Abstract
Streptomyces spp. are robust producers of medicinally-, industrially- and agriculturally-important small molecules. Increased resistance to antibacterial agents and the lack of new antibiotics in the pipeline have led to a renaissance in natural product discovery. This endeavor has benefited from inexpensive high quality DNA sequencing technology, which has generated more than 140 genome sequences for taxonomic type strains and environmental Streptomyces spp. isolates. Many of the sequenced streptomycetes belong to the same species. For instance, Streptomyces albus has been isolated from diverse environmental niches and seven strains have been sequenced, consequently this species has been sequenced more than any other streptomycete, allowing valuable analyses of strain-level diversity in secondary metabolism. Bioinformatics analyses identified a total of 48 unique biosynthetic gene clusters harboured by Streptomyces albus strains. Eighteen of these gene clusters specify the core secondary metabolome of the species. Fourteen of the gene clusters are contained by one or more strain and are considered auxiliary, while 16 of the gene clusters encode the production of putative strain-specific secondary metabolites. Analysis of Streptomyces albus strains suggests that each strain of a Streptomyces species likely harbours at least one strain-specific biosynthetic gene cluster. Importantly, this implies that deep sequencing of a species will not exhaust gene cluster diversity and will continue to yield novelty.
Introduction
More than two-thirds of all therapeutic small molecules used in medicine are derived or inspired from complex natural products produced by filamentous actinobacteria, most notably Streptomyces spp. [1]. Streptomyces spp. are predominantly known as filamentous soil bacteria that have a differentiating mycelial life-cycle, which begins with spore germination and outgrowth of a vegetative mycelium and ends with production of reproductive aerial hyphae and the formation of unigenomic spores [2]. Aerial hyphae production and sporulation is often accompanied by the production of secondary metabolites. These secondary metabolites are most likely used to outcompete neighbouring organisms [3]. Biotechnology has exploited many of these natural products as anticancer, antiviral, insecticidal, herbicidal, antibacterial, antifungal and immunosuppressive compounds [4].
Growing global concerns about resistance to antibacterial agents has led to a renaissance in bioprospecting and natural product discovery. The resurgence of interest in natural products is greatly aided by the relatively inexpensive cost to sequence genomes of strains that produce promising bioactive small molecules. One-hundred and forty-two streptomycete genomes are available in DDBJ/EMBL/Genbank. This dataset has made it abundantly clear that Streptomyces spp. only express a mere fraction of their biosynthetic genes under standard laboratory growth conditions. Activation of silent biosynthetic gene clusters and characterisation of their products represents a major potential source for new lead compounds for industry and is an area in which synthetic biology holds huge promise [5].
In order to capitalise on available genomic resources, systematic analyses of secondary metabolism are required. Doroghazi and Metcalf provided the first comparative analysis of secondary metabolism in organisms with closed genomes from the phylum Actinobacteria, which included eight Streptomyces species and revealed, for good reason, why this taxa has been the focus of rigorous genomic and biochemical analyses over the years [6]. Recently, Ziemert et al. performed a focused analysis of the secondary metabolism in 75 sequenced Salinispora species identified a total of 124 biosynthetic pathways encoded by the genus and provided insight into population-level genetic exchange of biosynthetic pathways in marine environments [7]. Doroghazi et al. recently developed a method for classification of gene clusters into families and used this approach to analyse the biosynthetic potential of 830 sequenced Actinobacteria, which they found to contain a total of 11,422 gene clusters comprising 4,122 gene cluster families [8]. More analyses of these type will be required in order to drive the fields of natural product discovery and synthetic biology forward and maximise the promise held by genome mining actinomycetes.
Streptomyces albus, which is one of the most widely geographically distributed streptomycetes and has been isolated from diverse environments including sponges, sea sediments and insects [9–14]. The archetype member of this species is S. albus J1074 which is a derivative of S. albus G in which the salI restriction system was deleted to better enable transformation [15]. S. albus J1074 has therefore been used as a host for heterologous expression of several natural product gene clusters, including cyclooctatin [16], fredericamycin [17], iso-migrastatin [18], moenomycin [19], napyradiomycin [20], steffimycin [21] and thiocoraline [22] and there has recently been renewed interest in further developing this expression platform because of its fastidious growth and naturally minimised genome [23]. The clear ability of S. albus J1074 to heterologously biosynthesise diverse and important natural products suggests strains of S. albus may encode important natural product gene clusters of their own, a question which genomics and genome mining is only just now beginning to address. As more researchers sequence closely related strains it makes necessary an understanding of strain-level diversity in secondary metabolism. With this view in mind, here I report a strain-level analysis of secondary metabolism for six sequenced S. albus strains. A total of 48 biosynthetic gene clusters were identified and approximately 18 specify the core secondary metabolome of S. albus, 14 are auxiliary gene clusters and 16 are strain-specific, indicating there is still appreciable chemical diversity to be discovered at the strain level.
