Abstract
Immunohistochemistry staining of p53 is a cheap and simple method to detect aberrant function of p53. However, there are some discrepancies between the result of immunohistochemistry staining and mutation analysis. This study attempted to find a new definition of p53 staining by its staining pattern. Immunohistochemistry staining of p53 and TP53 gene mutation analysis were performed in 148 gastric cancer patients. Also SNP-CGH array analysis was conducted to four cases. Positive staining of p53 was observed in 88 (59.5%) tumors. Tumors with positive p53 staining showed malignant features compared to negative tumors. Mutation of TP53 gene was observed in 29 (19.6%) tumors with higher age and differentiated type. In positive p53 tumors, two types could be distinguished; aberrant type and scattered type. With comparison to TP53 gene mutation analysis, all the scattered type had wild-type TP53 gene (P = 0.0003). SNP-CGH array showed that scattered-type tumors had no change in the structure of chromosome 17. P53-scattered-type staining tumors may reflect a functionally active nonmutated TP53 gene. In interpretation of p53 immunohistochemistry staining, distinguishing p53-positive tumors by their staining pattern may be important in gastric cancer.
Keywords: Gastric cancer, immunohistochemistry, mutation analysis, p53, staining pattern
Introduction
Gastric cancer still has the highest morbidity rate and the second highest mortality rate in Asian countries. Though many new remedy for gastric cancer have discovered, advanced gastric cancers are still difficult to treat. To conquer gastric cancer, understanding the gastric carcinogenesis may be important.
One of the causes for gastric cancer is Helicobacter pylori infection. It is reported that H. pylori counteracts the function of tumor suppressor p53 1,2. Also, the infection causes TP53 gene mutation 3. p53, the guardian of genome, is involved in many cellular function; cell cycle, apoptosis, restoration of DNA, aging, and angiogenesis 4,5. And breakdown of tumor suppressor p53 pathway results in the development of malignancies. Studying p53 is still important in gastric cancer.
To detect the aberrant p53, sequencing of the mutant TP53 gene is a general method. After the discovery of the first mutation in TP53 gene 6, the study on TP53 gene was done vastly. The results from these studies revealed that ∼75% of the mutant TP53 gene had missense mutation 7. This finding was well-characterized for TP53 gene mutation as other tumor suppressor genes such as APC or BRCA1/2 frequently have nonsense mutation or frameshift mutation. By investigating the impact of each mutation on p53 function, we may understand the role of TP53 gene mutation in carcinogenesis.
To find the aberration of TP53 gene in everyday clinic, a simple and cheap method is used. That is immunohistochemistry staining of p53. In normal condition, p53 is unstable because its half-life is no longer than 20 min owing to cleavage through Mdm2 8, which cannot be detected by staining. When p53 is aberrant, the nucleus of the cell is stained and aberrant p53 can be detected.
Many studies are done on immunohistochemistry staining of p53. In lung cancer, meta-analysis study revealed that positive immunohistochemistry staining of p53 is an independent prognostic factor 9,10. Immunohistochemistry staining of p53 is a useful tool to understand the biology of cancer.
However, the result of immunohistochemistry staining of p53 reflects the aberrant function of p53 or not is controversial. Some reports showed discrepancies between the results of immunohistochemistry and those of mutation analysis 11–13.
Here, we report a new discrimination on immunohistochemistry staining of p53 by its staining pattern. This discrimination may reflect the result of mutation analysis. Also, we introduced SNP-CGH array method to demonstrate the accuracy of this discrimination.
Material and Methods
Tissue samples
This study included 148 unselected Japanese patients with primary gastric cancer. All of the patients underwent gastrectomy between 1994 and 2006 at the Department of Surgery and Science, Graduate School of Medical Sciences, Kyushu University Hospital, Fukuoka, Japan. Informed consent was obtained from all patients, and those who did not agree to the study were excluded. A thorough histological examination was carried out with hematoxylin- and eosin-stained tissue preparations, and classification was made according to the general rules established by the Japanese Gastric Cancer Association 14. No patients who were treated preoperatively with cytotoxic drugs were included in this study.
