Abstract
Objective
Infection with Helicobacter pylori is the strongest known risk factor for adenocarcinoma of the stomach. Tumorigenic transformation of gastric epithelium induced by H. pylori is a highly complex process driven by an active interplay between bacterial virulence and host factors, many aspects of which remain obscure. In this work, we investigated the degradation of p53 tumour suppressor induced by H. pylori.
Design
Expression of p53 protein in gastric biopsies was assessed by immunohistochemistry. Gastric cells were co-cultured with H. pylori strains isolated from high-gastric risk and low-gastric risk areas and assessed for expression of p53, p14ARF and cytotoxin-associated gene A (CagA) by immunoblotting. siRNA was used to inhibit activities of ARF-BP1 and Human Double Minute 2 (HDM2) proteins.
Results
Our analysis demonstrated that H. pylori strains expressing high levels of CagA virulence factor and associated with a higher gastric cancer risk more strongly suppress p53 compared with low-risk strains in vivo and in vitro. We found that degradation of p53 induced by bacterial CagA protein is mediated by host HDM2 and ARF-BP1 E3 ubiquitin ligases, while the p14ARF protein counteracts H. pylori-induced signalling.
Conclusions
Our results provide novel evidence that tumorigenicity associated with H. pylori infection is linked to inhibition of p53 protein by CagA. We propose a model in which CagA-induced degradation of p53 protein is determined by a relative level of p14ARF. In cells in which p14ARF levels were decreased due to hypermethylation or deletion of the p14ARF gene, H. pylori efficiently degraded p53, whereas p53 is protected in cells expressing high levels of p14ARF.
INTRODUCTION
Helicobacter pylori is a bacterial pathogen that infects approximately half of the world’s population. During its long co-evolution with human hosts, H. pylori developed the ability to persist in the gastric niche, mostly causing asymptomatic inflammation. In some individuals, however, H. pylori infection may result in the development of mucosa associated lymphoid tissue (MALT) lymphoma and gastric cancer. Accumulating data suggest that interplay between bacterial virulence and host factors underlies abnormal activation of multiple oncogenic pathways (Wnt/β-catenin, epidermal growth factor receptor/phosphoinositide 3-kinase/AKT, Rat Sarcoma Viral Oncogene Homolog (RAS) and others) and gastric tumorigenesis. The best-characterised bacterial virulence factors are the vacuolating cytotoxin A and the cytotoxin-associated gene A (cagA). The cagA gene is located within a 40-kb DNA fragment known as the cag pathogenicity island that encodes a type IV secretion system (T4SS), which H. pylori uses for the injection of bacterial components directly into host cells. CagA protein, which is injected through the T4SS, behaves as a bacterial oncoprotein. CagA is able to induce anchor-independent growth of gastric epithelial cells in soft agar.1 Its transgenic expression in mice induces gastric tumour.2 Genetic and functional differences in virulence factors have been suggested to affect the ability of H. pylori strains to induce cancer as has been found using animal models. Infection of Mongolian gerbils with the human clinical isolate B128 leads to successful colonisation of the gerbil stomach, but does not eventuate in the development of gastric tumours, whereas its oncogenic derivative, strain 7.13, which was generated by a serial passage of H. pylori in rodents, induces gastric tumours in 4–8 weeks.3 The H. pylori genetic composition may also significantly contribute to gastric cancer incidence between geographical regions as has been shown for the state of Nariño in Colombia, South America. H. pylori strains isolated from the inhabitants of the high altitude Andes, which have a high incidence rate of gastric cancer, are phylogenetically different from strains isolated from inhabitants of a low-incidence region located on the Pacific coast.4
In normal cells, aberrant activation of oncogenes is counteracted by tumour suppressor mechanisms. The p53 protein plays a key role in this process by limiting abnormal cellular proliferation and eliminating transformed cells that otherwise may cause tumour development. The p14ARF tumour suppressor functions upstream of p53 and is required for accumulation of p53 under oncogenic stress. The role of p14ARF is to inhibit proteasomal degradation of p53 by sequestering the Human Double Minute 2(HDM2) protein in the nucleoli and inhibiting its E3 ligase activity.5 During gastric tumorigenesis, both p14ARF and p53 have been shown to be frequently inactivated. Promoter hypermethylation and deletions of the p14ARF gene have been found in approximately 30% of gastric tumours, while p53 is primarily inactivated by mutations in 40%–50% of patients with gastric cancer.6 H. pylori infection has been reported to enhance mutagenesis of the p53 gene.7 However, emerging evidence suggests that H. pylori compromises p53 function by mutation-independent mechanism analogous to a number of oncogenic viruses, which have developed specific strategies to circumvent cellular control mediated by p53. We have previously reported that H. pylori inhibits p53 protein in gastric epithelial cells, although little is currently known about its mechanism and functional role.8 Here, we built upon these findings to explore in detail how the p53 pathway is regulated by H. pylori.
