Summary
Reconstructed volumes generated by tilt-image electron-microscope tomography offer the best spatial resolution currently available for studying cell structures in situ. Analysis is often accomplished by creating surface models that delineate grayscale contrast boundaries. Here, we introduce a specialized and convenient sequence of segmentation operations for making such models that greatly improves their reliability and spatial resolution as compared to current approaches, providing a basis for making accurate measurements. To assess the reliability of the surface models, we introduce a spatial uncertainty measurement based on grayscale gradient scale length. The model generation and measurement methods are validated by applying them to synthetic data, and their utility is demonstrated by using them to characterize macromolecular architecture of active zone material at the frog’s neuromuscular junction.
Introduction
Tilt-image electron-microscope tomography (EMT) can be used to generate 3D reconstructions of sections from stained, plastic-embedded biological tissue samples (Frank, 1992; Harlow et al., 2001; He et al., 2003; Horowitz et al., 1994; Ladinsky et al., 1999; Lenzi et al., 1999; Martone et al., 1999; McEwen et al., 1986, 1993; Sedzik et al., 1992; Taylor et al., 1999; Woodcock et al., 1991). The volume reconstructions make it possible to study the structure of cellular components within the depth of the sections, acquiring information about sizes, shapes, and relationships that cannot be obtained in any other way. However, analysis is often hindered by the abundance of structures, the complexity of their shapes and relationships, staining inhomogeneity, and noise. To alleviate these problems, it is useful to form 3D models of the components-of-interest within the reconstructions. Surface models, which represent the boundary between components and the space surrounding them, are commonly used.
Because of the complexity and noise, most research in EMT has made use of manual surface-model generation methods. Typically, virtual slices are formed through a reconstruction and the boundary of each component-of-interest is traced in the slices in which it appears. The traces are then interpolated to create a 3D surface (Frank et al., 1996; Hessler et al., 1992; Kremer et al., 1996; Li et al., 1997; Marko and Leith, 1996; Perkins et al., 1997). Although this method reveals the general shape and organization of logically distinct cellular components, the tedious and subjective nature of manual tracing makes it difficult to generate models that reliably represent the components at the full spatial resolution of the reconstructed volume.
More sophisticated model-generation methods are also available for EMT. One EMT software package, IMOD (Kremer et al., 1996), permits the creation of a model contour at a particular isodensity level on a series of virtual slices. General purpose commercial software applications such as Amira (ZIB, Indeed-Visual Concepts GmbH, Berlin, Germany) and AVS (Advanced Visual Systems Inc., Waltham, MA) offer a plethora of alternative segmentation, visualization, and analysis methods. While these software packages each have their advantages and limitations, there remains a substantial need for additional approaches that offer improvements in speed, convenience, and accuracy.
Here, we present a set of 3D image processing ways-and-means, specifically designed for EMT data volumes, which provide convenient and reliable generation of full-resolution surface models. Independently controllable, 3D surface models of distinct structural components are created using a two-step, dual-resolution iso-density volume-of-interest (IVOI) approach. The first step is a low-resolution slice-by-slice segmentation that produces small volumes-of-interest (VOIs) that enclose, but do not precisely delineate, individual components. The second step forms an isodensity surface as a full-resolution model of the structural component enclosed by each VOI. Because the initial IVOI segmentation step is error tolerant, it can be performed manually, or by using interactive, semiautomatic methods. To assist both manual and automatic segmentation, we describe a parametric-spline path generation method. We also introduce an active-contour method designed to automatically segment the membranous structures that are common in tissues imaged using EMT. Finally, we introduce a measurement, spatial uncertainty, which quantifies the spatial reliability of a surface model based on the local grayscale noise and gradient scale length.
Our methods were validated using a synthetic volume that simulated a typical EMT reconstructed tissue section. The simulated volume contained cylinders, spheroids, and a folded sheet which represent the most common shapes of cellular components: for example filaments, vesicles, and a plasma membrane, respectively. Surface texture was added to these simulated structures to assess the accuracy of the model generation and measurement methods. Various amounts of noise were also added to the volume to assess the performance of the segmentation methods for a range of experimental data quality, and to quantify the accuracy of the spatial uncertainty measurement. We also show how our methods were used to generate and analyze surface models from EMT reconstructions of tissue sections. The sections were from fixed, stained, and plastic-embedded active zones, i.e., neurotransmitter release sites, in axon terminals of the frog’s neuromuscular junction. The active zones are composed of filamentous cytoplasmic macromolecules, vesicles, and plasma membrane. We have briefly described elsewhere (Harlow et al., 2001) the use of our methods for model generation in the analysis of active zone structures in these sections. Here, we provide a full account of the model-generation methods, use the spatial-uncertainty metric to verify the reliability of our models, and apply some example measurements to IVOI models of active zone components that confirm our understanding of synaptic structure.
Methods and Results
Segmentation
In the dual-resolution IVOI approach, the goal of segmentation is to create VOIs that enclose distinct structures, including both stained structures and adjacent background to facilitate subsequent boundary detection. All segmentation is performed using virtual slices through the reconstructed volume at various user-defined positions and orientations. Controls for segmentation and slice display are provided by a 2D GUI (Figure 1A).
Figure 1. 2D and 3D Graphical User Interfaces (GUI).
(A) The 2D GUI shows orthogonal slices through the volume along the three principal axes. The location of the cut planes is indicated by the red crosshairs and can be changed using a mouse. The volume can be rotated arbitrarily to optimize the contrast of stained structures on the cut-plane images.
(B) The 3D GUI provides control of the generation and display of the models. The display window offers real-time control of the orientation and scaling of the models. The line graphics and associated controls on the left side of the GUI are used to adjust the isodensity value for each surface model.