Immunohistochemical staining of p53
Formalin-fixed, paraffin-embedded tissue specimens were used for immunohistochemical staining. A paraffin block contained both cancerous and adjacent noncancerous tissue, and cancerous tissue that invaded the deepest area of the stomach wall was selected in all cases. Sections 5 μm thick from paraffin-embedded blocks were deparaffinized in xylene and rehydrated in a graded series of ethanol. Procedures for immunohistochemical staining have been described previously 15. The sections were pretreated with autoclaving at 121°C for 15 min in 0.01 mol/L citrate-buffered saline (pH 6.0) for antigen retrieval. Endogenous peroxidase activity was blocked by incubation with 3% H2O2 for 30 min at room temperature. The sections were incubated with 10% normal goat serum for 1 h to block nonspecific binding of the immunological reagents. After incubation with mouse monoclonal antibodies against p53 (Clone DO-7; Dako Cytomation, Glostrup, Denmark) at 4°C overnight, streptavidin–biotin complex and horseradish peroxidase were applied, and reaction products were visualized using the Histofine SAB-PO (M) immunohistochemical staining kit (Nichirei, Tokyo, Japan), according to the manufacturer's instructions. The peroxidase labeling was developed by incubation of the sections in diaminobenzidine tetrahydrochloride for 1 min. Finally, nuclear counterstaining was done using Mayer's hematoxylin solution. Two blinded observers (K. A. and Y. Z.) independently examined the immunostained sections. When >10% of nuclear-stained cancer cells were included in the section, the tumor was considered to be p53 positive 16,17.
DNA preparation
DNA was extracted as described previously 18,19. Briefly, the frozen samples were incubated in a lysis buffer (0.01 mol/L Tris-HCl, pH 8.0; 0.1 mol/L ethylenediaminetetraacetic acid (EDTA), pH 8.0; 0.5% sodium dodecyl sulfate) containing proteinase K (100 mg/mL) at 37°C for 2 h. The samples were extracted twice in phenol, then once in phenol/chloroform and once in chloroform. Following ethanol precipitation, the samples were diluted in TE (0.01 mol/L Tris-HCl, pH 8.0; 0.01 mol/L EDTA, pH 8.0) buffer.
The TP53 gene mutation analysis
The TP53 gene, exon 5 to exon 9 including exon–intron junctions, were amplified by polymerase chain reaction (PCR) using “p53 primers” (Nippon Gene) and Ex Taq DNA polymerase with 3′ exonuclease activity (TaKaRa Bio Inc., Tokyo, Japan). The PCR products were purified and used as templates for cycle sequencing reactions with Big Dye Terminator Cycle Sequencing Kit Ver.1.0 (Applied Biosystems, Foster City, CA). Mutations found in a PCR product were verified by reverse sequencing and reconfirmed in two independently amplified PCR products.
SNP-CGH analysis
Four surgically resected gastric cancer specimens and their corresponding noncancerous tissues were genotyped by using 1,140,419 autosomal SNPs (HumanOmni1-Quad BeadChip; Illumina Inc., San Diego, CA). Copy number variation was analyzed with GenomeStudio V2009.1 (Illumina Inc.) as described previously 20. Two transformed parameters, the log-normalized intensity ratio (log R ratio) and B allele frequency, were plotted along the entire genome for all SNPs on the array in the single sample analysis mode.
Statistical analysis
Statistical analysis was performed by using JMP 9.0 software (SAS institute, Cary, NC). The χ2 test, Fisher's exact test, and one-way ANOVA were used as appropriate. A P < 0.05 was considered significant.
Results
Patients and tumors
Of the 148 patients, 65.5% (n = 97) were male and 34.5% (n = 51) were female. The mean age was 63.4 ± 11.6, ranging from 29 to 86.
p53 staining in gastric cancer
It is widely recognized that, tumors with aberrant function of p53, p53 can be stained immunohistochemically 21. Consistent with previous studies, the staining of positive p53 was seen in the nuclei of tumor cells (Fig.1A) 16,22. On the other hand, negative p53 staining tumors showed no signal of p53 (Fig.1B). Of 148 tumors, 88 (59.5%) had positive staining of p53. With clinicopathological analysis (Table1), p53-positive tumors had more lymph node and liver metastases compared with tumors with negative p53 staining (P = 0.02, P = 0.02, respectively). Also, more vascular involvement was seen in aberrant p53-positive tumors (P = 0.03). p53-positive tumors tended to have deeper invasion (P = 0.06). Tumors with positive staining of p53 had malignant features.
Figure 1.