Results and Discussion
A multilocus phylogeny of Streptomyces spp. reveals significant redundancy in sequenced organisms
Many of the 142 genome sequences available for Streptomyces spp. originate from so-called environmental isolates and their taxonomic classification remains enigmatic. A multilocus phylogeny was reconstructed in order to infer a taxonomic relationship among sequenced Streptomyces spp. and assess redundancy in the genomic database. Multiple loci were used to infer phylogenetic relationships because of well recognised problems with the use of solely the 16S rRNA gene as a phylogenetic marker, as it only provides an accurate and reliable classification to the genus level of streptomycetes [24] likely due to extensive recombination in the evolutionary past [25]. The loci selected for this study were those employed by previous multilocus phylogenies of streptomycetes: 16S rDNA, aptD (ATP synthase), gyrA (DNA gyrase subunit A), recA (recombination protein), rpoB (RNA pol subunit) and trpB (tryptophan biosynthesis) [26,27]. 16S rDNA sequences could not be identified in some draft genome sequences. This is presumably a result of an inadequacy with DNA assembly software to process the multiple copies (five to seven copies) of the ribosomal RNA locus streptomycetes are known to harbour. The partial 16S rDNA sequences (variable region IV) that were retrieved had a maximum pairwise divergence of ∼5% over 292 nt (determined by blast analysis). With the motivation to include as many genome sequences in this analysis as possible, the decision was therefore made to exclude the 16S rRNA gene as a phylogenetic marker for this study. Partial DNA sequences for atpD, gyrA, recA, rpoB and trpB, corresponding to regions targeted by well established oligonucleotide primer sequences employed in phylogenetic analyses [26,27] were retrieved from Genbank (see methods). Due to the poor quality of some of the genome sequences and/or the absence of some of these genes entirely, ∼14% (20 genomes) were excluded from this analysis. Redundant genomes for type-strains were also excluded, namely S. bottropensis ATCC 25435 ([Genbank:AOCF00000000]), S. clavuligerus ATCC 27064 ([Genbank:ADGD00000000]) and S. albus J1074 ([Genbank:ABYC00000000]).
An approximately maximum-likelihood phylogenetic tree based on concatenated aptD-gyrB-recA-rpoB-trpB gene fragments (2566 nt in total) was constructed (Fig. 1). Overall, there was good separation and statistical support for most of the branches in the tree. Interestingly, the tree suggested that many Streptomyces species have been sequenced more than once. To further analyse this, the concatenated aptD-gyrB-recA-rpoB-trpB gene fragments were next binned into operational taxonomic units (OTUs) with a shared identity threshold of 97%, which is a widely used threshold for species-level classification [28]. Approximately 70% (82 out of 120) of the sequenced streptomycetes analysed here correspond to a unique species of Streptomyces (S1 Table). The most (over-)represented species for which a genome sequence is available is Streptomyces albus (seven sequences in total). The availability of multiple genome sequences for a single species enables valuable analyses of the diversity and distribution of secondary metabolism which have only now become possible and will help inform and direct bioprospecting efforts in Streptomyces spp.
Figure 1. An approximately maximum likelihood phylogenetic tree of sequenced Streptomyces species.
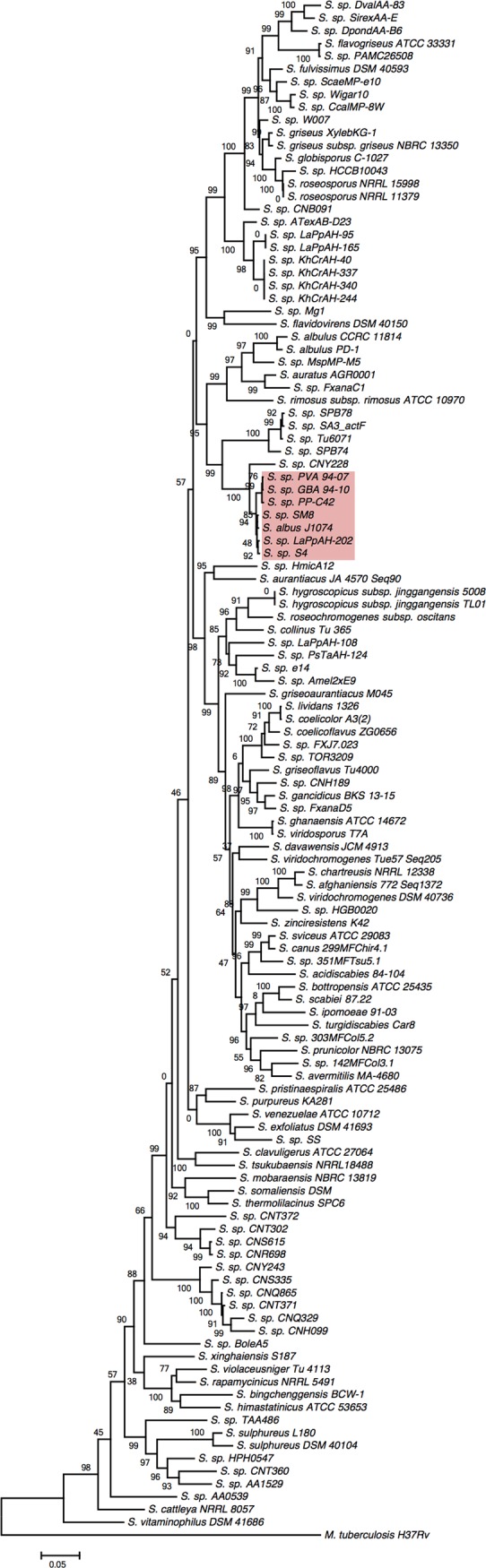
A phylogeny was inferred for Mycobacterium tuberculosis and 120 sequenced streptomycetes based on concatenated partial sequences of aptD-gyrB-recA-rpoB. FastTree local support values (expressed as a whole number) are indicated at each node. Streptomyces albus strains are highlighted in red. The scale bar indicates 5% estimated sequence divergence.
Secondary metabolism in S. albus
The archetype member of the S. albus clade is S. albus J1074 [15] commonly used as a heterologous expression host [16–22]. The six additionally sequenced strains of S. albus were identified more recently and their isolation was motivated, at least in part, by bioprospecting in unexploited microbial niches and include: S. sp. PVA-94-07, S. sp. GBA 94-10, S. sp. SM8, S. sp. PP-C42, S. sp. LaPpAH-202 and S. sp. S4. Details of S. albus strains are summarised in Table 1. The poor quality of the genome sequence available for S. sp. PP-C42 (>7,000 contigs) prevented its inclusion in this analysis so therefore a total of six S. albus genomes were analysed here.
