Abstract
We have used electron paramagnetic resonance (EPR) to examine the structural impact of oxidizing specific methionine (M) side chains in calmodulin (CaM). It has been shown that oxidation of either M109 or M124 in CaM diminishes CaM regulation of the muscle calcium release channel, the ryanodine receptor (RyR), and that mutation of M to Q (glutamine) in either case produces functional effects identical to those of oxidation. Here we have used site-directed spin labeling and double electron-electron resonance (DEER), a pulsed EPR technique that measures distances between spin labels, to characterize the structural changes resulting from these mutations. Spin labels were attached to a pair of introduced cysteine residues, one in the C-lobe (T117C) and one in the N-lobe (T34C) of CaM, and DEER was used to determine the distribution of interspin distances. Ca binding induced a large increase in the mean distance, in concert with previous x-ray crystallography and NMR data, showing a closed structure in the absence of Ca and an open structure in the presence of Ca. DEER revealed additional information about CaM’s structural heterogeneity in solution: In both the presence and absence of Ca, CaM populates both structural states, one with probes separated by ~4 nm (closed) and another at ~6 nm (open). Ca shifts the structural equilibrium constant toward the open state by a factor of 13. DEER reveals the distribution of interprobe distances, showing that each of these states is itself partially disordered, with the width of each population ranging from 1.5 to 3 nm. Both mutations (M109Q and M124Q) decrease the effect of Ca on the structure of CaM, primarily by decreasing the closed-to-open equilibrium constant in the presence of Ca. We propose that Met oxidation alters CaM’s functional interaction with its target proteins by perturbing this Ca-dependent structural shift.
1. Introduction
1.1 Muscle aging, disease, and methionine oxidation
Reactions that use oxygen to drive cellular respiration create highly reactive oxygen species (ROS) that are potentially damaging to the cell. Biological aging and degenerative disease are strongly influenced by the resulting oxidative stress, causing post-translational modification of DNA, lipids, and proteins. Protein oxidation is strongly associated with loss of strength in both skeletal and cardiac muscle, and is proposed to play a major role in aging [1, 2, 3], muscular dystrophy [4], and heart failure [5, 6]. Understanding the initiation and progression of muscle aging and disease requires identification and characterization of ROS targets.
The sulfur-containing amino acids, cysteine (Cys) and methionine (Met), are the prime cellular targets of biological oxidants [7, 8, 9]. In particular, Met oxidation and subsequent reduction by Met sulfoxide reductase have far-reaching implications in metabolic, cardiovascular, neurological, and immune related dysfunction [10, 11, 12]. We have identified specific Met residues in proteins as targets of oxidation in muscle contractile and regulatory proteins [13, 14, 15]. Met oxidation has been proposed as a mechanism through which the muscle cell responds to oxidative stress by modulating metabolism and energy utilization [16]. Met oxidation can perturb local secondary structure, induce conformational disorder, and disrupt key hydrophobic interactions [17, 18, 19]. However, Met oxidation in the context of protein structure has only been systematically evaluated for a handful of proteins [13, 20, 21, 22]. Here, we have linked the oxidation of particular functionally sensitive Met residues to discrete and measurable changes in protein structure in order to understand how the oxidation of a single protein side chain can contribute to altered regulatory interactions in muscle.
1.2 Methionine oxidation alters CaM regulation of target proteins
We seek a molecular structural explanation for how oxidative modifications impact muscle protein function, focusing on the ubiquitous Ca signaling protein calmodulin (CaM). CaM plays a central role in Ca-mediated regulation of muscle contraction. Among its hundreds of target proteins, CaM acts as a feedforward activator of calcium pumps, a feed-back inhibitor of calcium channels, and an activator of a multitude of CaM-dependent kinases [23]. CaM has unusually high Met content, including 46% of the hydrophobic residues in the binding pockets, which are crucial for CaM’s interactions with over 400 diverse target proteins [24]. CaM containing oxidized Met residues has been isolated from both skeletal muscle and the brain of aged animals [25, 26]. Met oxidation impairs CaM’s ability to regulate the ryanodine receptor calcium channel (RyR) [27, 28], the plasma membrane Ca2+ ATPase (PMCA) [17, 21, 29, 30] and numerous other targets [31, 32, 33]. As a central node in the calcium signaling network, CaM is in an ideal position to orchestrate redox control of cellular homeostasis.