Membranes are segmented using an interactive, partially automated method that is a novel form of active-contour (“snake”) segmentation combining grayscale morphology, thinning, and path formation using parametric splines. Segmentation is initiated by choosing a slice and slice-orientation that most clearly shows the structure of interest as a contrast boundary. Two topologies of membranes are permitted: open (extending beyond the slice boundaries, e.g., plasma membranes) and closed (e.g., synaptic vesicle membranes). The user marks points along the membrane using a mouse or trackball (Figure 2A). As each point is marked by clicking the input device button, a cubic-spline path is interpolated parametrically between the points, that is, the x and y coordinates of the path are separately interpolated together with single-pixel precision. This anchor path can be edited by adding or deleting points. Open-topology anchor paths are completed by extending them to the closest image boundary. Closed-topology anchor paths are completed by replicating the initial point as the end-point.
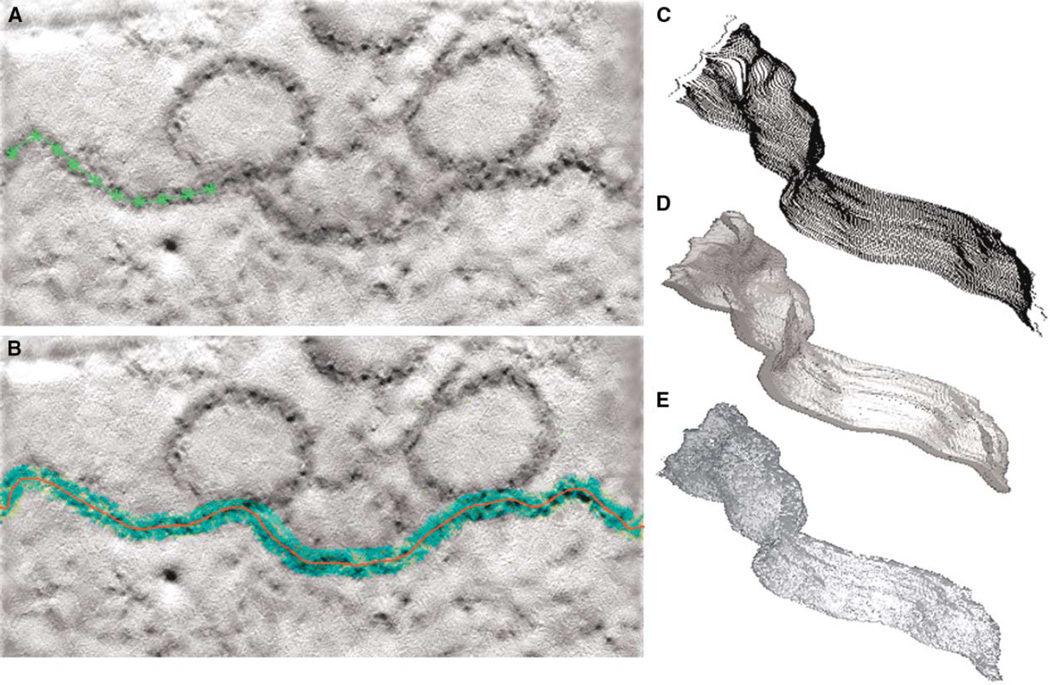
Figure 2. Steps in the Generation of a Surface Model Using a Semiautomatic IVOI Approach.
(A) A user chooses a slice where a structural component-of-interest is clearly distinguished (here the presynaptic plasma membrane of a frog neuromuscular junction). Points are marked along its length and interpolated together to form a continuous path (green points and line).
(B) The blue region shows the pixels corresponding to the user-defined grayscale range within the search-region width of the anchor path; the red line shows the calculated segmentation path.
(C) The complete segmentation is a 3D ensemble of points produced by propagating the segmentation path through the volume.
(D) The volume-of-interest (VOI) is produced by dilation of these points.
(E) An isodensity surface model is created from the stained structure within the VOI.
In the next stage of segmentation, the user selects a grayscale range and a search-region width. For convenience, the code sets default values for the grayscale range based on the mean and standard deviation of the grayscale values along the anchor path. A search is then performed in the vicinity of the anchor path for pixels that are within the specified width; pixels within the range are shown graphically using a false-color overlay (Figure 2B). The grayscale range and search-width are set so that the well-stained, high-confidence regions of the entire membrane are selected.
Next, the selected-pixel regions of the membrane are thinned to find the image vertices that lie along their median axis. To create a new path, we now identify each of the anchor path vertices with its nearest neighbor within the set of thinned vertices. If multiple thinned vertices correspond to a single anchor-point vertex, the mean thinned-vertex position is chosen. If multiple anchor-path vertices correspond to a single thinned vertex, only that correspondence that occurs first along the anchor path is retained, and the others are discarded. The same parametric cubic-spline method described above is then used to interpolate this new set of points into the segmentation path. To enforce a degree of stiffness in the segmentation, this path is smoothed parametrically by convolution with a smoothing kernel applied independently to the x and y vertex coordinate values; the degree of smoothing can be adjusted by the user (Figure 2B).
The user now initiates automatic propagation of the segmentation. Starting with the slice containing the initial segmentation, the segmentation path becomes the anchor path on an adjacent slice, and the subsequent pixel search and path movement described above are repeated to produce a new segmentation path for that slice. The process is repeated in both directions to deter-mine segmentation paths for all slices in the volume. The propagation in either direction is terminated when the pixel search fails. The results of the propagation can be subsequently edited by trimming segmented slices from either direction of propagation, and by setting additional anchor paths and repropagating the new paths along a new range of orthogonal slices.
The resulting set of segmentation paths is a collection of points that define a smoothed representation of the geometric center of the membrane (Figure 2C). This is converted to a VOI by dilating all these points using a spherical 3D structuring element with a user-definable diameter (default value is the search-region width for the segmentation). Thus, the VOI encloses both the well-stained membrane and some adjacent unstained or lightly stained background (Figure 2D).
For lightly stained objects exhibiting complex topologies, such as macromolecular filaments, we use a segmentation method in which a closed path is manually created to define the VOI on each slice. The path can be marked by parametric cubic-spline interpolation, as before, or by piecewise-linear interpolation. The user’s goal is to enclose both the stained structure and some amount of the adjacent background around it. The path on the first slice can be propagated without modification to an adjacent slice where it can be edited by adding or deleting points. The VOI is literally defined by combining the interior regions of all marked slices.