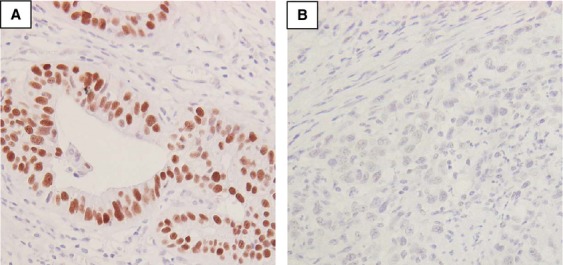
Staining pattern of p53 in gastric cancer. (A) Positive case. Staining of p53 is seen in the nuclei (magnification, 400×). (B) Negative case. No staining of p53 is seen in the nuclei (magnification, 400×).
Table 1.
P53 expression and clinicopathological factors in gastric cancer patients.
| p53 expression (%) |
|||
|---|---|---|---|
| Factors | Negative (n = 60) | Positive (n = 88) | P-value |
| Age (mean ± SD) | 63.5 ± 11.2 | 63.4 ± 12.1 | 0.95 |
| Gender | |||
| Male | 38 (63.3) | 59 (67.1) | 0.64 |
| Female | 22 (36.7) | 29 (32.9) | |
| Differentiation | |||
| Differentiated | 25 (41.7) | 35 (39.8) | 0.81 |
| Undifferentiated | 35 (58.3) | 53 (60.2) | |
| Vascular involvement | |||
| V0 | 36 (60.0) | 44 (50.0) | 0.03* |
| V1 | 16 (26.7) | 24 (27.3) | |
| V2 | 8 (13.3) | 12 (13.6) | |
| V3 | 0 (0.0) | 9 (9.1) | |
| Lymphatic involvement | |||
| Ly0 | 19 (31.7) | 26 (29.6) | 0.21 |
| Ly1 | 17 (28.3) | 19 (21.6) | |
| Ly2 | 18 (30.0) | 23 (26.1) | |
| Ly3 | 6 (10.0) | 20 (22.7) | |
| Depth of invasion | |||
| M, SM | 10 (16.7) | 6 (6.8) | 0.06 |
| MP, SS, SE, SI | 50 (83.3) | 82 (93.2) | |
| Lymph node metastasis | |||
| Negative | 26 (43.3) | 23 (26.1) | 0.02* |
| Positive | 34 (56.7) | 65 (73.9) | |
| Liver metastasis | |||
| Negative | 60 (100) | 83 (94.3) | 0.02* |
| Positive | 0 (0) | 5 (5.7) | |
| Stage | |||
| I + II | 32 (53.3) | 37 (42.1) | 0.17 |
| III + IV | 28 (46.7) | 51 (57.9) | |
M, mucosa; SM, submucosa; MP, muscularis propria; SS, subserosa; SE, penetration of serosa; SI, invasion of adjacent structures.
P < 0.05.
TP53 gene mutation analysis in gastric cancer
Of 11 exons that exist in TP53 gene, exon5 to exon8 is a major spots for mutation; R175H, G245S, R248Q, R248W, R249S, R273C, R273H, and R282H 23. In this study we investigated the mutations of TP53 gene in exon5 to exon9. As shown in Table2, of 148 tumors, 29 (19.6%) had mutant TP53 gene. Of those, six tumors (20.7%) showed nonsense mutation and other 23 tumors (79.3%) showed missense mutation. TP53 gene mutations were found more frequently in differentiated-type tumors than in undifferentiated type (P = 0.009). This result was consistent to previous report 24. Relation with other clinicopathological factors showed that, TP53 gene mutations were found in higher age and positive vascular involvement tumors (P = 0.02, P = 0.01, respectively).
Table 2.
TP53 gene status and clinicopathological factors in gastric cancer patients.