CaM is a dumbbell-shaped protein, with two globular domains (lobes) connected by a flexible α-helical linker (Fig. 1). CaM’s C-terminal lobe (C-lobe) Met residues are particularly susceptible and functionally sensitive to oxidation. Oxidation of Met 144 and Met 145 prevents CaM from fully activating the PMCA [30]. For the RyR, CaM binding is linked to profound changes in the Ca dependence of both activation and inactivation [35, 36]. Specific Met residues within the C-lobe are critical for CaM-mediated regulation of the RyR [27, 28]. Met-to-Gln (M-to-Q) mutations designed to mimic Met oxidation were used to determine site-specific contributions of C-lobe Met oxidation to changes in CaM regulation of RyR. It was found that M124Q induces a two-fold increase in the concentration of CaCaM required for half-maximal inhibition, while M109Q attenuates maximal RyR activation by apoCaM [27, 28]. M109Q and M124Q mutations were also found to uniquely block activation of smooth muscle myosin light chain kinase, CaM-dependent protein kinase Ila, and CaM-dependent protein kinase IV [37]. Here we pursue a structural explanation for the observed functional impact of oxidation of M109 and M124, focusing on changes in the structural transition that accompanies Ca binding. We have linked oxidation of specific Met to measurable changes in protein structure by (1) mimicking oxidation of particular amino acids through mutagenesis and (2) using spectroscopic distance measurements to resolve subtle changes in protein structure and dynamics.
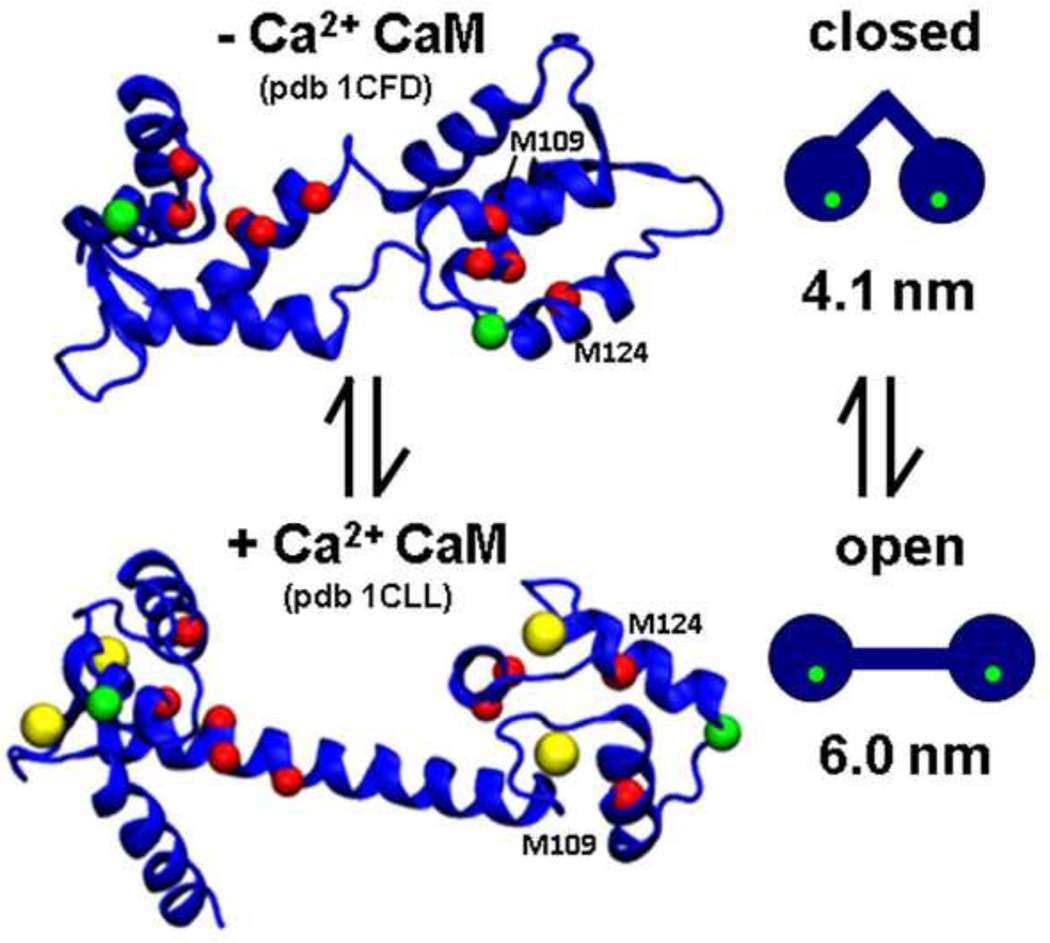
Fig. 1. CaM structural model.
