosphate) to the liposomes did not affect the efficiency of binding for either Rvs161/Rvs167 or Rvs162/Rvs167-3.
FIG 7.
Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimers bind membranes in vitro. Coomassie-stained SDS-PAA gels used in lipid cosedimentation experiments are shown. Purified Rvs162(+H0)/Rvs167-3(−H0) (A) and Rvs161(−H0)/Rvs167(−H0) (B) heterodimers were incubated at 5.3 μM and 1.7 μM, respectively, for 30 min at room temperature with Folch liposomes containing PS (15%) or PS and PI(4,5)P2 (5%) lipid species and then ultracentrifuged to separate the liposome-bound fraction (P) from the unbound fraction (S). An asterisk (*) indicates the 6His-MBP contaminant in the Rvs167-3/Rvs162 preparation. Similar results were obtained for the Rvs161/Rvs167 heterodimer at a higher protein concentration (see Fig. S6 in the supplemental material).
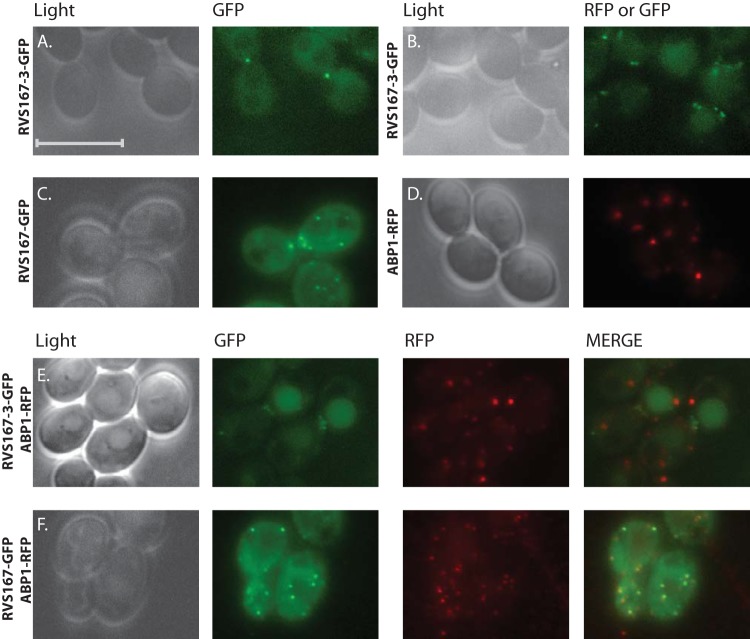
Rvs167-3 and Rvs167 localize to different cortical structures.
The ability of both BAR heterodimers to bind liposomes implies a function for these proteins at a membrane (either the plasma membrane or an intracellular membrane). To assess their localization in living cells, we C-terminally tagged Rvs167 and Rvs167-3 with GFPγ and C-terminally tagged Abp1, a clathrin-mediated endocytosis marker protein, with yEmRFP. In addition, we constructed double-tagged strains expressing both a GFP-tagged Rvs protein and Abp1-RFP. All genes were expressed from their endogenous promoter, and control experiments suggested that the GFP tag does not interfere with Rvs167 or Rvs167-3 function (see Fig. S7 in the supplemental material). Live-cell imaging revealed a dot-like fluorescent signal in the vicinity of the plasma membrane for RVS167-3-GFP, RVS167-GFP, and the endocytic marker Abp1-RFP (Fig. 8A to D). In addition, Rvs167 showed overall diffuse staining well above background levels, suggesting that there may have been a cytosolic pool of Rvs167. The observed punctate pattern at the cell surface for all three proteins is very reminiscent of the previously reported localization of ScRvs167 and ScAbp1 to cortical actin patches, which are sites of endocytosis (47, 48). To determine if CaRvs167 and CaRvs167-3 localize to sites of endocytosis, we performed colocalization experiments with Abp1 (Fig. 8E and F). While Rvs167 strictly colocalized with Abp1, we could not observe any colocalization of Rvs167-3 with sites of endocytosis marked with Abp1. Together, these results show that Rvs167 and Rvs167-3 localize to different cortical membrane structures and suggest that Rvs167-3 may not be involved in clathrin-mediated endocytosis in C. albicans.
