Abstract
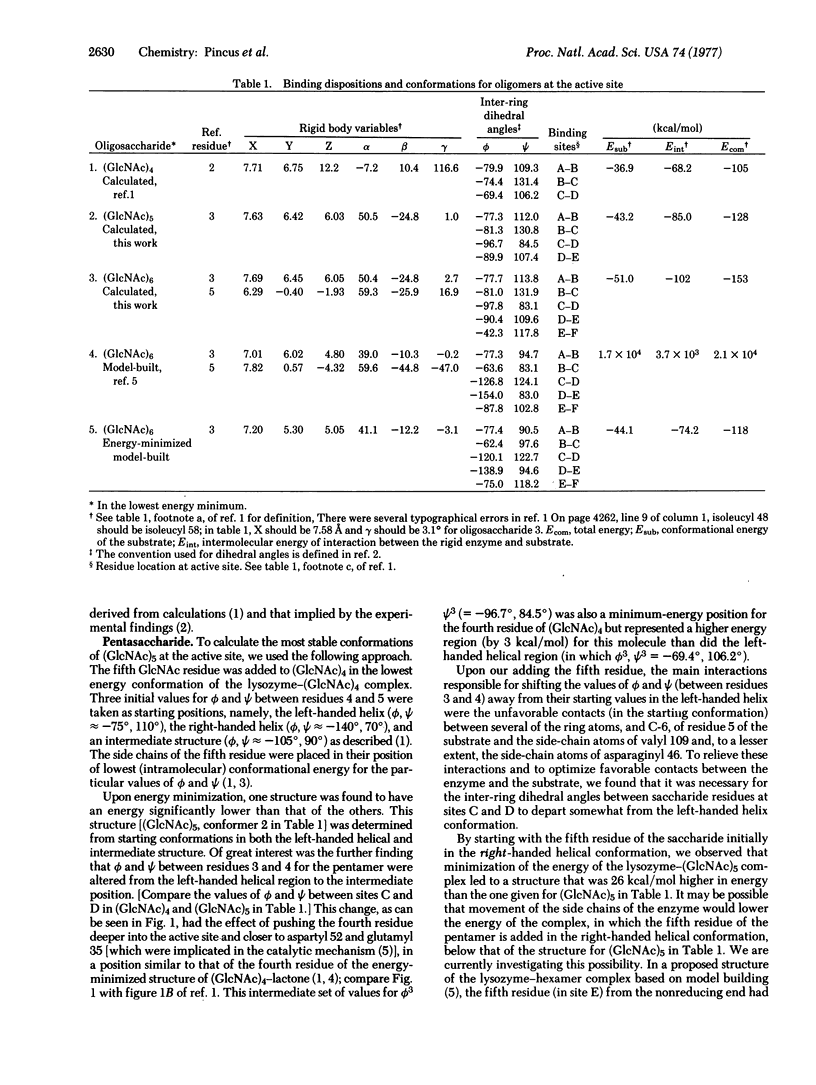
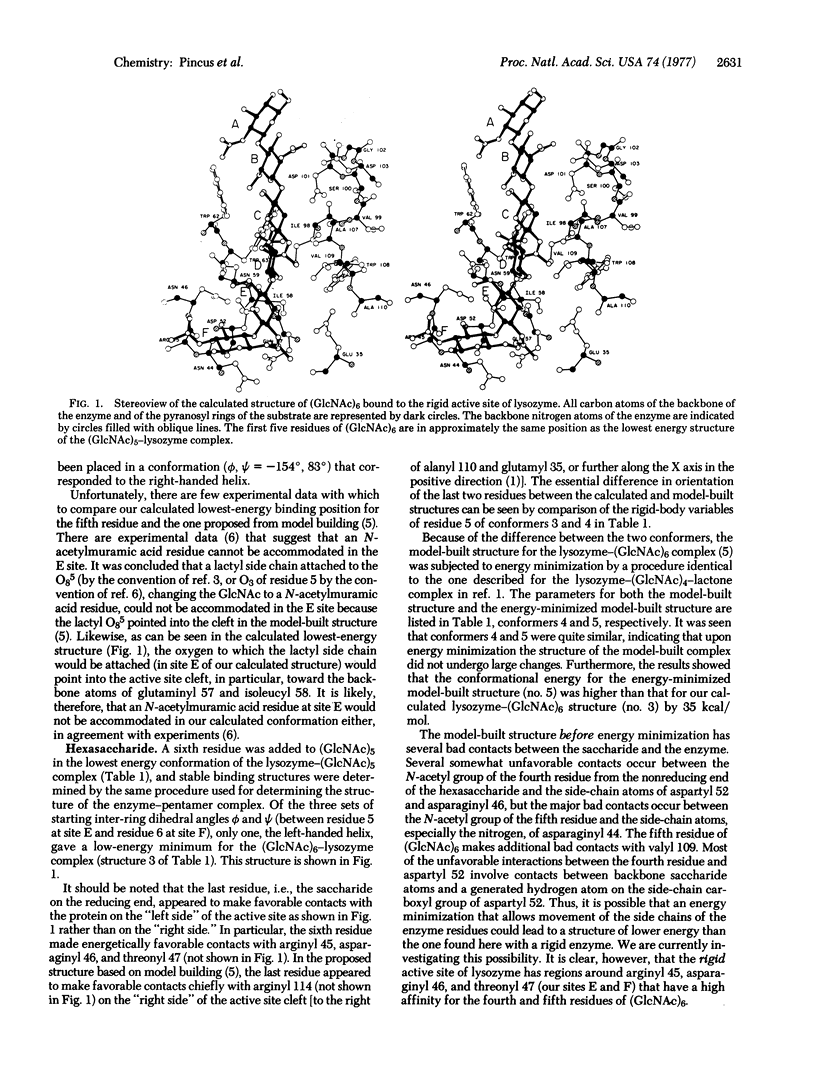
Conformational energy calculations were used to determine the binding structures of two oligosaccharides (GlcNAc)n, in which n = 5 and 6, in the rigid active site of lysozyme (mucopeptide N-acetylmuramoylhydrolase, EC 3.2.1.17). Starting with the lowest energy binding structures of (GlcNAc)4 as determined in a previous publication, we added a fifth GlcNAc residue to this tetramer in three different conformations, corresponding to the left-handed and right-handed helical structures and an intermediate structure, and the energy of each complex was minimized. The most stable binding conformation of the fifth residue of the pentamer was closest to the left-handed helical one. During energy minimization, the fourth residue of the pentamer moved from its initial position near the surface of the active site farther into the active site cleft at binding site D. Binding structures of (GlcNAc)6 were then examined by addition of a residue to the lowest energy structure of (GlcNAc)5, and it was found that the sixth residue of the hexamer binds in a conformation again close to the left-handed helical one. Stable binding regions of the rigid active site for the fifth and sixth residues were found to be near arginyl 45 and asparaginyl 46, on the opposite side of the active site cleft from arginyl 114. When the calculated structure of the lysozyme-(GlcNAc)4 complex (used here as the starting structure for addition of the fifth and sixth residues) is compared with recent experimental data, it is found that the calculated structure is a reasonable one. Of all binding regions available to the saccharide residues, the C site binds GlcNAc with the lowest energy, in agreement with experiments.
Keywords: GlcNAc, binding regions, conformational energy, active site