Segmentations can also be created from the logical union or intersection between or among any number of previously created VOIs. These operations enable analysis of relationships among structures contained within the VOIs. Specifically, union allows the combination of multiple structures to form groups or wholes, and intersection facilitates identification of portions of structures that meet or contact one another.
Surface-Model Generation
To create a representation of the surface of a stained object within a VOI, vertex points and a connectivity graph are calculated at a particular gray level. This surface provides a grayscale driven representation of the stained structure that maximizes spatial resolution, completing the IVOI approach to model generation.
The default choice of gray level is obtained using the VOI’s grayscale cumulative-distribution function. The cumulative-distribution function, normalized to unity, represents the fraction of VOI voxels that will be interior to the corresponding interpolated surface. By default, we choose a surface that encloses 60% of the VOI volume (Figure 2E).
The surface gray level can also be adjusted using graphical controls. In general, there is a plainly visible range of appropriate values for the stain density: if the level is too high (too light), the surface becomes clipped at the edges of the VOI; too low, and the surface appears to disintegrate. The most accurate representation of the biological structure is somewhere between these two extremes. The surface gray level is usually adjusted to minimize the spatial uncertainty, as described below.
Visualization
The IVOI surface models can be visually analyzed using an extensive collection of tools provided by the 3D GUI (Figure 1B). For example, models can be interactively rotated, scaled, and translated. Moreover, each model can be assigned a different name, color, and opacity. To make their spatial relationships more obvious, the user can choose to render all or only a subset of the models. The GUI also provides a flexible lighting model and spin animation about an arbitrary axis to assist in visual analysis, and the results of appropriate quantitative analysis steps can be superimposed on the rendered surfaces (Figure 3B-3D).
Figure 3. IVOI Surface Models with Measurements for Biological Data.
(A) Schematic of the active zone at the frog’s neuromuscular junction showing the presynaptic membrane (gray), docked synaptic vesicles (blue), and the most superficial 15 nm of the AZM, consisting of beams (brown gold), ribs (yellow gold), and pegs (orange gold). Cation channels in the presynaptic membrane are shown in green.
(B) Spatial uncertainty of ribs and beams. An ensemble of models of ribs and beams in UC-1, showing the spatial uncertainties as a color overlay.
(C) Spatial uncertainty of ribs and pegs. A portion of the active zone in MPI-10 is shown in the transverse plane to reveal the spatial relationships among a single rib (yellow gold), its pair of associated pegs (orange gold), and the vesicle and presynaptic membranes (blue and gray, respectively). Inset shows spatial uncertainty overlay of the rib-peg assembly (same scale as Figure 3B).
(D) Spatial proximity of rib-vesicle contacts with the presynaptic membrane. Models for the pair of docked vesicle in MPI-9 are marked with a vertex color overlay indicating spatial proximity to the presynaptic membrane. Gold patches show contact regions among the vesicles and the ribs. About three-fourths of each vesicle was included in the tissue section.
(E) Method used to determine the longitudinal and transverse spacings of the pegs. Horizontal view of the of the pegs (orange gold) upon the cytoplasmic surface of presynaptic membrane (gray) in MPI-9. The red spots indicate the peg centroids. Black line segments illustrate the longitudinal distances between each pair of pegs; blue segments illustrate the transverse distances.
Measurement
To quantify the accuracy of surface models, we calculate the spatial uncertainty at each vertex. Our approach takes the view that uncertainty in the grayscale value drives a spatial uncertainty in the physical position of each isodensity surface model vertex. For example, if noise were to make the gray values larger near some vertex of a surface model, that vertex would move inward, and vice versa. The sensitivity of each vertex location to the noise is proportional to the normal component of the local spatial gradient. If the gradient is large, a noise-induced shift in position will be relatively small, and vice versa. The calculation therefore begins by determining the grayscale spatial gradients, ∇⃗S, in the neighborhood of each vertex location, x⃗. ∇⃗S is calculated by convolving three 3×3×3 kernels, each containing nine replicas of the vector {−0.5 1 −0.5} oriented along the x, y, and z axes, to generate the respective components of the vector gradient. This is the smallest symmetric kernel available to calculate the gradient; larger kernels did not perform as well, tending to underestimate the magnitude of the local gradient. The grayscale noise, σg, is estimated by calculating the standard deviation in a relatively blank (transmissive) portion of volume, and this noise is assumed to be stationary throughout.
The isodensity surface calculation produces the outward vector normal, n⃗ (x⃗), at every vertex. Altogether, we define the spatial uncertainty as:
This equation can be rewritten to emphasize its geometrical interpretation. The denominator term is related to the gradient scale length of the grayscale density along the normal to the surface model, ln = S/(n⃗•∇⃗S). If we combine this relationship with the signal-to-noise ratio, R = S/σg, we get the form, σn = ln/R. Thus, our spatial uncertainty measure is simply the normal component of the gradient scale length divided by the signal-to-noise ratio.
To visualize the spatial uncertainty, the uncertainty values are scaled onto a user-selectable color map and displayed as an overlay on the surface (Figure 3B and 3C). The user can choose different color tables and scaling parameters, so that only a subset range of uncertainty values is spanned by the overlay. Generally, a small subset of the vertices span a region of the volume where the gradient points inward rather than outwards because of noise or structural complexity. These vertices are marked with a user-definable color.
To quantify the spatial relationships among surface models, we calculate the nearest-neighbor distances between their vertices. This calculation begins by defining one object as a reference and one or more other objects as destinations. For every vertex on a destination object, we calculate the distance to all of the reference-object vertices; the smallest distance is then associated with that destination vertex. The ensemble of vertex distances are then scaled onto a color table and overlaid onto the destination object (Figure 3D).
To measure the positions, surface areas, and volumes of stained structures, we calculate various geometric moments. Specifically, the position of the model is defined as the centroid of its vertices (Figure 3E). The area of a surface model is calculated by summing the area of each of its constituent polygons. For closed surfaces, the volume is calculated by counting the number of interior voxels.