The positions of coordinated calcium ions (yellow spheres), T34C and T1 17C labeling sites (green spheres), and all nine methionine residues (red spheres) are indicated. The two methionine residues of interest are labeled. 1CLL (top, based on NMR in solution) and 1CFD (bottom, from crystallography) were rendered using VMD [34].
1.3 Structural model to be tested
Although CaM is highly dynamic, most crystal and NMR structures can be assigned to one of two broad categories, the "open" or "closed" state (Fig. 1). The open structural state, which is stabilized by Ca binding, is defined by (a) a perpendicular orientation of pairs of EF hand helices, (b) an exposed patch of hydrophobic residues on each lobe, (c) an outward rotation of the lobes that elongates the entire molecule, and (d) a stable α-helical linker connecting the lobes [38]. Exposure of hydrophobic patches is thought to facilitate target protein binding and occurs upon the reorientation of the EF hand helices with Ca binding. The closed structure has been more difficult to characterize because of crystallization problems at low Ca. The solution NMR structure of apoCaM [39, 40] (Fig. 1, top) serves as a model for the closed structural state and is characterized by (a) EF hand helices in a tight four-helix bundle, (b) buried hydrophobic patches, (c) inward rotated lobes yielding a compact molecular shape, and (d) a discontinuous α-helical linker connecting the lobes.
The static nature of the models presented in Fig. 1 is an oversimplification. There is probably not a rigid coupling in solution between CaM structural state (open or closed) and biochemical state (low or high Ca), since there is an instance of Ca-loaded CaM crystallized in the closed structural state [41]. There are several lines of evidence suggesting that CaM undergoes conformational exchange in solution, particularly in the linker helix [42] and in the C-lobe [43, 44]. Indeed, NMR relaxation measurements and single-molecule FRET studies detected the presence of open and closed CaM states on the millisecond timescale [45, 46]. The existence of conformational equilibrium has been proposed as a mechanism through which target protein binding occurs through "selection" of pre-existing CaM conformations [47]. CaM’s promiscuity in binding interactions might very well stem from CaM’s broad intrinsic dynamics [24].
Here we have used spectroscopic distance measurements to better define the relationship between CaM Ca binding, Met oxidation, and the structural dynamics of the open and closed structural states. We chose to use the increasingly popular pulsed EPR technique DEER, over fluorescence techniques such as FRET, because it allows for the use of smaller, identical probes and provides superior resolution of distinct conformational states, mole fractions, and disorder [48, 49]. DEER is also much more effective than NMR in resolving the kind of long-range structural changes and conformational heterogeneity predicted by Fig. 1 [48, 49]. The results provide new insight into CaM structural dynamics and function.
2. Materials and Methods
2.1 Sample preparation and characterization
Mammalian calmodulin mutants with Cys substitutions for spin-labeling (T34C and T34C.T117C) and Met to Gln substitutions (T34C.T117C.M124Q, and T34C.T117C.M109Q), were prepared by site-directed mutagenesis, expressed, and purified as previously described [20], then dialyzed overnight at 4°C against CaM buffer (10mM NaCl, 10mM Tris, pH 7.0). CaM concentration was determined by UV absorption [28. Spin-labeled CaM concentration was determined using the BCA assay (Pierce, Rockford, IL). The sites for Cys substitution were chosen because they are in stable α helices in both crystal structures, and the predicted interspin distances are within the sensitivity range of DEER (~2 to 6 nm,[48]) for both crystal structures (Fig. 1). Cys mutagenesis and spin labeling did not perturb CaM regulation of RyR [28]. The sites for Met to Gln mutagenesis were chosen because they have previously been shown to disrupt the regulation of RyR by CaM [28]. Samples were flash-frozen in liquid nitrogen and stored at −80°C with 10% glycerol added as a cryoprotectant. Maleimide spin label (MSL, N-(1-oxyl-2,2,5,5-tetramethyl pyrrolidinyl) maleimide, Toronto Research Chemicals, Canada) was prepared as a 0.3 M stock in dimethylformamide (DMF). CaM (120 µM) was pre-treated with 1 mM TCEP for 20 minutes at 37°C to reduce disulfide bonds between Cys, MSL was added to a final concentration of 2 mM at 22°C for two h, followed by exhaustive dialysis against CaM buffer. Electrospray mass spectrometry and EPR spin counting both showed that all samples were fully spin-labeled. Some samples were treated with H2O2 as previously described, resulting in complete and selective oxidation of methionine side chains, as verified by mass spectrometry [20].