Table 1. Accessions and genomic features of Streptomyces albus strains.
| Organism | Accession | Genome size (Mb) | Number of contigs | Source | Reference |
|---|---|---|---|---|---|
| S. albus J1074 | NC_020990 | 6.83 | Closed | Unknown | [23] |
| S. sp. PVA-94-07 | ASHE00000000 | 7.10 | 20 | Nordic fjords | [9] |
| S. sp. GBA 94-10 | ASHF00000000 | 7.22 | 34 | Nordic fjords | [9] |
| S. sp. PP-C42 | AEWS00000000 | 6.46 | 7,074 | Baltic Sea sediment | [11] |
| S. sp. SM8 | AMPN00000000 | 7.15 | 513 | North Sea sponges | [10] |
| S. sp. LaPpAH-202 | ARDM00000000 | 7.00 | 36 | Plant-ants | [14] |
| S. sp. S4 | CADY00000000 | 7.61 | 269 | Leaf-cutting ants | [12] |
Gene clusters encoding putative secondary metabolites were identified using antiSMASH 2.0 [29] and, crucially, were edited to best reflect published experimental data. Three independent analyses of secondary metabolism in S. albus J1074 have been conducted in this year [9,23,30]. These analyses disagree with regard to the total number of putative biosynthetic gene clusters encoded by S. albus J1074. Briefly, these analyses were hindered by using the draft version of the S. albus J1074 genome sequence [30], use of an earlier version of antiSMASH [23] and not taking into consideration experimental data [9,23].
S. albus strains encode between 25–30 biosynthetic gene clusters with S. albus J1074 encoding the least (25) and S. sp. PVA-94-07 encoding the most gene clusters (30) (Table 2). A pairwise comparison of gene clusters revealed significant redundancy in the putative secondary metabolites produced by S. albus strains. Importantly, the pairwise comparison also revealed that between 3 and 21% of gene clusters harboured by an individual strain are in fact strain-specific (Table 2), which suggests that gene cluster diversity may not be exhausted by deep-sequencing multiple strains of a single species, a prediction that was recently validated for the marine actinomycete, Salinispora spp. [7].
Table 2. Pairwise comparison of gene clusters encoding putative secondary metabolites from Streptomyces albus strains.
| S. albus J1074 | S. sp. PVA-94-07 | S. sp. GBA 94-10 | S. sp. SM8 | S. sp. LaPpAH-202 | S. sp. S4 | No. unique gene clusters | |
|---|---|---|---|---|---|---|---|
| S. albus J1074 | 25 (100%) | 20 (80%) | 19 (76%) | 20 (80%) | 23 (92%) | 21 (84%) | 2 (8%) |
| S. sp. PVA-94-07 | 30 (100%) | 28 (93%) | 20 (66%) | 21 (70%) | 21 (70%) | 1 (3%) | |
| S. sp. GBA 94-10 | 29 (100%) | 19 (65%) | 20 (68%) | 20 (68%) | 1 (3%) | ||
| S. sp. SM8 | 26 (100%) | 21 (80%) | 21 (80%) | 3 (11%) | |||
| S. sp. LaPpAH-202 | 27 (100%) | 22 (81%) | 3 (11%) | ||||
| S. sp. S4 | 28 (100%) | 6 (21%) |
The percentage in braces reflects the total number of gene clusters conserved in the pairwise comparison with respect to the strains listed vertically.
The core secondary metabolome of the S. albus clade
S. albus strains encode the production of a conserved set of 18 secondary metabolites (Table 3). Eight of these metabolites are produced by most (if not all) streptomycetes and include: desferrioxamine and aerobactin-like siderophores [31], ectoine osmolytes [32], hopanoid membrane components [33], carotenoid pigments [34], tetrahydroxynapthalenes [35], the morphologically-important lantipeptide, SapB [36] and volatile geosmins whose function is still obscure nearly 50 years after its discovery [37]. The remaining 10 gene clusters harboured by all S. albus strains are not as widely conserved at the genus level. These metabolites include: candicidin, a polyene antifungal compound [12,38], the respiratory chain inhibitor and anti-anti-apoptotic agent, antimycin [39,40], an antibacterial, similar to gramicidin [38], the volatile terpenoid antibacterial, albaflavenone [41], and the antifungal alteramide [30]. Interestingly, alteramide was first isolated from a sponge-associated Alteromonas spp. [42] and its production by S. albus J1074 was only observed after engineering its expression and is the first report of alteramide biosynthesis in the genus Streptomyces [30]. Additionally, the core secondary metabolome of S. albus includes five unknown products encoded by NRPS (2), Type I PKS (1) and bacteriocin (2) gene clusters (Table 3).
Table 3. The core secondary metabolome of Streptomyces albus.