MATERIALS AND METHODS
Cell lines, culture conditions and H. pylori strains
The human gastric cancer cell lines AGS, SNU-1, STKM2 (a gift from Dr Koshikawa, University of Tokyo, Japan) and HFE-145 harbouring wild type p53 protein and p53-null cell line, Kato III were grown at 37°C with 5% CO2 in Roswell Park Memorial Institute medium-1640 medium (Invitrogen, Grand Island, New York, USA) supplemented with 10% (v/v) foetal bovine serum. The reporter cell lines were generated by stable transfection of AGS cells with p53 luciferase reporters PG13-Luc and MG15-Luc following selection for p53 response. The following H. pylori strains were used: J166, B128 and 7.13; all strains were CagA-positive. H. pylori strain 7.13 was generated via in vivo adaptation of the B128 clinical isolate in gerbils as described previously.3 Additional H. pylori strains were isolated from antral mucosal biopsies from two groups of subjects in the state of Nariño, Colombia. One group consisted of individuals with intestinal metaplasia residing in the town of Túquerres at high altitude in the Andes mountains, where the incidence of gastric cancers is high (high-risk strains: PZ5056, PZ5074, PZ5086, PZ5091). Another group consisted of participants with non-atrophic gastritis from the Pacific coast town of Tumaco, where the incidence of gastric cancers is low (low-risk strains: PZ5001, PZ5009, PZ5010, PZ5018, PZ5024). H. pylori strains were grown in Brucella broth with 5% fetal bovine serum for 18 h, harvested by centrifugation, and added to gastric cells at a bacteria-to-cell ratio of 100:1. Heat-inactivated H. pylori were generated by heating the bacteria to 80°C for 10 min.
Plasmids, siRNA and antibodies
pcDNA3-myc-ARF (Addgene, Cambridge, Massachusetts, USA), pSP65SRα-CagA, MG15-Luc and PG13-Luc p53 luciferase reporters were described previously.8 shRNA vector against human p14ARF was kindly provided by Dr R Agami (Netherlands Cancer Institute, Netherlands). siRNA against HDM2 were obtained from Dharmacon (Lafayette, Colorado, USA), control siRNA was from Ambion (Grand Island, New York, USA), siRNA against ARF-BP1 was synthesised by Integrated DNA Technologies (Coralville, Iowa), using the following sequence: 5-GGAGGAAGAGG AACGGAAAGCUCGG-3.
The following antibodies were used: p53(DO-1), p21/Wild-type p53-activated fragment 1 (WAF1)(Ab-1) and ARF-BP1 from Calbiochem (San Diego, California, USA); β-Actin, p14ARF(4C6/4) and Phospho-Murine Double Minute 2 (MDM2)(Ser166) from Cell Signaling (Danvers, Massachusetts, USA); PMA-induced protein 1 (NOXA) from Imgenex (San Diego, California, USA); CDKN2A/p14ARF(ab470) from Abcam (Cambridge, Massachusetts, USA); MDM2(Ab-1) and phosphotyrosine from E. Merck, Darmstadt Millipore (Billerica, Massachusetts, USA); green fluorescent protein (GFP) from the Vanderbilt Monoclonal Antibody Core (Nashville, Tennessee, USA); CagA from Austral Biologicals (San Ramon, California, USA). Bacterial Heat Shock Protein B (HspB) antibody was a kind gift of Dr T Cover (Vanderbilt University).