FIG 8.
Rvs167-3 and Rvs167 localize to different cortical structures. Results of live-cell imaging of YPD-grown strains expressing Rvs167-3-GFPγ (A and B), Rvs167-GFPγ (C), or the endocytosis marker Abp1-yEmRFP (D) and of strains expressing both Rvs167-3-GFPγ and Abp1-yEmRFP (E) or both Rvs167-GFPγ and Abp1-yEmRFP (F) are shown. Note the almost complete colocalization of Rvs167-GFPγ with the endocytic marker Abp1-yEmRFP and the absence of colocalization of Rvs167-3 with sites of endocytosis marked by Abp1. Scale bar, 10 μm.
The rvs167-3Δ/Δ mutant is not deficient in endocytosis.
Douglas and coworkers (32) showed that CaRvs161 and CaRvs167, but not CaRvs162, are required for efficient endocytosis. To directly investigate the involvement of Rvs167-3 in endocytosis, we monitored the ability of the rvs167-3Δ/Δ mutant to internalize the lipophilic dye FM4-64 in a time course experiment (Fig. 9). The rates of FM4-64 internalization of the wild-type (control) strain and the rvs167-3Δ/Δ mutant were indistinguishable, as both strains showed staining of the vacuole and a few endocytic intermediates after 15 min of incubation. By 30 min, all FM4-64 localized in the vacuolar membrane in both strains, indicating that FM4-64 was efficiently endocytosed. Consistent with previous results (32), the rvs161 Δ/Δ mutant showed a severe endocytosis defect, as the FM4-64 staining was predominantly confined to the plasma membrane after 30 min of incubation, with no vacuolar staining.
FIG 9.
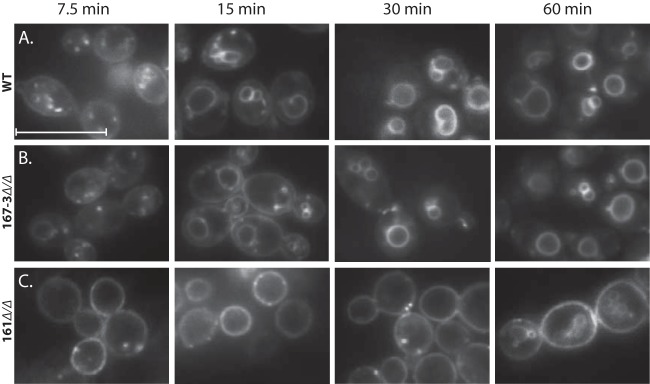
The Rvs167-3Δ/Δ mutant is not deficient in endocytosis. The indicated strains were incubated with FM4-64 (20 μM) on ice, washed, diluted in fresh medium, and incubated at 28°C for 7.5, 15, 30, and 60 min. Internalization of the stain was monitored by fluorescence microscopy. Note that the wild-type strain and the rvs167-3Δ/Δ mutant were able to efficiently internalize the stain as revealed by the predominant vacuolar staining and the absence of plasma membrane staining after 15 min of incubation, while the rvs161Δ/Δ mutant was severely affected in endocytosis, with little or no internalization of the stain after 30 min. Scale bar, 10 μm.
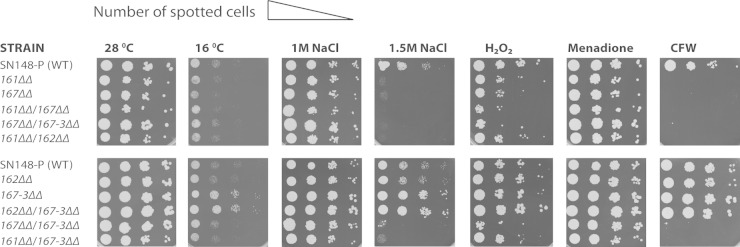
Phenotypic analysis reveals a genetic interaction between Rvs167-3 and the Rvs161/Rvs167 heterodimer.