| Predicted biosynthetic system | Putative product | S. albus J1074 | S. sp. PVA-94-07 | S. sp. GBA 94-10 | S. sp. SM8 | S. sp. LaPpAH-202 | S. sp. S4 |
|---|---|---|---|---|---|---|---|
| Hopene / Squalene synthase | Hopanoids | 308626..335220 | 647179..673767 | 608447..635035 | Multiple contigs*** | NZ_KB890705.1 522286..548874 | CADY01000122.1 571..33079 |
| NRPS-independent siderophore synthase | Desferrioxamine | 4740450..4752270 | 4783002..4794822 | 4778566..4790386 | NZ_AMPN01000107.1 27451..38705 | NZ_KB890704.1 497713..509533 | CADY01000052.1 71864..83684 |
| NRPS-independent siderophore synthase | Aerobactin-like | 1268164..1283196 | 1461319..1476318 | 1419952..1434951 | Multiple contigs*** | NZ_KB890708.199818..114849 | CADY01000147.1 1..13852 |
| Ectoine synthase | Ectoine | 5635346..5645744 | 5646629..5657027 | 5645248..5655646 | NZ_AMPN01000262.1 1748..15421 | NZ_KB890727.1 65721..76119 | CADY01000033.1 3246..13644 |
| Phytoene / polyprenyl synthetase | Carotenoids | 6401161..6430221 | 6435309..6461258 | 6426764..6452713 | NZ_AMPN01000036.1 9070..37323 | NZ_KB890733.1 196543..229868 | CADY01000098.1 59202..91885 |
| Terpene synthase | Geosmin | 1531759..1554059 | 1713992..1736292 | 1672644..1694944 | NZ_AMPN01000357.1 6256..26511 | NZ_KB890732.1 367671..389971 | CADY01000157.1 6106..28406 |
| Terpene | Albaflavenone | 1865241..1887220 | 2122786..2144075 | 2082541..2103830 | NZ_AMPN01000386.1 17700..38989 | NZ_KB890732.1 22700..43989 | CADY01000162.1 47721..69010 |
| Type III PKS | Tetrahydroxynapthalene | 6520374..6561471 | 6555445..6596542 | 6545260..6586357 | NZ_AMPN01000015.1 1..12125 | NZ_KB890733.1 61315..102412 | CADY01000095.1 47527..88624 |
| Type I PKS | Candicidin | 6566408..6721648 | 6586760..6741995 | 6576564..6731817 | Multiple contigs*** | Multiple contigs*** | Multiple contigs*** |
| Type I PKS* | Unknown | 6776680..6838639 | 6797026..6858963 | 6786847..6849240 | Multiple contigs*** | NZ_KB890710.1 1..60636 | Multiple contigs*** |
| Hybrid NRPS / PKS* | Antimycin | 6730563..6755198 | 6750919..6775544 | 6740741..6765365 | NZ_AMPN01000430.1 1.7480 | Mutiple contigs*** | CADY01000091.1 16873..41495 |
| Hybrid NRPS / PKS | Alteramide | 224752..275005 | 560646..610065 | 522971..572390 | NZ_AMPN01000189.1 1..20256 | NZ_KB890705.1 438436..488689 | CADY01000120.1 1..40866 |
| Hybrid NRPS / PKS** | Unknown | 6755272..6776675 | 6775619..6797021 | 6765440..6786842 | Multiple contigs*** | NZ_KB890710.1 60641..82030 | CADY01000091.1 1..16795 |
| NRPS | Gramicidin-like | 3877105..3982798 | 3970595..4076299 | 3946534..4052238 | Multiple contigs*** | Multiple contigs*** | CADY01000205.1 1..90613 |
| NRPS | Unknown | 4469477..4514441 | 4546088..4590455 | 4542814..4587181 | NZ_AMPN01000006.1 1..22395 | NZ_KB890704.1 738253..783217 | CADY01000068.1 1..34880 |
| Bacteriocin | Unknown | 415649..425903 | 740656..750895 | 701944..712183 | NZ_AMPN01000269.1 2352..8553 | NZ_KB890705.1 614326..624580 | CADY01000124.1 50376..61254 |
| Bacteriocin | Unknown | 879961..891289 | 1167101..1178414 | 1125708..1137021 | NZ_AMPN01000026.1 11838..21948 | NZ_KB890709.1 5759..17087 | CADY01000132.1 5648..13695 |
| Lantipeptide | SapB | 2713288..2735999 | 2912898..2935531 | 2877890..2900523 | NZ_AMPN01000379.1 313..18317 | NZ_KB890715.1 87381..116434 | CADY01000176.1 1574..29018 |
*denotes genomic coordinates were edited manually
** denotes a gene cluster which was not annotated by antiSMASH 2.0
***denotes a gene cluster which is spread over multiple contigs (refer to S2 Table).
Auxiliary biosynthetic capabilities of Streptomyces albus
Beyond the core metabolome, S. albus harbours 14 ‘auxiliary’ biosynthetic gene clusters. Auxiliary biosynthetic gene clusters are conserved to varying extents by S. albus strains, the details of which are summarized in Table 4. NRPS gene clusters were the most abundant class of biosynthetic system (7 out of 14 gene clusters) followed by hybrid NRPS / PKS systems (2 out of 14). As to be expected, the overwhelming majority of auxiliary gene clusters encode the production of unknown products (Table 4). Thus far, only one product of an auxiliary gene cluster has been elucidated, indigoidine. Indigoidine is a blue NRPS-derived pigment produced by S. albus J1074 and S. sp. LaPpAH-202. Interestingly, biosynthesis of indigoidine, at least in S. albus J1074 is repressed under normal laboratory growth conditions, and indigoidine production was only achieved by knocking-in the ermE* promoter upstream of core biosynthetic genes [30]. Although production of only one auxiliary metabolite has been analysed, bioinformatics analyses suggest that both S. sp. PVA 94-07 and S. sp. GBA 94-10 possess gene clusters coding for the biosynthesis of enterocin and a compound related to kijanamycin, which are both antibacterial agents [9].