|
TP53 gene status (%) |
|||
|---|---|---|---|
| Factors | Wild (n = 119) | Mutant (n = 29) | P-value |
| Age (mean ± SD) | 62.5 ± 12.1 | 67.2 ± 9.3 | 0.02* |
| Gender | |||
| Male | 75 (63.0) | 22 (75.9) | 0.18 |
| Female | 44 (37.0) | 7 (24.1) | |
| Differentiation | |||
| Differentiated | 42 (35.3) | 18 (62.1) | 0.009* |
| Undifferentiated | 77 (64.7) | 11 (37.9) | |
| Vascular involvement | |||
| V0 | 70 (58.8) | 10 (34.5) | 0.01* |
| V1 | 32 (26.9) | 8 (27.6) | |
| V2 | 14 (11.8) | 6 (20.7) | |
| V3 | 3 (2.5) | 5 (17.2) | |
| Lymphatic involvement | |||
| Ly0 | 34 (28.6) | 11 (37.9) | 0.06 |
| Ly1 | 25 (21.0) | 11 (37.9) | |
| Ly2 | 36 (30.2) | 5 (17.2) | |
| Ly3 | 24 (20.2) | 2 (6.9) | |
| Depth of invasion | |||
| M, SM | 11 (9.2) | 5 (17.2) | 0.23 |
| MP, SS, SE, SI | 108 (90.8) | 24 (82.8) | |
| Lymph node metastasis | |||
| Negative | 41 (34.4) | 8 (27.6) | 0.47 |
| Positive | 78 (65.6) | 21 (72.4) | |
| Liver metastasis | |||
| Negative | 116 (97.5) | 27 (93.1) | 0.28 |
| Positive | 3 (2.5) | 2 (6.9) | |
| Stage | |||
| I + II | 53 (44.5) | 16 (55.2) | 0.3 |
| III + IV | 66 (55.5) | 13 (44.8) | |
M, mucosa; SM, submucosa; MP, muscularis propria; SS, subserosa; SE, penetration of serosa; SI, invasion of adjacent structures.
P < 0.05.
Relation between p53 immunohistochemistry and TP53 gene mutation
Then relation between p53 immunohistochemistry staining and TP53 gene mutation analysis was investigated. Twenty percent of p53-negative tumors had mutant TP53 gene (12/60 tumors) and 19% of p53-positive tumors had mutant TP53 gene (17/88 tumors) (P = 0.91). This result showed that there is no significant relation between p53 immunohistochemistry staining and TP53 gene mutation.
We then sought for a new definition for p53 immunohistochemistry staining pattern.
TP53 gene mutation in p53-negative tumors reflect both wild-type and nonsense mutation
First we examined the TP53 gene mutation status in 60 tumors with p53-negative staining. Of 60 tumors, 48 (80%) tumors had wild-type TP53 gene. And as shown above, 6 (10%) tumors had nonsense mutation. The rest six tumors had missense mutation. Negative staining of p53 may reflect wild-type TP53 gene or nonsense mutation which needs sequence analysis.
Two staining pattern of p53-positive tumors; scattered type and aberrant type
In this study, 88 tumors had positive staining of p53. In these tumors, 10% or more nuclei were stained with p53. According to previous report 25, we also found that in these positive tumors, two subtypes could be distinguished by its staining pattern and percentage of stained nuclei; “aberrant type” and “scattered type.” Aberrant type (Fig.2A) had widespread staining of 70% or more nuclei in the sample. Of 88 tumors, this type was seen in 63 (71.6%) tumors. On the other hand, “scattered type” (Fig.2B) had staining of single cells throughout the tumor sample. In these tumors 20–50% of nuclei were stained. Of 88 tumors, 25 (28.4%) tumors were this type. Compared to p53-negative tumors, “scattered type” tumors had deeper invasion and tend to have more lymph node metastases (Table3). This result showed that p53-negative tumors and “scattered type” tumors have different features. There was no relation with clinicopathological factors between scattered type and aberrant type (data not shown). However, a significant relation was observed with TP53 gene mutations (Table4). Interestingly, all scattered-type tumors were wild-type TP53 gene. And all mutant TP53 gene tumors, which were missense, had aberrant-type staining of p53 (P = 0.0003). It seemed that “scattered type” reflects wild-type p53.
Figure 2.
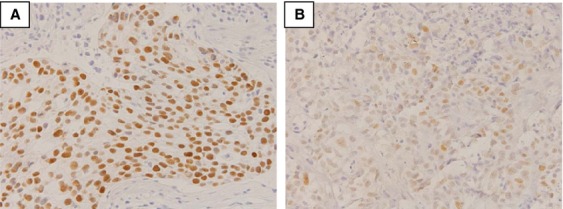
Staining pattern of “aberrant type” and “scattered type” in positive staining group. (A) Aberrant type. Widespread staining of all nuclei of the tumor sample is seen (magnification, 400×). (B) Scattered type. Staining of single cells scattered throughout the tumor sample (magnification, 400×).
Table 3.