To understand the capabilities of our modeling methods, we made estimates of spatial resolution and noise for the experimental data. The resolution of the reconstructed volume was measured by characterizing the contrast profile of relatively continuous segments of trilaminar membrane measured along a surface normal obtained from its corresponding IVOI model. By treating the outer leaflets of the membranes as a pair of sharp edges, we could calculate an upper limit for the full-width-at-half-maximum resolution by measuring their separation and the contrast between the edge and center regions and assuming a Gaussian form for the local line-spread function. Specifically, if we define the unattenuated gray value as S0, the gray value corresponding to the leaflet as S1, the gray value between the two leaflets as S2, and the measured spacing along the normal between the two leaflets as d, the resolution is
(data not shown). Resolution varied depending upon orientation, ranging from <1 nm in the plane of the tilt axis to 2–4 nm perpendicular to the tilt axis, as expected from the theory of tilt-image tomography (Frank, 1992). In our recent data sets, where 141 tilt images are typically taken for a 50–60 nm thick sample, the spatial resolution was at least 2 nm throughout the volume. To estimate the noise in the sample, we chose a small VOI within a fully reconstructed portion of the volume that is as bright and featureless as possible, and within this VOI we calculated the mean, Gbright, and standard deviation, σg of the gray values. We also obtained a measure of minimum gray value, Gdark, by picking off the value corresponding to 1% on the cumulative distribution function of the volume. The noise-to-contrast ratio was then defined as NCR ≅ σg/(Gbright - Gdark). Using this method, we observe NCR levels in the range of 5–20%.
Simulations
We created a simulated volume containing representations of synaptic vesicles, a plasma membrane, and macromolecular filaments (see Figure 4A). Voxel size was set to 1 nm, a reasonable value for actual reconstructed EMT volumes. Vesicles were simulated by creating isodensity surfaces from the sum of a few low-order spherical harmonics to create a spheroidal shape with roughly 50 nm diameter. To assess the ability of our surface models to delineate fine-scale structure, surface texture was added by summing additional high-order spherical harmonics with amplitudes in the range of 2–4 nm. Superposing two such spheroids with radii offset by 5 nm simulated the trilaminar character of stained membranes. A plasma membrane was simulated using very similar methods but with Fourier rather than spherical harmonics. Macromolecular filaments were simulated using simple, homogeneous solid cylinders. The gray level of each vesicle and the membrane were set to a small value (e.g., 10% of maximum) to simulate the dark stain typically observed in tissue volumes. In typical reconstructed EMT volumes, stain levels vary significantly from object-to-object. Accordingly, gray values were randomly varied for the vesicle and plasma membranes over a range of ±20%. The filamentous components were given lighter gray value (e.g., 40% of maximum), with 50% component-to-component random variability. The entire simulated volume was smoothed slightly (3×3×3 boxcar, repeated twice) to reduce quantization artifacts. For clarity, we did not project and then reconstruct this volume before applying our analysis procedures. This was necessary to permit the quantitative interpretation of the segmentation results, avoiding confusion between segmentation errors and reconstruction artifacts.
Figure 4. Qualitative Performance of the IVOI Approach with Synthetic Data.
Models were generated from an artificial volume simulating components at the active zone of a neuromuscular junction. The volume contained a folded sheet (presynaptic membrane), four spheroids (vesicles), and several lower-contrast cylinders (AZM). Membrane surface texture was 2–4 nm. Left images (A, C, and E) show an oblique 3D rendering of the IVOI surface models; right images (B, D, and F) show a 2D cross-section of the grayscale volume with a red overlay marking vertices of the surface models. The noise-free panels (A and B) show all of the simulated structures and surface texture. The panels corresponding to 20% noise (C and D), an upper limit for most biological data, are similar to the noise free case, although the surface texture and lightly stained AZM are somewhat distorted. The panels corresponding to 50% noise (E and F), an unusually large value for biological data, continue to show the gross structures, but the surface texture and some of the AZM are obscured.
Various amounts of normally distributed noise were added to simulate different reconstructed volume qualities. We chose to model the noise as white, although actual noise in tomographic reconstructions of EMT data has a more complex character. In projection imaging methods, the sampling density varies linearly with spatial frequency, so we expect noise levels to increase with spatial frequency. However, other noise mechanisms have different spectral properties; for example, stain inhomogeneity would have a dominantly low spatial-frequency character. In general, these mechanisms do not have well-characterized statistics, but white noise is a reasonable choice to reflect the statistical character of a superposition of many noise processes.
Models of Simulated Structures
The synthetic volume contained a plasma membrane, four synaptic vesicles, and two pairs of solid, cylindrical connecting rods that simulated macromolecular filaments (Figure 4). When we applied the IVOI approach to create surface models of these various elements, the simulated structures were accurately modeled (Figure 4A and 4B). Each surface model followed the contrast boundary at the edges of each object, e.g., the darkly stained laminae of the simulated membranes.
We added a controlled amount of noise to the simulated volume and repeated the segmentations. In our simulated data with a NCR of 20%, the largest value generally observed in experimental data, all of the gross structure and some of the fine surface texture were still evident on the membranous structures (Figure 4C and 4D). Because the various elements within the volume were assigned different “stain” values, their corresponding surface models are affected differently by the noise. Two of the manually segmented connecting rods were given very low contrast, so they are particularly distorted by the noise. At an unusually high noise level (NCR = 50%), the segmented surface models of the membranous structures reveal only their gross features, while the superimposed fine structure is no longer evident (Figure 4E and 4F). The two lightly stained connecting rods were completely obscured by the noise and could no longer be segmented from the volume.
To quantify the errors in the IVOI surface models produced by the additive noise, we used the proximity-mapping method to calculate the nearest-neighbor separation between vertices in a noisy surface model and the corresponding noise-free surface model. For one object, the upper left vesicle in Figure 4, the errors produced by 20% additive noise are shown as a vertex color map overlay in Figure 5A. The root-mean-square (rms) errors are shown as a function of NCR in Figure 5C. At a noise level of 20%, the error is 0.6 pixels, low enough to resolve some of the fine-scale detail. At a noise level of 50%, the rms error rises to 1.5 pixels, obscuring all of the fine-scale detail.