Table 4. Auxiliary secondary metabolites produced by Streptomyces albus.
| Predicted biosynthetic system | Predicted product | S. albus J1074 | S. sp. PVA-94-07 | S. sp. GBA 94-10 | S. sp. SM8 | S. sp. LaPpAH-202 | S. sp. S4 |
|---|---|---|---|---|---|---|---|
| Hybrid NRPS / PKS | Unknown | 3011..61711 | 288401..349562 | — | NZ_AMPN01000108.1 1..17827 | NZ_KB890705.1 211618..272818 | Multiple contigs** |
| NRPS | Unknown | 1136316..1199422 | — | — | — | Multiple contigs** | — |
| Lantipeptide | Unknown | 2376688..2409159 | 2603848..2636308 | 2566778..2599136 | — | NZ_KB890736.1 70394..102866 | CADY01000169.1 12631..53654 |
| NRPS | Unknown | 3553726..3604015 | — | — | Multiple contigs** | NZ_KB890716.1 353451..403740 | Multiple contigs** |
| NRPS | Indigoidin | 6336942..6381213 | — | — | — | NZ_KB890733.1 244920..289191 | — |
| Terpene | Unknown | — | 274831..297419 | 284406..306988 | NZ_AMPN01000169.1 36623..53502 | NZ_KB890705.1 195282..217864 | CADY01000116.1 145638..168220 |
| Hybrid NRPS / PKS | Kijanimycin-like | — | 397572..502620 | 366365..471413 | — | — | — |
| Type I PKS | Unknown | — | 771534..818373 | 731491..778330 | — | — | — |
| NRPS | Unknown | — | 3152743..3211020 | 3129401..3187816 | — | — | — |
| NRPS | Unknown | — | 3750249..3812487 | 3726395..3788633 | — | — | — |
| NRPS | Unknown | — | 4898187..4960375 | 4891468..4953657 | — | — | — |
| NRPS* | Unknown | 1..49621 (6957897..7007517) | 2794..56173 (6969866..7023245) | ||||
| Type I PKS-butyrolactone* | Unknown | 30737..134245 (6888273..6976781) | 37289..140797 (6900242..6988750) | ||||
| Type II PKS | Enterocin | — | 6259612..6302316 | 6251573..6294277 | — | — | — |
* Denotes a gene cluster harboured in duplicate; genomic coordinates for the additional copy are provide in braces.
**Denotes a gene cluster spread over multiple contigs, which are presented in S2 Table.
Strain-specific metabolites produced by Streptomyces albus
In addition to core and auxiliary metabolites, S. albus strains harbour a total of 17 strain-specific gene clusters whose putative products comprise all of the major classes of secondary metabolites (Table 5). Each S. albus strain specifies at least one strain-specific gene cluster, which is consistent with Salinispora arenicola, S. pacific and S. tropica strains each encoding the production of ∼1.0 strain-specific polyketide or non-ribosomal peptide [7]. S. sp. PVA 94-07 and S. sp. GBA 94-10 harbour a single strain-specific gene cluster apiece, which is the fewest number specified out of all strains (Tables 2 and 5). However, eight gene clusters with unknown products are shared between S. sp. PVA 94-07 and S. sp. GBA 94-10 and are not harboured by other S. albus strains, suggesting that despite this, S. sp. PVA 94-07 and S. sp. GBA 94-10 produce a significant amount of novel chemistry. S. sp. S4 harbours six strain-specific gene clusters whose products represent 21% of its secondary metabolome, which is the most of any S. albus strain (Table 2) and may reflect its possible role as a defensive symbiont of fungus-growing ants [12]. Paulomycin, the product of a hybrid NRPS/PKS gene cluster encoded by S. albus J1074 is the only analysis of a strain-specific gene cluster thus far [30,43]. Although chemical analysis is required for confirmation, there is strong bioinformatics support to suggest that products of two of the strain-specific gene clusters encoded by S. sp. S4 are the hybrid type I / type III polyketide kendomycin and the type II polyketide fredericamycin [38]. The remaining 13 biosynthetic gene clusters harboured by S. albus strains are unknown. The antiSMASH 2.0-implementation of MultiGeneBlast [44] was used to identify the closest relative for each strain-specific gene cluster. Organisms harbouring putative orthologous gene clusters and the associated MultiGeneBlast score are reported in Table 5. A possible orthologue was identified for all but one strain-specific gene cluster specifying a bacteriocin harboured by S. albus S4 (Table 5).
Table 5. Strain-specific gene clusters encoded by Streptomyces albus.