2.2 EPR spectroscopy
DEER was performed on doubly spin-labeled CaM samples, prepared by dialyzing 150 µM CaM into CaM buffer, with either 5 mM CaCl2 or 5 mM EGTA, and 10% glycerol as a cryoprotectant. Samples were loaded into quartz capillaries (1.1 mm ID, 1.6 mm OD, 20 µL sample volume) (Wilmad glass, Buena NJ), flash-frozen in liquid nitrogen, and stored at −80C until use. A Bruker E580 spectrometer (Billerica, MA) was used, operating at Q-band (34 GHz) with a EN5107 resonator, using a 4-pulse DEER protocol [48]. The π/2 pulse width was 12 ns, and the ELDOR pulse width was 24 ns. The static field (observe position) was set near the high-field resonance, with the ELDOR frequency (pump position) set to the maximum of the nitroxide absorption spectrum. Temperature was maintained at 65° K during acquisition, which lasted 4–24 h. The background-corrected DEER decay, whose shape is explicitly determined by the ensemble of distances between the pair of spin labeled sites on the protein, was analyzed using the model-free Tikhonov regularization method provided in the software Deer Analysis 2013.2 [50] to determine the distribution of distances present in the frozen sample. As results based on Gaussian distance distributions are more useful for discussing models based on discrete conformational states, we fit the resulting distance distribution to a model assuming a sum of Gaussians [51]:
| Eq. 1 |
| Eq. 2 |
The 3n-1 variable parameters in the fit were xi (mole fraction), ri (center distance), and σi (standard deviation), where the full width at half maximum is given by 2.355 σ. In all cases the dominant distance distributions reported by Tikhonov regularization were well fit by two Gaussians (n = 2).
3. Results
3.1 DEER resolves open and closed structural states of CaM
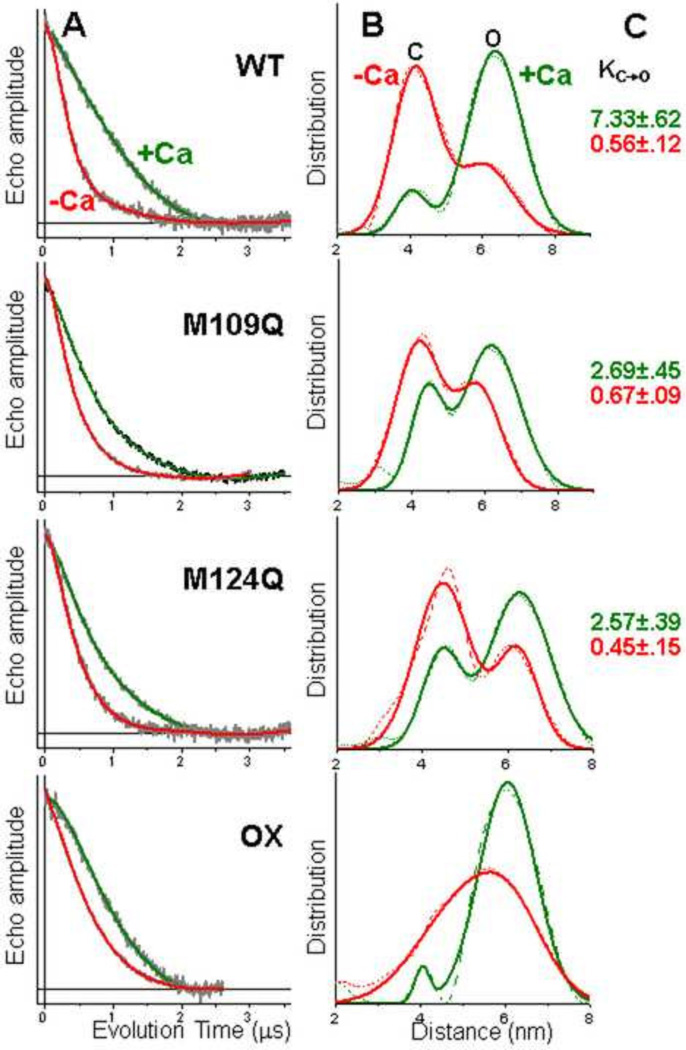
The shape of the time-resolved DEER decay reports the distances between the spin labeled protein domains, with fast decays representing shorter distances and slow decays representing longer distances. The decay is also encoded with information about distribution widths (disorder) and the relative mole fraction of distributions in the case of multiple distance populations. Ca substantially slows the DEER decay for spin-labeled WT CaM (Fig. 2A, top), indicating a substantial increase in the distance between the two lobes of CaM. At first glance, this is quite consistent with the model based on crystal and NMR structures, in which the structure is closed (~4 nm probe separation) and open (~6 nm probe separation) in the absence and presence of Ca, respectively (Fig. 1). However, closer inspection of the distance distributions derived from analysis of the DEER decay (Fig. 2B, top) reveals a more complex picture; both closed and open structural states are present simultaneously in both the presence and absence of Ca. Thus both biochemical states of CaM (−Ca and +Ca) populate closed (C) and open (O) states simultaneously. The mole fractions from the two-component fits of the distance distributions provide a quantitative determination of the equilibrium constant for the closed-to-open transition: 0.56 (−Ca) and 7.33 (+Ca) (Fig. 2C, top), giving a value of 13.1 for the ratio of equilibrium constants in the presence and absence of Ca. Each of the two structural states exhibited widths of 1.5 to 3 nm, too large to be explained by disorder of spin-labeled side chains. We conclude that the CaM structural states have intrinsic backbone disorder (probably dynamic flexibility) on the order of 1 –2 nm.