Figure 5. Quantitative Performance of the IVOI Approach with Synthetic Data.
(A) Proximity map showing errors of a surface model obtained from a noisy (20%) simulated volume.
(B) Spatial uncertainty map on the same object.
(C) The dependence of mean proximity error and median uncertainty as a function of noise level. The median uncertainty tracks and somewhat exceeds the actual mean error, confirming its utility as a reliability metric.
We also applied the spatial uncertainty calculation to the upper left vesicle in the simulated volume (Figure 4). At 20% noise levels, the spatial uncertainties show the same spatial pattern as the actual errors, lower near the poles and larger near the equators (Figure 5B). The greater uncertainty and error near the equator are caused by the larger amount of fine surface detail that was imposed in this region, thus reducing the local grayscale spatial gradients. The spatial uncertainty calculation inevitably produces a few very large values where the surface model is interpolated through a region of small grayscale gradients, and some negative values where the gradients point inward. Consequently, we use the median of the positive values as a more robust metric for the error across the entire surface model. As expected, the median spatial uncertainty increases with the noise level (Figure 5C). The median uncertainty is always somewhat greater than the mean error. Thus, the median spatial uncertainty provides a conservative measure of the reliability of a surface model.
To test the effects of user subjectivity on the semiautomatic version of the IVOI approach, we had three users independently generate surface models of the vesicles and plasma membrane in two of the noisy simulated volumes (10% and 20% NCR). All of the resulting surface models were remarkably similar. Using the proximity mapping method to quantify the differences between the surfaces, we found them to be very low, ≪1 pixel rms.
Models of the Active Zone Components
The active zone of the frog’s neuromuscular junction, when viewed in aldehyde-fixed, heavy-metal stained tissue sections, is composed of three gross structures (Figure 3A): the presynaptic plasma membrane of the axon terminal; a 100 × 1000 nm patch of filamentous macromolecules (active-zone material, AZM), which is attached to the cytoplasmic surface of the plasma membrane and extends about 75 nm into the cytoplasm; and a row of 50 nm diam synaptic vesicles docked on (i.e., held at) the presynaptic membrane on each side of the active zone material. Freeze-fracture replicas from the portion of the plasma membrane adjacent to the AZM reveal linear arrays of macromolecules, which include cation (calcium and calcium-activated potassium) channels paralleling the rows of docked vesicles (Figure 3A) (Heuser and Reese, 1981; Heuser et al., 1979; Robitaille et al., 1993). All of the above structures are believed to be essential for synaptic transmission of the nerve impulse, and they are typically found at synapses throughout the nervous system, although their configurations vary (Harlow et al., 2001; Heuser and Reese, 1977; Katz, 1969).
In our previous study (Harlow et al., 2001), we used the modeling methods described above to examine the macromolecular architecture of the first 15 nm of AZM internal to the presynaptic membrane in three reconstructed active zones designated MPI-9, MPI-10, and UC-1. By applying the semiautomatic IVOI method for individually segmenting the presynaptic membrane and synaptic vesicles, and the manual IVOI method for individually segmenting the more complicated macromolecules in the AZM, we determined from the resulting surface models that there are three classes of AZM macromolecular components based on their position and orientation; we termed these beams, ribs, and pegs (Figure 3A). We also learned that the beams, ribs, and pegs have specific connections. Beams are connected to each other and to ribs, ribs are connected to docked vesicles and pegs, and pegs are connected to the presynaptic membrane in a way suggesting they are linked to the cation channels within the membrane. These structures are evident in the schematic of Figure 3A and in the ensemble of IVOI models in Figure 1B (horizontal view of the active zone: presynaptic membrane, gray; synaptic vesicles, blue; beam, brown gold; ribs, yellow gold; pegs, orange gold).
The manual segmentations in our earlier work (Harlow et al., 2001) were tedious and difficult because the edges of structures were highly irregular and there was significant noise. These factors could have led to significant errors in the models. To obtain an objective measure of the models’ reliability, we have now calculated the spatial uncertainty for the same models using the method described above. The color-map overlay in Figure 3B shows the spatial uncertainty obtained for the 19 ribs and four beams in UC-1. The median uncertainty for all of these components was 0.8 nm, and 90% of the vertices had spatial uncertainties < 2.0 nm. The spatial uncertainties in MPI-9 and MPI-10 were higher because their contrast-to-noise ratio was lower, typically ~5 as compared to ~20 in UC-1. Figure 3C shows a rib and the two pegs linked to it from MPI-10; the median uncertainty for the three structures was 1.4 nm, and 90% of the vertices had spatial uncertainties < 3.3 nm. Median spatial uncertainties for the assemblies of beams, ribs, and pegs were 1.9 nm for MPI-9 (2 beams, 8 ribs, 16 pegs), and 1.6 nm for MPI-10 (2 beams, 8 ribs, 14 pegs). Thus, the spatial uncertainty in all three data sets was sufficiently low to validate the existence of the beam-rib-peg assemblies reported previously.
In our earlier work (Harlow et al., 2001), we observed that each rib contacted the vesicle membrane near the presynaptic membrane, but the curvature of the vesicles and undulations in the presynaptic membrane made it difficult to precisely quantify their separation. We have now performed proximity calculations to measure this separation for the 12 rib-vesicle contact regions in the four docked vesicles in MPI-9 and MPI-10 (Figure 3D). IVOI models of the contact regions were obtained from the intersections of the rib and vesicle VOIs. For each model, proximity values were obtained at every vertex, and the mean value recorded. The mean value for all the rib-vesicle contact regions was 7.5 ± 3.0 nm (SD). Thus, the proximity calculation demonstrated that for MPI-9 and MPI-10, the ribs contacted each vesicle in a narrow band paralleling the presynaptic membrane, and the separation of the band from the membrane was nearly the same from vesicle-to-vesicle.