| Predicted biosynthetic system | Predicted product | Coordinates | Closest relative (Accession, cumulative MultiGeneBlast Score) |
|---|---|---|---|
| S. albus J1074 | |||
| NRPS / Oligosaccharide | Paulomycin | 684407..718548 | Streptomyces pristinaespiralis ATCC 25486, NZ_CM000950.1, 3720 |
| Bacteriocin | Unknown | 2560714..2571226 | Streptomyces sp. SPB74, NZ_GG770539.1, 683 |
| S. sp. PVA-94-07 | |||
| Lantipeptide | Unknown | 1862908..1885125 | Streptomyces sp. SPB74, NZ_GG770539.1, 4533 |
| S. sp. GBA 94-10 | |||
| Other* | Unknown | CM002272.1 609..41190 | Streptomyces sp. W007, NZ_AGSW01000123.1, 3201 |
| S. sp. SM8 | |||
| Type II PKS | Unknown | NZ_AMPN01000020.1 1..23214 | Streptomyces pristinaespiralis ATCC 25486, NZ_CM000950.1, 8748 |
| Butyrolactone | Unknown | NZ_AMPN01000075.1 188..10692 | Streptomyces lavendulae, AB434932.1, 1901 |
| Bacteriocin | Unknown | NZ_AMPN01000145.1 80715..91067 | Streptomyces hygroscopicu s, NZ_GG657754.1, 555 |
| S. sp. LaPpAH-202 | |||
| NRPS | Unknown | NZ_KB890705.1 132033..188388 | Streptomyces sp. ATCC 700974, FN545130.1, 8976 |
| NRPS | Unknown | NZ_KB890711.1 46489..98964 | Streptomyces hygroscopicus ATCC 53653, NZ_GG657754.1, 6631 |
| Type I PKS** | Unknown |
NZ_KB890711.1 107544..132946 NZ_KB890725.1 1..5945 NZ_KB890733.1 334378..366455 NZ_KB890733.1 366455..396056 |
Streptomyces tsukubaensis NRRL18488, AJSZ01000000, *** |
| S. sp. S4 | |||
| Other | Unknown | CADY01000053.1 1..9842 | Frankia sp. EUN1f, NZ_ADGX01000038.1, 551 |
| Type I PKS / Type III PKS | Kendomycin | CADY01000062.1 1..35064 | Streptomyces griseus XylebKG-1, NZ_GL877172.1, 4372 |
| NRPS | Mannopeptimycin-like | CADY01000178.1 54079..109040 | Streptomyces venezuelae ATCC 10712, FR845719.1, 5568 |
| Butyrolactone | Unknown | CADY01000186.1 1..29569 | Streptomyces sp. W007, NZ_AGSW01000147.1, 1300 |
| Bacteriocin | Unknown | CADY01000195.1 1..2452 | — |
| Type II PKS | Fredericamycin | CADY01000200.1 57212..86518 | Streptomyces griseus, AF525490.2, 11189 |
*Denotes a gene cluster encoded on a plasmid
**Denotes a gene cluster composed of partial antiSMASH gene clusters that likely represent a single cluster according to NaPDoS analysis
***AntiSMASH did not report a cumulative MultiGeneBlast score, because the gene cluster is spread over multiple contigs
Conclusions and perspectives
The genomes of S. albus isolates have been sequenced more than any other species of Streptomyces. The putative biosynthetic capabilities of six S. albus strains were analysed here, which identified a core secondary metabolome specified by 18 biosynthetic gene clusters as well as 14 auxiliary gene clusters and 16 strain-specific gene clusters. The products of 29 of the 48 gene clusters identified in this analysis are unknown, representing an attractive reservoir of compounds that may have useful medicinal or industrial applications or may otherwise comprise a chemically interesting scaffold. The flurry of recent analyses investigating secondary metabolism of S. albus strains have collectively resulted in assigning products to 15 of the 25 gene clusters encoded by S. albus J1074, rivaling what is known about S. coelicolor which has been rigorously studied for over half a century [45]. Robust and thorough bioinformatics approaches that prioritise taxonomic uniqueness of producing organisms and novel gene clusters will drive the discovery of new compounds. However, many of the gene clusters encoded by streptomycetes are not expressed under normal laboratory growth conditions. In order to therefore maximally exploit the biosynthetic potential of these organisms the regulation of biosynthetic systems must be refactored in the native host or cloned and heterologously expressed variants whose expression has been engineered. These efforts are aided by recent advances in the selective cloning of large genomic DNA inserts [46,47] and will be further aided by the decreasing price of custom DNA synthesis and the ability to assemble these fragments in yeast [48].
Materials and Methods
Phylogenetic analyses
The Genomic Blast service hosted by NCBI was used to query all complete and draft genomic sequences from bacteria taxonomically classified as Streptomyces spp. (taxid = 1883) with partial DNA sequences for atpD, gyrA, recA, rpoB and trpB, which corresponded to the sequences targeted by oligonucleotide primers used by [26,27] to infer a multilocus phylogeny. FASTA sequence files for relevant accession numbers were downloaded from Genbank using Batch Entrez and BedTools 2.19.0 [49] was used to extract nucleotide sequence ranges reported in the blast search into a multifasta file. The BioPerl [50] script shortenID.pl (http://nebc.nox.ac.uk/scripts/parse/shortenID.pl ) written by Bela Tiwari, NERC Environmental Bioinformatics Centre,was used to shorten headers for FASTA entries and the BioPerl script split_multifasta.pl (http://iubio.bio.indiana.edu/gmod/genogrid/scripts/split_multifasta.pl ) written by the Genome Informatics Lab at Indiana University was used to generate individual FASTA files from the resulting multifasta output from BedTools. DNA sequences were aligned using eight iterations of the MEGA 5.2.2 implementation of Muscle [51] and were trimmed to the same length (including gaps) and subsequently concatenated in the order: aptD-gyrB-recA-rpoB-trpB. Phylogenetic relationships were inferred from the concatenated sequences by approximate maximum likelihood analysis using FastTree 2.1.7 [52]. Mycobacterium tuberculosis H37Rv was used as an outgroup and MEGA 5.2.2 was used to visualise and edit the tree. Concatenated aptD-gyrB-recA-rpoB-trpB sequences were grouped into operational taxonomic units (OTUs) using the MacQiime v1.80 implementation of UCLUST [28,53] with a shared identity threshold of 97%.