To further investigate the in vivo function of the Rvs proteins in C. albicans, we constructed strains with single or double homozygous deletions of the four RVS genes and performed an extensive phenotypic screen (Fig. 10; see also Table S3 in the supplemental material). As reported previously (32), rvs161Δ/Δ and rvs167Δ/Δ cells displayed increased sensitivity to CFW (calcofluor white) and SDS, suggesting that the mutants have a defect in cell wall biosynthesis. Rvs161Δ/Δ and rvs167Δ/Δ cells also showed increased sensitivity to LiCl and oxidative stress (H2O2 and menadione). In contrast, the rvs167-3Δ/Δ and rvs162Δ/Δ strains did not show a phenotype in any of these conditions. These data corroborate previous findings for strain rvs162Δ/Δ and indicate that the Rvs162/Rvs167-3 heterodimer does not play a role in cell wall integrity pathways. Interestingly, we found marked growth differences between the deletion strains in the presence of high salt concentrations (1 M and 1.5 M) and, to a lesser extent, at low temperature (16°C). In contrast to what has been reported, the rvs161Δ/Δ and rvs167Δ/Δ strains but not the rvs162Δ/Δ strain were very sensitive to high salt. The rvs167-3Δ/Δ strain showed an increased tolerance of high salt concentrations and growth at low temperature (16°C) compared to the wild-type strain. Deletion of either RVS161 or RVS167 suppressed the high-salt-concentration- and low-temperature-tolerance phenotype of the rvs167-3Δ/Δ strain, implying a genetic interaction between Rvs167-3 and the Rvs161/Rvs167 heterodimer.
FIG 10.
Phenotypic analysis reveals a genetic interaction between Rvs167-3 and the Rvs161/Rvs167 heterodimer. Logarithmically grown wild-type cells and rvs single- or double-mutant cells were serially diluted and spotted on YPD agar plates containing the following addition: NaCl (1 M or 1.5 M), H2O2 (6 mM), menadione (80 μM), or CFW (20 μM). Plates were incubated at 28°C for 2 days or at 16°C for 4 days.
DISCUSSION
The N-BAR-containing Rvs proteins and their membrane binding and bending properties play an essential role in endocytosis in yeast (23). Here we focus on the N-BAR Rvs family in the human fungal pathogen C. albicans and identify a novel family member, Rvs167-3 (orf19.1861). The domain organization of Rvs167-3 is similar to that of the previously identified CaRvs167 and CaRvs167-2 proteins, harboring an N-terminal N-BAR domain and a C-terminal SH3 domain (Fig. 1). While CaRvs167-3 shows a relatively low level of sequence similarity with CaRvs167, ScRvs167, and Schizosaccharomyces pombe Rvs167 (SpRvs167) (Hob1) and contains several insertions, a prediction of its secondary structure (http://bioinf.cs.ucl.ac.uk/psipred//?program=psipred) revealed striking structural conservation, with three predicted α-helices and a short N-terminal amphipathic α-helix, thus identifying Rvs167-3 as an N-BAR protein (see Fig. S2 in the supplemental material). The relatively low sequence similarity with Rvs167 proteins of other yeast species may explain why CaRvs167-3 was not previously recognized as an N-BAR protein. A BLAST search of the sequenced fungal genomes at the Broad Institute (http://www.broadinstitute.org/annotation/genome/FGI_Blast/) revealed that four other closely related fungal species (C. tropicalis, C. lusitaniae, C. dubliniensis, and Debaryomyces hansenii) contain both the canonical Rvs167 and an ortholog of the CaRvs167-3 protein (Fig. S2 and data not shown), suggesting that they, like C. albicans, express two distinct BAR heterodimers (see below).