In our earlier work (Harlow et al., 2001), we suggested that pegs are linked to the linear arrays of cation channels in the presynaptic membrane based on visual inspection of the models from MPI-9 and MPI-10. The peg-membrane contacts were arranged in linear arrays having a particular longitudinal (parallel to the beams) and transverse (perpendicular to the beams) spacings, and the overall configuration of these contacts was similar to that of the macromolecules observed in freeze-fracture replicas from the active-zone’s presynaptic membrane. We have now performed moment calculations to test this hypothesis, measuring the center-to-center spacings of the peg centroids in the longitudinal and transverse directions for data sets MPI-9 and MPI-10 (Figure 3E). These data were compared to similar measurements made on the presynaptic membrane macromolecules in images of freeze-fracture replicas from muscles fixed in the same way as those used for EMT (McMahan and Slater, 1984). The mean longitudinal spacing for the 28 pegs was 19.8 ± 5.0 nm, while the mean transverse spacing was 17.3 ± 6.2 nm (SD). Despite the small sample size, these values are in good agreement with our measurements on a much larger sample of presynaptic-membrane macromolecules: 17.2 ± 3.6 and 17.3 ± 2.5 nm, respectively.
Discussion
We described two methods within a dual-resolution IVOI approach to segmentation. The first was the use of parametric cubic splines to create a smooth interpolation of a sparse set of points. Application of this well-known method provided two advantages: first, as a convenient scheme to interactively define a path, and second, as a critical component of the active-contour segmentation scheme. The second method was a novel approach to membrane segmentation that combines an active-contour concept with thinning. In addition, we described a spatial uncertainty calculation that quantifies the reliability of isodensity surface models generated from these segmentations. Altogether, these methods offer a convenient means to reliably generate high-resolution models of structure from reconstructed EMT volumes.
Previous segmentation approaches have not been satisfactory for high resolution studies of EMT volumes. Active-contour segmentation (or, more generally, level-set segmentation) has seen extensive use in biomedical image segmentation (Klemencic et al., 1998; Kozerke et al., 1999; Suri et al., 2002; Yezzi et al., 1997), but it tends to fail in EMT volumes because of complexity and noise; the segmentation contours are driven from the target surface by the many actual interconnections or by noise. Watershed schemes have also been used extensively to find contours in biomedical images (Grau et al., 2004; Lin et al., 2003; Ortiz de Solorzano et al., 1999; Volkmann, 2002), but these schemes fail in EMT data for similar reasons; complex structure and noise cause oversegmentation or poor boundary detection. The success of our hybrid scheme suggests that it may be possible to devise a level-set method that incorporates a particular topological preference (e.g., 50 nm diam spheroidal vesicles) into the artificial forces that guide the contour’s trajectory. Another useful direction may be the development of 3D skeletonization algorithms (Bajaj et al., 2003; Cooper et al., 2003; Wu and Bourland, 2000). Both of these approaches could be effective for the error-tolerant segmentation step of the IVOI approach.
Models can also be generated from reconstructed EMT volumes using various methods available in commercial 3D image processing software packages such as Amira (ZIB, Indeed-Visual Concepts GmbH, Berlin, Germany) or AVS/Express (Advanced Visual Systems Inc., Waltham, MA). These packages are capable of generating dual-resolution IVOI models, but only by manual means. They also provide interactive automatic segmentation tools designed to find grayscale boundaries. There are several reasons why these tools are often unsatisfactory for high-resolution EMT analysis. Most important, they are not designed to operate in the dual-resolution fashion described above. They attempt to find a boundary based on grayscale level and gradient operations, thus creating VOIs that define structural components rather than enclose them. Consequently, the models generated from these VOIs do not precisely delineate surface boundaries in high-resolution data. Also, the packages are designed for general-purpose 3D image processing. They offer tools optimized for quick visualization and measurement using proprietary methods. By comparison, high-resolution structural analysis of EMT data requires precise, standardized, and extensive metrology of structural features. In general, specific, high-precision segmentation and 3D measurement problems concerning biological systems have required specialized software packages; examples include the segmentation of brain structures from magnetic-resonance images (Teo et al., 1997; Van Essen et al., 2001; Zavaljevski et al., 2000) and cytometry (Bocker et al., 1999; van Vliet et al., 1990). We devised our dual-resolution IVOI approach as a standardized and repeat-able method to generate full-resolution structural models from EMT data.
Application of our methods to simulated data demonstrated their performance. The resulting IVOI models accurately delineated the contrast boundaries of structural components. Noise distorted the boundaries, but both gross structure and 2–4 nm surface texture were evident at noise levels typically observed in our biological EMT data. Virtually identical models were formed by three different users, showing the power of the IVOI approach. Using the simulations, we also demonstrated that the spatial uncertainty metric could effectively assess the reliability of the surface models, providing a conservative estimate that tracks the actual errors. We now routinely use a form of the spatial uncertainty measurement as an automatic optimization metric during segmentation and model generation of biological data (Ress et al., 2003).
The conversion of local values of uncertainty into spatial uncertainty may be useful in the analysis of 3D structural data other than that produced by EMT. For example, in single-particle analysis of isolated macro-molecules a model volume is generated that includes both a mean gray scale density and its uncertainty. These data could be combined to calculate spatial uncertainty for a surface model constructed from the volumetric data. The spatial reliability method could be also useful in evaluating the similarity of models produced by different 3D structural analysis methods (Ofverstedt et al., 1997; Wriggers and Chacon, 2001). For instance, one could evaluate the spatial reliability of an EMT model of a particular macromolecule that appears to be a best visual fit to a model obtained by X-ray crystallography (He et al., 2003). Indeed, the spatial uncertainty calculation may provide a general method to quantify spatial resolution at a contrast boundary in 3D structural biology.