Fig. 2.
Representative DEER waveforms (A) and resulting distance distributions (B) for CaM samples spin-labeled at T34C and T1 17C, for wildtype (WT), M109Q and M124Q mutants, and H2O2-treated WT (OX). For the DEER waveforms (A), background-corrected data (grey) is overlaid with the best-fit simulation (red −Ca, green +Ca). For distance distributions (B), the same red/green color scheme applies. Model-independent Tikhonov analysis is shown as a dashed curve, and the best-fit two-Gaussian function is shown as a solid curve. (C) shows the equilibrium constant K(C→O) for the closed-to-open transition, calculated from the DEER-determined mole fractions (Eq. 1). No equilibrium constants are given for OX (bottom right), because the 2 components were not clearly resolved in that sample.
3.2 Mutations mimicking Met oxidation shift CaM’s structural distribution, reducing the magnitude of the Ca effect
CaM DEER decays were affected by Met-to-Gln substitutions, and by peroxide oxidation. In all cases, the effect was to decrease the magnitude of the Ca effect (Fig. 2A, note decrease in the red-green difference). The most significant change was an increase in the rate of decay for +Ca (Fig. 2A, green), indicating a shift toward the closed state (Fig. 2B, green). Thus the equilibrium constant K for the C-to-O transition in the presence of Ca is decreased by about a factor of 3 for both M109Q and M124Q (Fig. 2C, green). These mutations did not significantly change the widths of the distributions. The peroxide-treated WT sample, in which all nine Met residues were oxidized, was qualitatively like the M-to-Q mutants (smaller effect of Ca, Fig. 2A), but there was no longer sufficient resolution to resolve the closed and open states (Fig. 2B). We conclude that full oxidation induces changes more severe that those of single-site modification.
4. Discussion
We have used DEER’s ability to resolve protein distance distributions and the mole fractions of these structural states to better define the relationship in CaM between Ca binding, Met oxidation, and the structural dynamics of the open and closed structural states. In both −Ca and +Ca conditions, DEER indicates that CaM is distributed over at least two major structural states, closed and open (Fig. 1). Thus there is not a tight coupling between CaM’s structural states (closed and open) and biochemical states (−Ca and +Ca). In the absence of Ca, this structural equilibrium favors the closed state by about a factor of 2 (Kc→o = 0.56, Fig. 2C, top), while Ca shifts this equilibrium toward the open state by a factor of 13 (Kc→o = 7.33, Fig. 2C, top). The magnitude of this Ca-dependent shift in Kc→o is decreased by about a factor of 3 (Fig. 2C) by either of two specific Met-to-Gln mutations (M109Q and M124Q) that were previously shown to partially mimic the effects of Met oxidation on the Ca-dependent regulation of the muscle calcium release channel (RyR) [27, 28] and other CaM targets [37]. While the effects of these point mutations on CaM structure equilibrium are qualitatively similar to those produced by oxidizing all 9 Met side chains, decreasing the magnitude of the Ca effect, the effect of complete oxidation is greater than that of either M-to-Q mutation (Fig. 2). This is not surprising, since the functional effect is also much greater [27, 28].