We provide several lines of evidence to support the model that C. albicans expresses two functionally distinct BAR heterodimers: Rvs161/Rvs167 and Rvs162/Rvs167-3. First, we show a strong interaction between the BAR domains of Rvs161 and Rvs167 and of Rvs162 and Rvs167-3 both in yeast two-hybrid assays and in vitro (Fig. 3 and 4). Second, we show that Rvs161 coimmunoprecipitates with Rvs167 and Rvs162 with Rvs167-3 (Fig. 5). Third, in line with previous observations in S. cerevisiae (18, 46, 49), we show for the Rvs161/Rvs167 heterodimer that each RVS gene is required for the in vivo stability of its Rvs protein partner. While deletion of RVS162 resulted in a strong reduction of Rvs167-3 levels, we did not observe a reduction of Rvs162 protein levels when RVS167-3 was deleted (Fig. 5). Control experiments showed that Rvs162 protein levels were also not affected in a strain harboring a deletion of RVS167-2 or RVS167-2 and RVS167-3 (see Fig. S5 in the supplemental material), suggesting that while Rvs167-2 can interact with Rvs162 in yeast two-hybrid assays, this protein is not involved in the in vivo stabilization of Rvs162. Why Rvs162 is stable in the absence of its (known) partner proteins remains to be investigated.
Previous studies on mammalian amphiphysins and the amphiphysin homologs Rvs161 and Rvs167 in yeast have shown their capability to bind to liposomes in vitro (4, 18, 50). Here we report for the first time that C. albicans Rvs161/Rvs167 and Rvs162/Rvs167-3 heterodimers, purified from a heterologous system, bind to liposomes (Fig. 7). A significantly higher percentage of membrane binding (up to 50% binding) was found in the lipid cosedimentation assay for Rvs162/Rvs167-3 than for Rvs161/Rvs167 (up to 20% binding). As the N-terminal amphipathic helix (H0) in N-BAR proteins contributes to membrane binding activity (4, 51) a likely explanation for this observation is that the constructs used to purify the Rvs161/Rvs167 heterodimer both lack the N-terminal helix whereas in the Rvs162/Rvs167-3 heterodimer, one of the subunits, Rvs162, still contains the N-terminal helix. Due to solubility issues with constructs harboring the N-terminal amphipathic helix, we have been unable to further address its contribution to the membrane binding activity of the Rvs heterodimers. The two Rvs heterodimers bound equally well to standard Folch lipids and to Folch lipids containing 5% PI(4,5)P2, suggesting that, like amphiphysins (4), the Rvs heterodimers bind to membranes in a PI(4,5)P2-independent manner.
The function of the Rvs161/Rvs167 heterodimer in endocytosis and actin polarization is well documented both in yeast (18, 46) and, more recently, in C. albicans (32, 33). Consistent with this, we show that CaRvs167 localizes to sites of endocytosis marked by Abp1 (Fig. 8) and that deletion of RVS167 or its partner RVS161 results in increased sensitivity to high salt concentrations and cell-wall-disturbing compounds (Fig. 10). These data extend previous findings (32, 33) and underscore the role of Rvs161/Rvs167 in endocytosis. In contrast, Rvs167-3-GFP foci do not colocalize with the endocytic marker protein Abp1 (Fig. 8) and the rvs167-3Δ/Δ mutant did not exhibit an endocytosis deficiency (Fig. 9). Also, phenotypic analysis of the rvs167-3Δ/Δ strain did not provide any evidence for a role of Rvs167-3, or its partner Rvs162, in endocytosis, as neither rvs167-3Δ/Δ cells nor rvs162-Δ/Δ cells showed increased sensitivity to high salt concentrations or cell wall stressors such as CFW and Congo red. Intriguingly, the rvs167-3Δ/Δ strain showed increased tolerance of high salt concentrations and cell wall stressors compared to wild-type cells (Fig. 10), a phenotype that was completely reversed when either RVS161 or RVS167 was also deleted in the rvs167-3Δ/Δ strain. These data suggest a genetic interaction between Rvs167-3 and Rvs167/Rvs161. The nature of this genetic interaction remains to be investigated.