Clinicopathological factors in gastric cancer patients with negative p53 and scattered type.
| p53 expression (%) |
|||
|---|---|---|---|
| Factors | Negative (n = 60) | Scattered (n = 25) | P-value |
| Age (mean ± SD) | 63.5 ± 11.2 | 64.9 ± 12.6 | 0.62 |
| Gender | |||
| Male | 38 (63.3) | 15 (60.0) | 0.77 |
| Female | 22 (36.7) | 10 (40.0) | |
| Differentiation | |||
| Differentiated | 25 (41.7) | 7 (28.0) | 0.22 |
| Undifferentiated | 35 (58.3) | 18 (72.0) | |
| Vascular involvement | |||
| V0 | 36 (60.0) | 14 (56.0) | 0.19 |
| V1 | 16 (26.7) | 9 (36.0) | |
| V2 | 8 (13.3) | 1 (4.0) | |
| V3 | 0 (0.0) | 1 (4.0) | |
| Lymphatic involvement | |||
| Ly0 | 19 (31.7) | 9 (36.0) | 0.11 |
| Ly1 | 17 (28.3) | 3 (12.0) | |
| Ly2 | 18 (30.0) | 6 (24.0) | |
| Ly3 | 6 (10.0) | 7 (28.0) | |
| Depth of invasion | |||
| M, SM | 10 (16.7) | 0 (0.0) | 0.006* |
| MP, SS, SE, SI | 50 (83.3) | 25 (100) | |
| Lymph node metastasis | |||
| Negative | 26 (43.3) | 6 (24.0) | 0.08 |
| Positive | 34 (56.7) | 19 (76.0) | |
| Stage | |||
| I + II | 32 (53.3) | 9 (36.0) | 0.14 |
| III + IV | 28 (46.7) | 16 (64.0) | |
M, mucosa; SM, submucosa; MP, muscularis propria; SS, subserosa; SE, invasion of adjacent structures.
P < 0.05.
Table 4.
Scattered staining reflects TP53 gene wild type in p53 positive gastric cancer.
| p53 expression (%) |
|||
|---|---|---|---|
| Scattered (n = 25) | Aberrant (n = 63) | P-value | |
| TP53 gene status | |||
| Wild (n = 71) | 25 (100) | 46 (73.0) | 0.0003 |
| Mutation (n = 17) | 0 (0) | 17 (27.0) | |
SNP-CGH array analysis revealed a different pattern of chromosome 17 between “aberrant” and “scattered” type
For further analysis on p53, we performed SNP-CGH array analysis in four gastric cancer samples. Two “aberrant type” and two “scattered type” were analyzed. For aberrant type, one TP53 gene wild-type tumor (Fig.3A left) and one mutant-type tumor (Fig.3A right) were analyzed. With regard to chromosome 17, data from two “aberrant type” specimen showed that there was a deflection in the log R ratio (Fig.3A). In these two specimens, including the TP53 gene wild-type tumor (Fig.3A left), the heterozygous state split into two clusters in the B allele frequency was also observed, especially in the short arm containing the p53 locus. These results showed p53 was aberrant from the chromosomal state in “aberrant type.”
Figure 3.
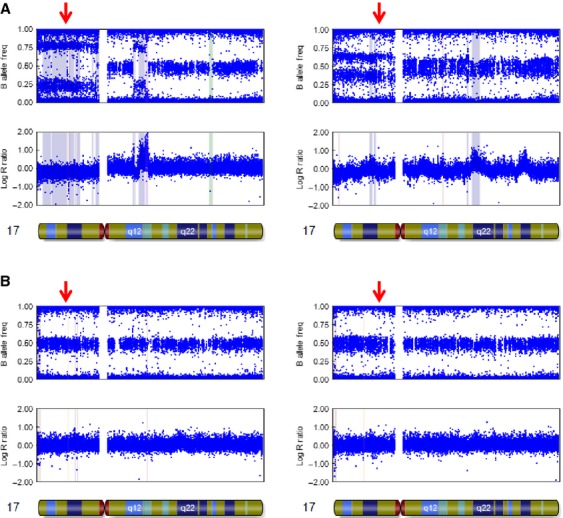
SNP-CGH array analysis of chrmosome17 in aberrant type and scattered type. (A) Analysis in aberrant type. Abnormal chromosome is seen. Especially, aberrant allele is observed in short arm. (B) Analysis in scattered type. The chromosome 17 is normal. The red arrow shows the locus of p53.
On the contrary, in “scattered type” no deflection in the log R ratio was observed. And, the heterozygotes were clustered around t0.5 in the B allele frequency (Fig.3B), which was also observed in the noncancerous tissue (data not shown). As previously shown, TP53 gene is wild type in these two tumors.
These results suggested that “scattered type” staining had normal structure of p53 from the chromosomal state.