Application of our methods to make models of components of the active zone of the frog’s neuromuscular junction has shown its ability to reveal unique in situ structural information. Our earlier work, published elsewhere without full methodological details (Harlow et al., 2001), revealed macromolecular components and associations that had not been detected by any other imaging method. The discovery of beams, ribs, and pegs and their connections to each other, synaptic vesicles and the presynaptic membrane is now significantly strengthened by this full exposition. Our demonstration here that the spatial uncertainty at the large majority of the model vertices was <2 nm shows that the models represented the AZM’s structure at the full 2 nm resolution of the reconstructed volumes.
Our surface models of the AZM components led to specific hypotheses as to its structure and function (Harlow et al., 2001). Comprehensive tests of these hypotheses will rely on the precise measurement of structural relationships of models from preparations of normal AZM and after its exposure to various experimental conditions. We show here that these accurate models provide a basis for making such measurements. Centroid measurements on the transverse and longitudinal spacings between the pegs in the two data sets that were analyzed were similar to those on the transverse and longitudinal spacings of macromolecules in the presynaptic membrane as observed in freeze fracture replicas, consistent with our hypothesis that the pegs are connected to the macromolecules. Proximity measurements on the separation of the presynaptic membrane from the contact points between ribs and synaptic vesicles in the same two data sets revealed that the contact points on each of the four synaptic vesicles were not only very near the presynaptic membrane but that there was also little variability in such separation from one contact point to the next. Thus, our model-generation methods can provide a quantitative map of the macro-molecular architecture of the normal active zone.
Our methods are designed to generate surface models at contrast boundaries regardless of the contrast mechanism. In this report, we have presented IVOI models of cellular components where contrast was obtained by conventional heavy metal staining. However, there is also limited contrast in unstained cells imaged by cryo-EM techniques (Hsieh et al., 2002; Medalia et al., 2002). Accordingly, the methods described here should be effective in generating surface models of such unstained components in cryosamples at the full resolution of the reconstructed volume, and the spatial uncertainty metric should provide a measure of their reliability.
Most structures detected in cells by conventional 2D electron microscopy have dimensions smaller than the thinnest tissue sections that can be routinely cut as do the beams, ribs, and pegs of the AZM. Our current understanding of cell biology leads to the prediction that such structures in general are components of an extensive network of macromolecules distributed throughout a cell’s interior, and that the functions of each component is partially dependent on its position and associations within the network. Accordingly, making accurate 3D maps of the configuration of macromolecules in cells using EMT is an essential step toward understanding the molecular basis of cell function and disease. Our methods, which generate surface models at the full resolution of EMT reconstructions, together with the measurement methods enabled by these accurate models, provide the opportunity for structural biologists to take a systematic and standardized approach to mapping the conformation and organization of cellular components with macromolecular spatial resolution.
The methods presented here are part of an integrated EMT software package called EM3D©, available free-of-charge at http://em3d.stanford.edu. It also contains projection-alignment, reconstruction, and extensive 3D-visualization and measurement tools that allow complete and efficient analysis of EMT data from raw data to quantitative structural models.
Acknowledgments
This research was funded by the National Institute of Mental Health (Human Brain Project/Neuroinformatics, MH068065), and the National Institute of Neurological Disorders and Stroke (NS014506 and NS007158).
References
- Bajaj C, Yu Z, Auer M. Volumetric feature extraction and visualization of tomographic molecular imaging. J. Struct. Biol. 2003;144:132–143. doi: 10.1016/j.jsb.2003.09.037. [DOI] [PubMed] [Google Scholar]
- Bocker W, Rolf W, Bauch T, Muller WU, Streffer C. Automated comet assay analysis. Cytometry. 1999;35:134–144. [PubMed] [Google Scholar]
- Cooper DM, Turinsky AL, Sensen CW, Hallgrimsson B. Quantitative 3D analysis of the canal network in cortical bone by micro-computed tomography. Anat. Rec. 2003;274B:169–179. doi: 10.1002/ar.b.10024. [DOI] [PubMed] [Google Scholar]
- Frank J. Electron Tomography: Three-Dimensional Imaging with the Transmission Electron Microscope. New York: Plenum Press; 1992. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grau V, Mewes AU, Alcaniz M, Kikinis R, Warfield SK. Improved watershed transform for medical image segmentation using prior information. IEEE Trans. Med. Imaging. 2004;23:447–458. doi: 10.1109/TMI.2004.824224. [DOI] [PubMed] [Google Scholar]
- Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ. The architecture of active zone material at the frog’s neuromuscular junction. Nature. 2001;409:479–484. doi: 10.1038/35054000. [DOI] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Hessler D, Young SJ, Carragher BO, Martone ME, Lamont S, Whittaker M, Milligan RA, Masliah E, Hinshaw JE, Ellisman MH. Programs for visualization in three-dimensional microscopy. Neuroimage. 1992;1:55–67. doi: 10.1016/1053-8119(92)90007-a. [DOI] [PubMed] [Google Scholar]
- Heuser JE, Reese TS. Handbook of Physiology. Bethseda, MD: American Physiological Society; 1977. pp. 261–294. [Google Scholar]
- Heuser JE, Reese TS. Structural changes after transmitter release at the frog neuromuscular junction. J. Cell Biol. 1981;88:564–580. doi: 10.1083/jcb.88.3.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Reese TS, Dennis MJ, Jan Y, Jan L, Evans L. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 1979;81:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zigzag nucleosomal ribbon. J. Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CE, Marko M, Frank J, Mannella CA. Electron tomographic analysis of frozen-hydrated tissue sections. J. Struct. Biol. 2002;138:63–73. doi: 10.1016/s1047-8477(02)00034-5. [DOI] [PubMed] [Google Scholar]