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Johnson L. N., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Crystallographic studies of the activity of hen egg-white lysozyme. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):378–388. doi: 10.1098/rspb.1967.0035. [DOI] [PubMed] [Google Scholar]
- Bott R., Sarma R. Crystal structure of turkey egg-white lysozyme: results of the molecular replacement method at 5 A resolution. J Mol Biol. 1976 Oct 5;106(4):1037–1046. doi: 10.1016/0022-2836(76)90351-x. [DOI] [PubMed] [Google Scholar]
- Ford L. O., Johnson L. N., Machin P. A., Phillips D. C., Tjian R. Crystal structure of a lysozyme-tetrasaccharide lactone complex. J Mol Biol. 1974 Sep 15;88(2):349–371. doi: 10.1016/0022-2836(74)90487-2. [DOI] [PubMed] [Google Scholar]
- Holler E., Rupley J. A., Hess G. P. Productive and unproductive lysozyme-chitosaccharide complexes. Equilibrium measurements. Biochemistry. 1975 Mar 11;14(5):1088–1094. doi: 10.1021/bi00676a032. [DOI] [PubMed] [Google Scholar]
- Pincus M. R., Burgess A. W., Scheraga H. A. Conformational energy calculations of enzyme-substrate complexes of lysozyme. I. Energy minimization of monosaccharide and oligosaccharide inhibitors and substrates of lysozyme. Biopolymers. 1976 Dec;15(12NA-NA-770103-770104):2485–2521. doi: 10.1002/bip.1976.360151212. [DOI] [PubMed] [Google Scholar]
- Pincus M. R., Zimmerman S. S., Scheraga H. A. Prediction of three-dimensional structures of enzyme-substrate and enzyme-inhibitor complexes of lysozyme. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4261–4265. doi: 10.1073/pnas.73.12.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M., Assaf Y., Sharon N., Chipman D. M. Mechanism of lysozyme catalysis: role of ground-state strain in subsite D in hen egg-white and human lysozymes. Biochemistry. 1977 Feb 8;16(3):423–431. doi: 10.1021/bi00622a013. [DOI] [PubMed] [Google Scholar]
- Warshel A., Levitt M. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976 May 15;103(2):227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]