Discussion
To date, p53-positive tumors have been considered to have aberrant p53 function. However, there was some discrepancy between the result from immunohistochemistry and sequence analysis. In this study, we reevaluated the p53-positive tumors to two groups by its staining patterns. This evaluation showed us that even in p53-positive tumors, there were tumors with normal p53 function. Staining pattern may be important in interpreting the result of p53 immunohistochemistry.
Immunohistochemistry of p53 is said to be a cheap and rapid method to detect aberrant p53 function. In general, tumors with more than 10% positively stained cancer cells nuclei were defined as positive 16,17,26. In this study, 59.5% tumors had positive staining of p53. With clinicopathological analysis, p53-positive tumors had malignant features compared to negative tumors; more lymph node metastases and more liver metastases, which were consistent with previous reports 15–17,27. Immunohistochemistry of p53 might interpret the biology of the tumor.
On the other hand, about 20% of the tumor had mutant TP53 gene in this study. This result was consistent to previous reports 19,24. With clinicopathological analysis, tumors with mutant TP53 gene were frequently observed in higher age and differentiated cases. Differentiated gastric cancers are more frequently observed in older patients and follows multifocal atrophic gastritis, which is accompanied by intestinal metaplasia or dysplasia. In fact, multistep gastric carcinogenesis including a sequence of events that begins with H. pylori-induced superficial gastritis, progressing toward chronic atrophic gastritis, intestinal metaplasia, dysplasia, and finally cancer 28. It is reported that H. pylori infection induces point mutations of TP53 in 52% of gastritis 29. Also, Wei et al. reported that reported that H. pylori accelerates ubiquitination and proteasomal degradation of p53 in gastric epithelial cells 30. H. pylori infection and p53 may act in a synergistic fashion in gastric carcinogenesis 31. All these reports suggest that TP53 gene mutation has an important role in gastric carcinogenesis.
Immunohistochemistry staining of p53 has been proposed as an alternative to gene analytical method. However, in our study, only distinguishing positive or negative staining of p53 had no effect in detecting the mutation status of TP53 gene, similar to previous reports 11,13.
Kaserer et al. reported a study on staining patterns of p53 in colorectal cancer 25. They found that in positive p53 tumors, there were two types in staining pattern of p53; wide spread staining of all nuclei of the tumor cells and staining of single cells scattered throughout the tumor sample. In this study, we also classified p53-positive tumors in two groups according to their report; aberrant type and scattered type. “Scattered type” had staining of single cells throughout the tumor sample. In these tumors 20–50% of nuclei were stained. These tumors had deeper invasion and more lymph node metastases compared to p53-negative tumors which showed that “scattered type” tumors are different to p53-negative tumors. Also, we found that the entire “scattered type” staining group had wild-type TP53 gene. Even in p53-positive tumors, there were tumors with p53 normal function. Yemelyanova et al. reported an immunohistochemical and nucleotide sequence analysis of p53 in ovarian carcinomas 32. They found that combining two immunohistochemical labeling patterns associated with TP53 mutations (0% and 60–100% positive cells), correctly identified a mutation in 94% of cases (P < 0.001). They concluded that immunohistochemical analysis can be used as a robust method for inferring the presence of a TP53 mutation in ovarian carcinomas. In our study, we focused on “scattered type” and wild-type TP53. In Yemelyanova's report there were two cases with 20–50% positive cells, which is same to our “scattered type,” and these cases had wild-type TP53 gene. This result may reflect our results; “scattered type” tumor has wild-type TP53 gene.
From SNP-CGH array analysis, this finding was much clearer. The development of high-density SNP genotyping technology for genomic profiling represents a further advance, because simultaneous measurement of both signal intensity variations and changes in allelic composition makes it possible to detect chromosomal events 33. Data from two “scattered type” p53 staining specimens (Fig.3B) and all normal samples (data not shown) indicated no chromosomal alterations for the entire chromosome 17, especially on the p53 locus. This result also demonstrated that “scattered type” had normal p53 function.
Then why normal p53 can be stained; normal p53 should have short half-life that cannot be stained. Hall and Lane reported that the reason for normal p53 expression might be that wild-type p53 protein accumulates in response to spontaneous genetic errors occurring at a higher frequency in the tumor than in normal surrounding tissue 12. This “scattered type” staining of p53 may rather reflect an accumulation of wild-type p53 protein as a result of either a response to DNA damage, alterations in the normal degradation process, or the stabilization of the gene product by an interaction with viral or cellular proteins 34,35.