In conclusion, we show that C. albicans expresses two functionally distinct BAR heterodimers with membrane binding capabilities. While our data and those of others (32, 33) are consistent with a role of the Rvs161/Rvs167 heterodimer in clathrin-mediated endocytosis, we have no evidence to suggest that the Rvs162/Rvs167-3 heterodimer is involved in this process. Rvs162 and Rvs167-3 are expressed during vegetative growth, and the localization of Rvs167-3 suggests a role for this protein at the plasma membrane. Interestingly, orthologs of CaRvs167-3 are present in closely related species (Fig. S2 and data not shown), suggesting that Rvs167-3 function is conserved in the Candida clade. Two distinct features of the Rvs167-3 protein may help to unravel its function in the future. First, bioinformatic analysis (http://heliquest.ipmc.cnrs.fr/) revealed that the polar face of the amphipathic helix of Rvs167-3 (and of its orthologs in closely related yeast species) does not contain basic residues like Rvs167 but is enriched in polar residues (Ser/Thr/Asn/Gln) (see Fig. S8). It has been previously suggested that amphipathic helices play an important role in membrane curvature sensing (51, 52), and the different amino acid compositions and physicochemical properties of the Rvs167 and Rvs167-3 amphipathic helices may direct these proteins to membranes with distinct curvature. Second, in contrast to Rvs167, the SH3 domain of Rvs167-3 does not bind canonical Pro-rich ligands (A. Gkourtsa and B. Distel, unpublished data) but binds to noncanonical peptides lacking proline residues, suggesting that Rvs167-3 has different binding partners from Rvs167 in vivo. Identification of proteins interacting with the SH3 domain of Rvs167-3 may provide clues to the in vivo function of this protein in C. albicans and its close relatives.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Konopka for the pFA-GFPγCdHIS1 tagging plasmid and Candida strains, Suzanne M. Noble for Candida strains SN76 and SN148, Jürgen Wendland for the pFA plasmids, Neta Dean for the plasmid containing the yEmRFP, and Jan Stap for his expert help with fluorescence microscopy.
This work was partially funded by the EU-Marie Curie Research Training Network “Penelope” (MRTN-CT-2006-036076).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00282-14.
REFERENCES
- 1.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Farsad K, De Camilli P. 2003. Mechanisms of membrane deformation. Curr Opin Cell Biol 15:372–381. doi: 10.1016/S0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH, Boura E, Carlson L-A, Różycki B. 2010. Membrane budding. Cell 143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 5.Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Fütterer K. 2005. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J 24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henne WM, Kent HM, Ford MGJ, Hegde BG, Daumke O, Butler PJG, Mittal R, Langen R, Evans PR, McMahon HT. 2007. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure 15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Habermann B. 2004. The BAR-domain family of proteins: a case of bending and binding? EMBO Rep 5:250–255. doi: 10.1038/sj.embor.7400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mim C, Unger VM. 2012. Membrane curvature and its generation by BAR proteins. Trends Biochem Sci 37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda M, Mochizuki N. 2010. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin Cell Dev Biol 21:391–398. doi: 10.1016/j.semcdb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Pykäläinen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peränen J, Vihinen H, Jokitalo E, Salminen M, Ikonen E, Dominguez R, Lappalainen P. 2011. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol 18:902–907. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. 2012. Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campelo F, McMahon HT, Kozlov MM. 2008. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J 95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerberg J, Kozlov MM. 2006. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 14.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, McMahon HT. 2006. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gortat A, San-Roman MJ, Vannier C, Schmidt AA. 13 December 2011, posting date Single point mutation in Bin/Amphiphysin/RVS (BAR) sequence of endophilin impairs dimerization, membrane shaping, and SRC homology 3 domain-mediated partnership. J Biol Chem doi: 10.1074/jbc.M111.325837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT. 1997. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell 8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombardi R, Riezman H. 2001. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J Biol Chem 276:6016–6022. doi: 10.1074/jbc.M008735200. [DOI] [PubMed] [Google Scholar]
- 18.Youn J-Y, Friesen H, Kishimoto T, Henne WM, Kurat CF, Ye W, Ceccarelli DF, Sicheri F, Kohlwein SD, McMahon HT, Andrews BJ. 2010. Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell 21:3054–3069. doi: 10.1091/mbc.E10-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leprince C. 2003. Sorting nexin 4 and amphiphysin 2, a new partnership between endocytosis and intracellular trafficking. J Cell Sci 116:1937–1948. doi: 10.1242/jcs.00403. [DOI] [PubMed] [Google Scholar]