- Katz SB. The Release of Neural Transmitter Substances. Springfield, IL: CC Thomas; 1969. [Google Scholar]
- Klemencic A, Kovacic S, Pernus F. Automated segmentation of muscle fiber images using active contour models. Cytometry. 1998;32:317–326. doi: 10.1002/(sici)1097-0320(19980801)32:4<317::aid-cyto9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Kozerke S, Botnar R, Oyre S, Scheidegger MB, Pedersen EM, Boesiger P. Automatic vessel segmentation using active contours in cine phase contrast flow measurements. J. Magn. Reson. Imaging. 1999;10:41–51. doi: 10.1002/(sici)1522-2586(199907)10:1<41::aid-jmri6>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi D, Runyeon JW, Crum J, Ellisman MH, Roberts WM. Synaptic vesicle populations in saccular hair cells reconstructed by electron tomography. J. Neurosci. 1999;19:119–132. doi: 10.1523/JNEUROSCI.19-01-00119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Leith A, Frank J. Tinkerbell-a tool for interactive segmentation of 3D data. J. Struct. Biol. 1997;120:266–275. doi: 10.1006/jsbi.1997.3917. [DOI] [PubMed] [Google Scholar]
- Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry. 2003;56A:23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- Marko M, Leith A. Sterecon-three-dimensional reconstructions from stereoscopic contouring. J. Struct. Biol. 1996;116:93–98. doi: 10.1006/jsbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- Martone ME, Jones YZ, Young SJ, Ellisman MH, Zivin JA, Hu BR. Modification of postsynaptic densities after transient cerebral ischemia: a quantitative and three-dimensional ultrastructural study. J. Neurosci. 1999;19:1988–1997. doi: 10.1523/JNEUROSCI.19-06-01988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Radermacher M, Rieder CL, Frank J. Tomographic three-dimensional reconstruction of cilia ultrastructure from thick sections. Proc. Natl. Acad. Sci. USA. 1986;83:9040–9044. doi: 10.1073/pnas.83.23.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J. Cell Biol. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan UJ, Slater CR. The influence of basal lamina on the accumulation of acetylcholine receptors at synaptic sites in regenerating muscle. J. Cell Biol. 1984;98:1453–1473. doi: 10.1083/jcb.98.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Ofverstedt LG, Zhang K, Isaksson LA, Bricogne G, Skoglund U. Automated correlation and averaging of three-dimensional reconstructions obtained by electron tomography. J. Struct. Biol. 1997;120:329–342. doi: 10.1006/jsbi.1997.3915. [DOI] [PubMed] [Google Scholar]
- Ortiz de Solorzano C, Garcia Rodriguez E, Jones A, Pinkel D, Gray JW, Sudar D, Lockett SJ. Segmentation of confocal microscope images of cell nuclei in thick tissue sections. J. Microsc. 1999;193:212–226. doi: 10.1046/j.1365-2818.1999.00463.x. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Renken CW, Song JY, Frey TG, Young SJ, Lamont S, Martone ME, Lindsey S, Ellisman MH. Electron tomography of large, multicomponent biological structures. J. Struct. Biol. 1997;120:219–227. doi: 10.1006/jsbi.1997.3920. [DOI] [PubMed] [Google Scholar]
- Ress D, Harlow ML, Marshall RA, McMahan UJ. Optimization Method for Isodensity Surface Models Obtained with Electron Microscope Tomography Data; Paper presented at: Engineering in Medicine and Biology Society, 2003. Proceedings of the 25th Annual International Conference of the IEEE.2003. [Google Scholar]
- Robitaille R, Garcia ML, Kaczorowski GJ, Charlton MP. Functional colocalization of calcium and calcium-gated potassium channels in control of transmitter release. Neuron. 1993;11:645–655. doi: 10.1016/0896-6273(93)90076-4. [DOI] [PubMed] [Google Scholar]
- Sedzik J, Ofverstedt LG, Skoglund U. Three-dimensional reconstruction of bovine intradural spinal root myelin by electron microscope tomography. J. Neurosci. Res. 1992;31:387–393. doi: 10.1002/jnr.490310222. [DOI] [PubMed] [Google Scholar]
- Suri JS, Liu K, Singh S, Laxminarayan SN, Zeng X, Reden L. Shape recovery algorithms using level sets in 2-D/3-D medical imagery: a state-of-the-art review. IEEE Trans. Inf. Technol. Biomed. 2002;6:8–28. doi: 10.1109/4233.992158. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Schmitz H, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Poole K, Lucaveche C, Edwards RJ, et al. Tomographic 3D reconstruction of quick-frozen, Ca2+-activated contracting insect flight muscle. Cell. 1999;99:421–431. doi: 10.1016/s0092-8674(00)81528-7. [DOI] [PubMed] [Google Scholar]
- Teo PC, Sapiro G, Wandell BA. Creating connected representations of cortical gray matter for functional MRI visualization. IEEE Trans. Med. Imaging. 1997;16:852–863. doi: 10.1109/42.650881. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. J. Am. Med. Inform. Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet LJ, Young IT, Mayall BH. The Athena semi-automated karyotyping system. Cytometry. 1990;11:51–58. doi: 10.1002/cyto.990110107. [DOI] [PubMed] [Google Scholar]
- Volkmann N. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J. Struct. Biol. 2002;138:123–129. doi: 10.1016/s1047-8477(02)00009-6. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, McEwen BF, Frank J. Ultrastructure of chromatin. II. Three-dimensional reconstruction of isolated fibers. J. Cell Sci. 1991;99:107–114. doi: 10.1242/jcs.99.1.107. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Chacon P. Modeling tricks and fitting techniques for multiresolution structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- Wu QJ, Bourland JD. Three-dimensional skeletonization for computer-assisted treatment planning in radiosurgery. Comput. Med. Imaging Graph. 2000;24:243–251. doi: 10.1016/s0895-6111(00)00010-0. [DOI] [PubMed] [Google Scholar]
- Yezzi A, Jr, Kichenassamy S, Kumar A, Olver P, Tannen-baum A. A geometric snake model for segmentation of medical imagery. IEEE Trans. Med. Imaging. 1997;16:199–209. doi: 10.1109/42.563665. [DOI] [PubMed] [Google Scholar]
- Zavaljevski A, Dhawan AP, Gaskil M, Ball W, Johnson JD. Multi-level adaptive segmentation of multi-parameter MR brain images. Comput. Med. Imaging Graph. 2000;24:87–98. doi: 10.1016/s0895-6111(99)00042-7. [DOI] [PubMed] [Google Scholar]