However, a question remains; why are there still wild-type TP53 gene tumors in “aberrant type?” In this study we found 46 tumors with wild-type TP53 gene in 63 “aberrant type” tumors, which is a quite ratio. The staining pattern does not match with “scattered type.” We think that “aberrant type” represents a clonal expansion of cells accumulating p53. Clonal expansion of p53 accumulating cells in the presence of a wild-type TP53 gene has been observed previously 36. This was shown to represent stabilization of the protein by mechanisms other than inactivating mutations 36,37. The wild-type tumors in “aberrant type” may reflect this mechanism. From SNP-CGH array analysis in TP53 gene wild type of “aberrant type” tumor (Fig.3A left) may support this discussion. Even though this tumor had wild-type TP53 gene, its chromosome 17 had aberrant structure. In “scattered type,” chromosome 17 did not have aberrant structure. A further study is required to clarify the “aberrant type.”
Our study showed that positive staining of p53 has two types by its staining patterns in gastric cancer. “Scattered type” may reflect normal p53. In interpretation of p53 immunohistochemistry staining, discriminating p53-positive tumors by their staining pattern is important in gastric cancer.
Acknowledgments
We thank Ms. Y. Kubota for her preparation of the immunohistochemistry samples and Ms. T. Shishino for her preparation of the DNA samples. The authors have no conflict of interest to declare.
Conflict of Interest
None declared.
References
- Umehara S, Higashi H, Ohnishi N, Asaka M. Hatakeyama M. Effects of Helicobacter pylori CagA protein on the growth and survival of B lymphocytes, the origin of MALT lymphoma. Oncogene. 2003;22:8337–8342. doi: 10.1038/sj.onc.1207028. [DOI] [PubMed] [Google Scholar]
- Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- Lowe SW. Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Vousden KH. Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC. Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D. Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Hamajima N, Ogawa M. Takahashi T. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: a meta-analysis. Clin. Cancer Res. 2000;6:4055–4063. [PubMed] [Google Scholar]
- Steels E, Paesmans M, Berghmans T, Branle F, Lemaitre F, Mascaux C, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur. Respir. J. 2001;18:705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- Calzolari A, Chiarelli I, Bianchi S, Messerini L, Gallo O, Porfirio B, et al. Immunohistochemical vs molecular biology methods. Complementary techniques for effective screening of p53 alterations in head and neck cancer. Am. J. Clin. Pathol. 1997;107:7–11. doi: 10.1093/ajcp/107.1.7. [DOI] [PubMed] [Google Scholar]
- Hall PA. Lane DP. p53 in tumour pathology: can we trust immunohistochemistry?–Revisited! J. Pathol. 1994;172:1–4. doi: 10.1002/path.1711720103. [DOI] [PubMed] [Google Scholar]
- Hurlimann J, Chaubert P. Benhattar J. p53 Gene alterations and p53 protein accumulation in infiltrating ductal breast carcinomas: correlation between immunohistochemical and molecular biology techniques. Mod. Pathol. 1994;7:423–428. [PubMed] [Google Scholar]
- Japanese Gastric Cancer A. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011;14:101–112. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- Ando K, Oki E, Zhao Y, Ikawa-Yoshida A, Kitao H, Saeki H, et al. Mortalin is a prognostic factor of gastric cancer with normal p53 function. Gastric Cancer. 2013;17:255–262. doi: 10.1007/s10120-013-0279-1. [DOI] [PubMed] [Google Scholar]
- Kakeji Y, Korenaga D, Tsujitani S, Baba H, Anai H, Maehara Y, et al. Gastric cancer with p53 overexpression has high potential for metastasising to lymph nodes. Br. J. Cancer. 1993;67:589–593. doi: 10.1038/bjc.1993.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi Y, Kakeji Y, Egashira A, Mizokami K, Orita H. Maehara Y. Overexpression of hypoxia-inducible factor 1alpha and p53 is a marker for an unfavorable prognosis in gastric cancer. Clin. Cancer Res. 2006;12:5112–5117. doi: 10.1158/1078-0432.CCR-05-2382. [DOI] [PubMed] [Google Scholar]