How does CaM regulate such an impressively long list of target proteins with specificity? The binding interface between CaM and its known targets is quite variable, particularly in the spacing between the N-lobe and C-lobe binding sites. Existing structures of CaM-target peptide complexes sample a broad distribution of interlobe spacings [24]. Here, we find that under low and high Ca conditions, CaM’s opposing lobes intrinsically adopt a strikingly broad distribution of structures and that the distribution is sensitive to Ca-binding. The existence of multiple conformations has been proposed as a mechanism through which target protein binding occurs through “selection” of pre-existing CaM conformations [47]. Oxidation-induced change in the distribution of available CaM structures would therefore play a strong role in dictating which targets would bind CaM with highest affinity.
Typically, CaCaM binds to target proteins with higher affinity than apoCaM, so it is CaCaM that exerts regulatory influence. However, for the RyR channel, both apoCaM and CaCaM exert a regulatory role, with apoCaM activating RyR and CaCaM inhibiting RyR [35, 36]. M124Q and M109Q have slightly different effects on RyR regulation [28], with the effect of M109Q observable mainly at low Ca, and the effect of M124Q mainly at high Ca. Our results do not show a significant difference in the effects of these mutations on the Ca-dependent conformational equilibrium (Fig. 2). Higher resolution studies (e.g., by NMR) will be needed to determine the structural basis of these differences.
Conclusion
We have used pulsed EPR (DEER) to resolve the closed and open structural states of calmodulin, in both the presence and absence of Ca. The relative populations of these states are sensitive to Ca binding and also to Met-to-Gln substitutions previously shown to partially mimic the functional effects of methionine oxidation. It is likely that the closed structural state is critical to CaM activation of RyR under low Ca conditions and that the open structural state is critical to RyR inhibition under high Ca conditions. Extension of these structural studies to complexes of CaM bound to RyR and other regulatory targets will be needed to test these hypotheses and refine molecular models of regulation.
HIGHLIGHTS.
We measured the distance distribution between two spin labels on calmodulin by DEER.
Two structural states, open and closed, were resolved.at both low and high Ca.
Ca shifted the equilibrium toward the open state by a factor of 13.
Methionine oxidation, simulated by glutamine substitution, decreased the Ca effect.
These results have important implications for aging in muscle and other tissues.
Acknowledgements
This work was supported by grants to D.D.T. (NIH R37 AG26160) and J.C.K. (University of Wisconsin Faculty Development Grant). FN was supported by NIH Training Grant T32 AR07612. This project used the facilities of the Biophysical Spectroscopy Center, University of Minnesota. We thank Octavian Cornea for preparing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 2.Prochniewicz E, Thompson LV, Thomas DD. Age-related decline in actomyosin structure and function. Exp Gerontol. 2007;42:931–938. doi: 10.1016/j.exger.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraibar MA, Gueugneau M, Duguez S, Butler-Browne G, Bechet D, Friguet B. Expression and modification proteomics during skeletal muscle ageing. Biogerontology. 2013;14:339–352. doi: 10.1007/s10522-013-9426-7. [DOI] [PubMed] [Google Scholar]
- 4.Tidball JG, Wehling-Henricks M. The role of free radicals in the pathophysiology of muscular dystrophy. J Appl Physiol. 2007;102:1677–1686. doi: 10.1152/japplphysiol.01145.2006. [DOI] [PubMed] [Google Scholar]
- 5.Maack C, Kartes T, Kilter H, Schafers HJ, Nickenig G, Bohm M, Laufs U. Oxygen free radical release in human failing myocardium is associated with increased activity of rac1-GTPase and represents a target for statin treatment. Circulation. 2003;108:1567–1574. doi: 10.1161/01.CIR.0000091084.46500.BB. [DOI] [PubMed] [Google Scholar]