Analysis of secondary metabolite gene clusters
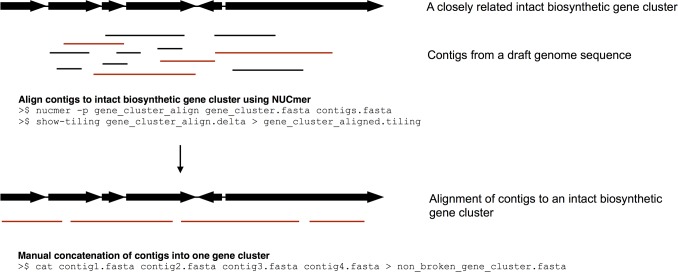
Genome sequences analysed here were downloaded from Genbank or EMBL (see Table 1 for accessions) and putative biosynthetic gene clusters for secondary metabolites were identified using the default settings in the web implementation of antiSMASH 2.0 [29] and the nucleotide sequence for each gene cluster was extracted from the outputted Genbank files using EMBOSS utility seqret [54]. The large number of contigs in some draft genomes caused antiSMASH 2.0 to identify numerous broken or incomplete gene clusters. This was a particular problem with polyketide synthase gene clusters. In order to minimise the impact of broken gene clusters on this analysis, the gene clusters identified from the fully sequenced genome of S. albus J1074 were used as a reference for NUCmer [55] alignments of gene clusters from draft genome sequences. A diagrammatic workflow of this approach is displayed in Fig. 2. Gene clusters from draft genomes that aligned to the same S. albus J1074 gene cluster were subsequently concatenated into a single FASTA file and considered a single gene cluster. A For gene clusters in which S. albus J1074 did not harbour a homologous cluster, NaPDoS [56] was used to identify and extract ketosynthase domains from gene clusters identified by antiSMASH 2.0. The resulting amino acid sequences were aligned by the Geneious 7.1.5 implementation of Muscle (eight iterations) and a neighbour-joining phylogenetic tree was inferred from the alignment using the Geneious 7.1.5 tree builder with a Jukes-Cantor distance model (not shown). A customised blast database was generated using Blast 2.2.29+ [57] and a combination of blast analysis and whole gene cluster alignments using Mauve 2.3.1 [58] were used to both further refine broken gene clusters in draft genome sequences and to ascertain the conservation of secondary metabolite gene clusters across the S. albus clade. Self vs. self blastn analyses were used to identify and remove duplicate gene clusters.
Figure 2. Diagrammatic workflow of the NUCmer approach used to piece together a biosynthetic gene clusters spread over more than one contig.
NUCmer is part of the MUMmer [55] and can be downloaded from http://sourceforge.net/projects/mummer/. NUCmer will align contigs from draft genomes to an intact gene cluster with high shared nucleotide identity. Commands used to perform an analysis of this type are given. Black arrows represent a biosynthetic gene cluster; black and red lines represent contigs in a draft genome sequence.
Supporting Information
(PDF)
(PDF)
Acknowledgments
I thank Matt Hutchings for his constructive comments of this work and two anonymous reviewers whose suggestions improved this manuscript. I also thank Jayne Louise Gifford for her support throughout this project.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The New Lecturer Startup Fund provided by the University of Leeds financially supported this work. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75: 311–335. 10.1021/np200906s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flärdh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Micro 7: 36–49. 10.1038/nrmicro1968 [DOI] [PubMed] [Google Scholar]
- 3. Davies J, Spiegelman GB, Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9: 445–453. 10.1016/j.mib.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 4. Challis GL, Hopwood DA (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA 100 Suppl 2: 14555–14561. 10.1073/pnas.1934677100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Medema MH, Breitling R, Bovenberg R, Takano E (2010) Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Micro 9: 131–137. [DOI] [PubMed] [Google Scholar]
- 6. Doroghazi JR, Metcalf WW (2013) Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14: 611 10.1186/1471-2164-14-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, et al. (2014) Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora . Proc Natl Acad Sci USA 111: E1130–E1139. 10.1073/pnas.1324161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR et al. (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10: 963–968. 10.1038/nchembio.1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ian E, Malko DB, Sekurova ON, Bredholt H, Rückert C, et al. (2014) Genomics of Sponge-Associated Streptomyces spp. Closely Related to Streptomyces albus J1074: Insights into marine adaptation and secondary metabolite biosynthesis potential. PLoS ONE 9: e96719 10.1371/journal.pone.0096719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viegelmann C, Margassery L, Kennedy J, Zhang T, O’Brien C, et al. (2014) Metabolomic Profiling and Genomic Study of a marine sponge-associated Streptomyces sp. Marine Drugs 12: 3323–3351. 10.3390/md12063323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan L, Liu Y, Li Z, Baumann HI, Kleinschmidt K, et al. (2011) Draft Genome Sequence of the Marine Streptomyces sp. Strain PP-C42, Isolated from the Baltic Sea. J Bacteriol 193: 3691–3692. 10.1128/JB.05097-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barke J, Seipke RF, Grüschow S, Heavens D, Drou N, et al. (2010) A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus . BMC Biol 8: 109 10.1186/1741-7007-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schoenian I, Spiteller M, Ghaste M, Wirth R, Herz H, et al. (2011) Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci USA 108: 1955–1960. 10.1073/pnas.1008441108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanshew AS, McDonald BR, Díaz Díaz C, Djieto-Lordon C, Blatrix R, et al. (2014) Characterization of Actinobacteria Associated with Three Ant–Plant Mutualisms. Microb Ecol: 1–12. 10.1007/s00248-014-0469-3 [DOI] [PubMed] [Google Scholar]
- 15. Chater KF, Wilde LC (1976) Restriction of a bacteriophage of Streptomyces albus G involving endonuclease SalI. J Bacteriol 128: 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S-Y, Zhao P, Igarashi M, Sawa R, Tomita T, et al. (2009) Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene Cyclase and two p450 hydroxylases. Chem Biol 16: 736–743. 10.1016/j.chembiol.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 17. Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, et al. (2005) Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus . J Am Chem Soc 127: 16442–16452. 10.1021/ja054376u [DOI] [PubMed] [Google Scholar]
- 18. Feng Z, Wang L, Rajski SR, Xu Z, Coeffet-LeGal MF, et al. (2009) Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorganic & Medicinal Chemistry 17: 2147–2153. 10.1016/j.bmc.2008.10.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Makitrynskyy R, Rebets Y, Ostash B, Zaburannyi N, Rabyk M, et al. (2010) Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37: 559–566. 10.1007/s10295-010-0701-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, et al. (2007) Molecular basis for chloronium-mediated meroterpene cyclization: cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J Biol Chem 282: 16362–16368. 10.1074/jbc.M611046200 [DOI] [PubMed] [Google Scholar]