- Egashira A, Morita M, Kakeji Y, Sadanaga N, Oki E, Honbo T, et al. p53 gene mutations in esophageal squamous cell carcinoma and their relevance to etiology and pathogenesis: results in Japan and comparisons with other countries. Cancer Sci. 2007;98:1152–1156. doi: 10.1111/j.1349-7006.2007.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki E, Zhao Y, Yoshida R, Egashira A, Ohgaki K, Morita M, et al. The difference in p53 mutations between cancers of the upper and lower gastrointestinal tract. Digestion. 2009;79(Suppl. 1):33–39. doi: 10.1159/000167864. [DOI] [PubMed] [Google Scholar]
- Saeki H, Kitao H, Yoshinaga K, Nakanoko T, Kubo N, Kakeji Y, et al. Copy-neutral loss of heterozygosity at the p53 locus in carcinogenesis of esophageal squamous cell carcinomas associated with p53 mutations. Clin. Cancer Res. 2011;17:1731–1740. doi: 10.1158/1078-0432.CCR-10-1996. [DOI] [PubMed] [Google Scholar]
- Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, et al. p53 mutations in colorectal cancer. Proc. Natl Acad. Sci. USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge RM, Clark GM, Fuqua SA, Yu YY. Allred DC. p53 protein accumulation detected by five different antibodies: relationship to prognosis and heat shock protein 70 in breast cancer. Cancer Res. 1994;54:3752–3757. [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki E, Tokunaga E, Nakamura T, Ueda N, Futatsugi M, Mashino K, et al. Genetic mutual relationship between PTEN and p53 in gastric cancer. Cancer Lett. 2005;227:33–38. doi: 10.1016/j.canlet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Kaserer K, Schmaus J, Bethge U, Migschitz B, Fasching S, Walch A, et al. Staining patterns of p53 immunohistochemistry and their biological significance in colorectal cancer. J. Pathol. 2000;190:450–456. doi: 10.1002/(SICI)1096-9896(200003)190:4<450::AID-PATH545>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Goncalves AR, Carneiro AJ, Martins I, de Faria PA, Ferreira MA, de Mello EL, et al. Prognostic significance of p53 protein expression in early gastric cancer. Pathol. Oncol. Res. 2011;17:349–355. doi: 10.1007/s12253-010-9333-z. [DOI] [PubMed] [Google Scholar]
- Zha Y, Cun Y, Zhang Q, Li Y. Tan J. Prognostic value of expression of Kit67, p53, TopoIIa and GSTP1 for curatively resected advanced gastric cancer patients receiving adjuvant paclitaxel plus capecitabine chemotherapy. Hepatogastroenterology. 2012;59:1327–1332. doi: 10.5754/hge12204. [DOI] [PubMed] [Google Scholar]
- Matysiak-Budnik T. Megraud F. Helicobacter pylori infection and gastric cancer. Eur. J. Cancer. 2006;42:708–716. doi: 10.1016/j.ejca.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Murakami K, Fujioka T, Okimoto T, Mitsuishi Y, Oda T, Nishizono A, et al. Analysis of p53 gene mutations in Helicobacter pylori-associated gastritis mucosa in endoscopic biopsy specimens. Scand. J. Gastroenterol. 1999;34:474–477. doi: 10.1080/003655299750026191. [DOI] [PubMed] [Google Scholar]
- Wei J, O'Brien D, Vilgelm A, Piazuelo MB, Correa P, Washington MK, et al. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;134:1412–1423. doi: 10.1053/j.gastro.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio-Preiser CM, Wang J, Stemmermann GN. Noffsinger A. TP53 and gastric carcinoma: a review. Hum. Mutat. 2003;21:258–270. doi: 10.1002/humu.10180. [DOI] [PubMed] [Google Scholar]
- Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih Ie M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod. Pathol. 2011;24:1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas IO, Mulder JW, Offerhaus GJ, Vogelstein B. Hamilton SR. An evaluation of six antibodies for immunohistochemistry of mutant p53 gene product in archival colorectal neoplasms. J. Pathol. 1994;172:5–12. doi: 10.1002/path.1711720104. [DOI] [PubMed] [Google Scholar]
- Kirsch DG. Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. J. Clin. Oncol. 1998;16:3158–3168. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- Burns JS, Blaydes JP, Wright PA, Lemoine L, Bond JA, Williams ED, et al. Stepwise transformation of primary thyroid epithelial cells by a mutant Ha-ras oncogene: an in vitro model of tumor progression. Mol. Carcinog. 1992;6:129–139. doi: 10.1002/mc.2940060208. [DOI] [PubMed] [Google Scholar]
- Lehman TA, Bennett WP, Metcalf RA, Welsh JA, Ecker J, Modali RV, et al. p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]