- 6.Avner BS, Shioura KM, Scruggs SB, Grachoff M, Geenen DL, Helseth DL, Jr, Farjah M, Goldspink PH, Solaro RJ. Myocardial infarction in mice alters sarcomeric function via post-translational protein modification. Mol Cell Biochem. 2012;363:203–215. doi: 10.1007/s11010-011-1172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies KJ. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 8.Kim G, Weiss SJ, Levine RL. Methionine oxidation and reduction in proteins. Biochim Biophys Acta. 2014;1840:901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drazic A, Winter J. The physiological role of reversible methionine oxidation. Biochim Biophys Acta. 2014;1844:1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Bigelow DJ, Squier TC. Thioredoxin-dependent redox regulation of cellular signaling and stress response through reversible oxidation of methionines. Mol Biosyst. 2011;7:2101–2109. doi: 10.1039/c1mb05081h. [DOI] [PubMed] [Google Scholar]
- 11.Cui ZJ, Han ZQ, Li ZY. Modulating protein activity and cellular function by methionine residue oxidation. Amino Acids. 2012;43:505–517. doi: 10.1007/s00726-011-1175-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim G, Weiss SJ, Levine RL. Methionine Oxidation and Reduction in Proteins. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein JC, Moen RJ, Smith EA, Titus MA, Thomas DD. Structural and functional impact of site-directed methionine oxidation in myosin. Biochemistry. 2011;50:10318–10327. doi: 10.1021/bi201279u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prochniewicz E, Lowe DA, Spakowicz DJ, Higgins L, O’Conor K, Thompson LV, Ferrington DA, Thomas DD. Functional, structural, and chemical changes in myosin associated with hydrogen peroxide treatment of skeletal muscle fibers. Am J Physiol Cell Physiol. 2008;294:C613–C626. doi: 10.1152/ajpcell.00232.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moen RJ, Cornea S, Oseid DE, Binder BP, Klein JC, Thomas DD. Redox-sensitive residue in the actin-binding interface of myosin. Biochemical and biophysical research communications. 2014 doi: 10.1016/j.bbrc.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigelow DJ, Squier TC. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim Biophys Acta. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Yao Y, Squier TC. Oxidatively modified calmodulin binds to the plasma membrane Ca-ATPase in a nonproductive and conformationally disordered complex. Biophysical journal. 2001;80:1791–1801. doi: 10.1016/S0006-3495(01)76149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Younan ND, Nadal RC, Davies P, Brown DR, Viles JH. Methionine oxidation perturbs the structural core of the prion protein and suggests a generic misfolding pathway. J Biol Chem. 2012;287:28263–28275. doi: 10.1074/jbc.M112.354779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J Biol Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balog EM, Lockamy EL, Thomas DD, Ferrington DA. Site-specific methionine oxidation initiates calmodulin degradation by the 20S proteasome. Biochemistry. 2009;48:3005–3016. doi: 10.1021/bi802117k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anbanandam A, Bieber Urbauer RJ, Bartlett RK, Smallwood HS, Squier TC, Urbauer JL. Mediating molecular recognition by methionine oxidation: conformational switching by oxidation of methionine in the carboxyl-terminal domain of calmodulin. Biochemistry. 2005;44:9486–9496. doi: 10.1021/bi0504963. [DOI] [PubMed] [Google Scholar]
- 22.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin target database. J Struct Funct Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 24.Yamniuk AP, Vogel HJ. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Yin D, Yao Y, Williams TD, Squier TC. Progressive decline in the ability of calmodulin isolated from aged brain to activate the plasma membrane Ca-ATPase. Biochemistry. 1998;37:9536–9548. doi: 10.1021/bi9803877. [DOI] [PubMed] [Google Scholar]
- 26.Boschek CB, Jones TE, Smallwood HS, Squier TC, Bigelow DJ. Loss of the calmodulin-dependent inhibition of the RyR1 calcium release channel upon oxidation of methionines in calmodulin. Biochemistry. 2008;47:131–142. doi: 10.1021/bi701352w. [DOI] [PubMed] [Google Scholar]
- 27.Balog EM, Norton LE, Bloomquist RA, Cornea RL, Black DJ, Louis CF, Thomas DD, Fruen BR. Calmodulin oxidation and methionine to glutamine substitutions reveal methionine residues critical for functional interaction with ryanodine receptor-1. J Biol Chem. 2003;278:15615–15621. doi: 10.1074/jbc.M209180200. [DOI] [PubMed] [Google Scholar]