- 20.Dislich B, Than ME, Lichtenthaler SF. 2011. Specific amino acids in the BAR domain allow homodimerization and prevent heterodimerization of sorting nexin 33. Biochem J 433:75–83. doi: 10.1042/BJ20100709. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Martin AC, Drubin DG. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell 11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Engqvist-Goldstein AEY, Drubin DG. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol 19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 23.Kaksonen M, Toret CP, Drubin DG. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 24.Kaksonen M, Toret CP, Drubin DG. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Kukulski W, Schorb M, Kaksonen M, Briggs JAG. 2012. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 26.Mayer BJ. 2001. SH3 domains: complexity in moderation. J Cell Sci 114:1253–1263. [DOI] [PubMed] [Google Scholar]
- 27.Prigent M, Boy-Marcotte E, Chesneau L, Gibson K, Dupré-Crochet S, Tisserand H, Verbavatz J-M, Cuif M-H. 2011. The RabGAP proteins Gyp5p and Gyl1p recruit the BAR domain protein Rvs167p for polarized exocytosis. Traffic 12:1084–1097. doi: 10.1111/j.1600-0854.2011.01218.x. [DOI] [PubMed] [Google Scholar]
- 28.Brizzio V, Gammie AE, Rose MD. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J Cell Biol 141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivadon P, Crouzet M, Aigle M. 1997. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett 417:21–27. doi: 10.1016/S0014-5793(97)01248-9. [DOI] [PubMed] [Google Scholar]
- 30.Balguerie A, Bagnat M, Bonneu M, Aigle M, Breton AM. 2002. Rvs161p and sphingolipids are required for actin repolarization following salt stress. Eukaryot Cell 1:1021–1031. doi: 10.1128/EC.1.6.1021-1031.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germann M. 2005. Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast Amphiphysin-like orthologs, Rvs161p and Rvs167p. J Biol Chem 280:4270–4278. doi: 10.1074/jbc.M412454200. [DOI] [PubMed] [Google Scholar]
- 32.Douglas LM, Martin SW, Konopka JB. 2009. BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect Immun 77:4150–4160. doi: 10.1128/IAI.00683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reijnst P, Walther A, Wendland J. 2010. Functional analysis of Candida albicans genes encoding SH3-domain-containing proteins. FEMS Yeast Res 10:452–461. doi: 10.1111/j.1567-1364.2010.00624.x. [DOI] [PubMed] [Google Scholar]
- 34.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strijbis K, van Roermund CWT, Hardy GP, van den Burg J, Bloem K, de Haan J, van Vlies N, Wanders RJA, Vaz FM, Distel B. 2009. Identification and characterization of a complete carnitine biosynthesis pathway in Candida albicans. FASEB J 23:2349–2359. doi: 10.1096/fj.08-127985. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Konopka JB. 2010. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot Cell 9:224–226. doi: 10.1128/EC.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keppler-Ross S, Noffz C, Dean N. 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strijbis K, van den Burg J, Visser WF, van den Berg M, Distel B. 2012. Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS Yeast Res 12:61–68. doi: 10.1111/j.1567-1364.2011.00761.x. [DOI] [PubMed] [Google Scholar]
- 39.Gola S, Martin R, Walther A, Dünkler A, Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 40.Schaub Y, Dünkler A, Walther A, Wendland J. 2006. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol 46:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- 41.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. [DOI] [PubMed] [Google Scholar]
- 42.Chevray PM, Nathans D. 1992. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci U S A 89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walther A, Wendland J. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet 42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl RH, Gietz RD. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 45.Friesen H, Murphy K, Breitkreutz A, Tyers M, Andrews B. 2003. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol Biol Cell 14:3027–3040. doi: 10.1091/mbc.E02-09-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friesen H, Humphries C, Ho Y, Schub O, Colwill K, Andrews B. 2006. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol Biol Cell 17:1306–1321. doi: 10.1091/mbc.E05-06-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, Galletta BJ, Schmidt KO, Chang FS, Blumer KJ, Cooper JA. 2006. Actin-based motility during endocytosis in budding yeast. Mol Biol Cell 17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boettner DR, Chi RJ, Lemmon SK. 2012. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol 14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. 2006. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev 70:37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. 2013. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep 4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegård P, Gether U, Stamou D. 2009. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J 28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drin G, Casella J-F, Gautier R, Boehmer T, Schwartz TU, Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol 14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.