- 21. Gullon S, Olano C, Abdelfattah MS, Brana AF, Rohr J, et al. (2006) Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl Environ Microbiol 72: 4172–4183. 10.1128/AEM.00734-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lombó F, Velasco A, Castro A, la Calle de F, Braña AF, et al. (2006) Deciphering the Biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. ChemBioChem 7: 366–376. 10.1002/cbic.200500325 [DOI] [PubMed] [Google Scholar]
- 23. Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A (2014) Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics 15: 97 10.1186/1471-2164-15-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, et al. (2011) Phylogenetic study of the species within the family Streptomycetaceae. Antonie van Leeuwenhoek 101: 73–104. 10.1007/s10482-011-9656-0 [DOI] [PubMed] [Google Scholar]
- 25. Doroghazi JR, Buckley DH (2010) Widespread homologous recombination within and between Streptomyces species. ISME J 4: 1136–1143. 10.1038/ismej.2010.45 [DOI] [PubMed] [Google Scholar]
- 26. Guo Y, Zheng W, Rong X, Huang Y (2008) A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol 58: 149–159. 10.1099/ijs.0.65224-0 [DOI] [PubMed] [Google Scholar]
- 27. Kaltenpoth M, Yildirim E, Gurbuz MF, Herzner G, Strohm E (2012) Refining the Roots of the beewolf-Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl Environ Microbiol 78: 822–827. 10.1128/AEM.06809-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7: 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, et al. (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41: W204–W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olano C, García I, González A, Rodriguez M, Rozas D, et al. (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7: 242–256. 10.1111/1751-7915.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imbert M, Béchet M, Blondeau R (1995) Comparison of the main siderophores produced by some species of Streptomyces . Curr Microbiol 31: 129–133. [Google Scholar]
- 32. Malin G, Lapidot A (1996) Induction of synthesis of tetrahydropyrimidine derivatives in Streptomyces strains and their effect on Escherichia coli in response to osmotic and heat stress. J Bacteriol 178: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seipke RF, Loria R (2009) Hopanoids are not essential for growth of Streptomyces scabies 87–22. J Bacteriol 191: 5216–5223. 10.1128/JB.00390-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takano H, Obitsu S, Beppu T, Ueda K (2005) Light-Induced Carotenogenesis in Streptomyces coelicolor A3(2): Identification of an Extracytoplasmic Function Sigma Factor That Directs Photodependent Transcription of the Carotenoid Biosynthesis Gene Cluster. J Bacteriol 187: 1825–1832. 10.1128/JB.187.5.1825-1832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Izumikawa M, Shipley PR, Hopke JRN, O’Hare T, Xiang L, et al. (2003) Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 30: 510–515. 10.1007/s10295-003-0075-8 [DOI] [PubMed] [Google Scholar]
- 36. Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, et al. (2004) The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor . Proc Natl Acad Sci USA 101: 11448–11453. 10.1073/pnas.0404220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gerber NN, Lechevalier HA (1965) Geosmin, an earthy-smelling substance isolated from actinomycetes. Appl Environ Microbiol 13: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seipke RF, Barke J, Brearley C, Hill L, Yu DW, et al. (2011) A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus . PLoS ONE 6: e22028 10.1371/journal.pone.0022028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seipke RF, Patrick E, Hutchings MI (2014) Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factor σ (AntA). PeerJ 2: e253 10.7717/peerj.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seipke RF, Hutchings MI (2013) The regulation and biosynthesis of antimycins. Beilstein J Org Chem 9: 2556–2563. 10.3762/bjoc.9.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gürtler H, Pedersen R, Anthoni U, Christophersen C, Nielsen PH, et al. (1994) Albaflavenone, a sesquiterpene ketone with a zizaene skeleton produced by a streptomycete with a new rope morphology. J Antibiot 47: 434–439. 10.7164/antibiotics.47.434 [DOI] [PubMed] [Google Scholar]
- 42. Shigemori H (1992) MA, Bae, Yazawa K., Sasaki T., Kobayashi Y.. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai J Org Chem 57: 4317–4320. [Google Scholar]
- 43. Braña AF, Rodríguez M, Pahari P, Rohr J, Gacía LA, et al. (2014) Activation and silencing of secondary metabolites in Streptomyces albus and Streptomyces lividans after transformation with cosmids containing the thienamycin gene cluster from Streptomyces cattleya. (2014). Arch Microbiol 196: 345–355. 10.1007/s00203-014-0977-z [DOI] [PubMed] [Google Scholar]
- 44. Medema MH, Takano E, Breitling R (2013) Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30: 1218–1223. 10.1093/molbev/mst025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Craney A, Ahmed S, Nodwell J (2013) Towards a new science of secondary metabolism. J Antibiot 66: 387–400. 10.1038/ja.2013.25 [DOI] [PubMed] [Google Scholar]
- 46. Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, et al. (2014) Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci USA 111: 1957–1962. 10.1073/pnas.1319584111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu J, Bian X, Hu S, Wang H, Huang F, et al. (2012) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30: 440–446. 10.1038/nbt.2183 [DOI] [PubMed] [Google Scholar]
- 48. Shao Z, Luo Y, Zhao H (2011) Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol Biosyst 7: 1056–1059. 10.1039/c0mb00338g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stajich JE (2002) The Bioperl Toolkit: Perl Modules for the Life Sciences. Genome Research 12: 1611–1618. 10.1101/gr.361602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol and Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Price MN, Dehal PS, Arkin AP. (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 54. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 55. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ziemert N, Podell S, Penn K, Badger JH, Allen E, et al. (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7: e34064 10.1371/journal.pone.0034064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- 58. Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5: e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.