- 28.Balog EM, Norton LE, Thomas DD, Fruen BR. Role of calmodulin methionine residues in mediating productive association with cardiac ryanodine receptors. Am J Physiol Heart Circ Physiol. 2006;290:H794–H799. doi: 10.1152/ajpheart.00706.2005. [DOI] [PubMed] [Google Scholar]
- 29.Yin D, Kuczera K, Squier TC. The sensitivity of carboxyl-terminal methionines in calmodulin isoforms to oxidation by H(2)O(2) modulates the ability to activate the plasma membrane Ca-ATPase. Chem Res Toxicol. 2000;13:103–110. doi: 10.1021/tx990142a. [DOI] [PubMed] [Google Scholar]
- 30.Bartlett RK, Bieber Urbauer RJ, Anbanandam A, Smallwood HS, Urbauer JL, Squier TC. Oxidation of Met144 and Met145 in calmodulin blocks calmodulin dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 2003;42:3231–3238. doi: 10.1021/bi026956z. [DOI] [PubMed] [Google Scholar]
- 31.Wolff J, Cook GH, Goldhammer AR, Berkowitz SA. Calmodulin activates prokaryotic adenylate cyclase. Proc Natl Acad Sci U S A. 1980;77:3841–3844. doi: 10.1073/pnas.77.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huhmer AF, Gerber NC, de Montellano PR, Schoneich C. Peroxynitrite reduction of calmodulin stimulation of neuronal nitric oxide synthase. Chem Res Toxicol. 1996;9:484–491. doi: 10.1021/tx950152l. [DOI] [PubMed] [Google Scholar]
- 33.Robison AJ, Winder DG, Colbran RJ, Bartlett RK. Oxidation of calmodulin alters activation and regulation of CaMKII. Biochem Biophys Res Commun. 2007;356:97–101. doi: 10.1016/j.bbrc.2007.02.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Tripathy A, Xu L, Mann G, Meissner G. Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor) Biophys J. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fruen BR, Bardy JM, Byrem TM, Strasburg GM, Louis CF. Differential Ca(2+) sensitivity of skeletal and cardiac muscle ryanodine receptors in the presence of calmodulin. Am J Physiol Cell Physiol. 2000;279:C724–C733. doi: 10.1152/ajpcell.2000.279.3.C724. [DOI] [PubMed] [Google Scholar]
- 37.Chin D, Means AR. Methionine to glutamine substitutions in the C-terminal domain of calmodulin impair the activation of three protein kinases. J Biol Chem. 1996;271:30465–30471. doi: 10.1074/jbc.271.48.30465. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 40.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 41.Fallon JL, Quiocho FA. A closed compact structure of native Ca(2+)-calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Qin Z, Squier TC. Calcium-dependent stabilization of the central sequence between Met(76) and Ser(81) in vertebrate calmodulin. Biophys J. 2001;81:2908–2918. doi: 10.1016/S0006-3495(01)75931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 44.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nat Struct Biol. 2001;8:990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 45.Johnson CK. Calmodulin, conformational states, and calcium signaling. A single-molecule perspective. Biochemistry. 2006;45:14233–14246. doi: 10.1021/bi061058e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malmendal A, Evenas J, Forsen S, Akke M. Structural dynamics in the C-terminal domain of calmodulin at low calcium levels. J Mol Biol. 1999;293:883–899. doi: 10.1006/jmbi.1999.3188. [DOI] [PubMed] [Google Scholar]
- 47.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 48.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 49.Lin AY, Prochniewicz E, James Z, Svensonn B, Thomas DD. Large-scale opening of utrophin’s tandem CH domains upon actin binding, by an induced-fit mechanism. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1106453108. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeschke G, Koch A, Jonas U, Godt A. Direct conversion of EPR dipolar time evolution data to distance distributions. Journal of magnetic resonance. 2002;155:72–82. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 51.Blackburn ME, Veloro AM, Fanucci GE. Monitoring inhibitor-induced conformational population shifts in HIV-1 protease by pulsed EPR spectroscopy. Biochemistry. 2009;48:8765–8767. doi: 10.1021/bi901201q. [DOI] [PubMed] [Google Scholar]