Tissue specimens and immunohistochemistry
Gastric tissue specimens used in this study were obtained from 20 H. pylori-infected subjects who underwent GI tract endoscopy in two public hospitals in the State of Nariño, Colombia that are located in two areas with contrasting gastric cancer risks. The individuals included in this study were randomly selected and represent part of a larger series of subjects that has been described previously.9 All participants provided informed consent. The protocol was approved by the Committees on Ethics of Universidad del Valle and Hospital Departmental in Nariño, Colombia and by the Institutional Review Board at Vanderbilt University. All personal identifiers were removed prior to receiving specimens and were coded.
Expression of p53/ARF was assessed by immunohistochemistry using p53 (DO-1) and p14ARF (4C6/4) antibodies and detected using EnVision+HRP kit (DakoCytomation, UK). Immunohistochemial staining was evaluated for intensity and staining frequency in the nuclear compartment of superficial gastric epithelium. The intensity of staining was graded as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). The frequency was graded according to the percentage of positive cells. Total scores were calculated by multiplying the intensity score by the percentage of positive cells.
RNA Extraction, nested real-time (RT-PCR), pyrosequencing and luciferase reporter analysis
RNA was isolated using the RNeasy Kit (Qiagen, Valencia, California, USA). Two micrograms total RNAwas reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, California, USA) following the manufacturer’s protocol. p14ARF gene transcription was assessed by qPCR using the following primers: GTTTTCGTGGTTCACATCCC and ACCAGCGTGTCCAGGAAG. Nested PCR was used for analysis of p14ARF gene transcription in cell lines, which have low levels of ARF mRNA. Primers used for the first round of PCR were CAGTTAAGGGGGCAGGAGTG and CACCAGCGTGTCCAGGAAG; nested primers were GTTTTCGTGGTTCACATCCC and ACCAGCGTGTCCAGGAAG. Hypoxanthine-Guanine Phosphoribosyltransferase mRNA expression was used as an internal control using primers described previously.8
Luciferase activity was assessed using the Luciferase Reporter Assay Kit (Promega, Madison, Wisconsin, USA), as described earlier10 and normalised to total protein levels. Pyrosequencing analysis of the p14ARF promoter was performed using the EZ DNA Methylation-Direct Kit D5020 (Zymo Research, Irvine, California, USA).
Statistical analysis
Statistical analysis was performed using the Student’s t-test. Results were expressed as mean values (±SEM unless otherwise noted). To assess correlations between p14ARF and p53 expression levels, linear regression was calculated using GraphPad Prism software (GraphPad Software, La Jolla, California, USA). Results were considered statistically significant if p<0.05.
RESULTS
H. pylori strains varied in their abilities to regulate p53
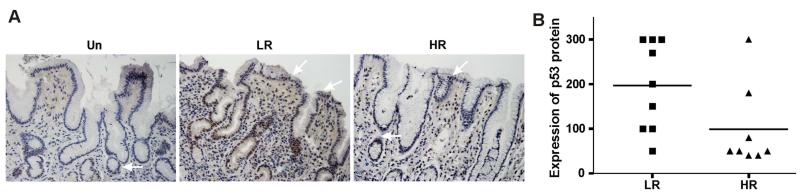
To investigate the regulation of p53 by H. pylori, we first examined the expression of p53 in patients with H. pylori infection who underwent GI tract endoscopy in two public hospitals in the State of Nariño, Colombia. The hospitals are located in two cities with contrasting gastric cancer risks: Túquerres in the Andes Mountains where gastric cancer incidence is high and Tumaco on the coast where gastric cancer incidence is low. Previous studies have reported that phylogenetically different groups of H. pylori bacteria with different tumorigenic potentials infect patients in these geographical areas contributing to the contrasting incidence rates of gastric tumours.4 Gastric biopsies from 20 patients with H. pylori infection with non-atrophic gastritis (10 patients in each group) were immunohistochemically stained with p53-specific antibody. p53 protein immunoreactivity was primarily found in epithelial cells in the foveolar compartment. Some mucosal inflammatory cells were also positive for p53 suggesting that tissue damage and inflammation associated with H. pylori infection may affect expression of p53. The expression of p53 was scored in the superficial gastric epithelium, a site where H. pylori primarily interacts with host cells. Our staining showed that p53 expression was consistently lower in patients from the high-risk gastric cancer area compared with the lower-risk counterparts (figure 1), suggesting that variability of H. pylori strains may differentially affect expression of p53.
Figure 1.
Expression of p53 protein varied between geographical areas with contrasting gastric cancer risk in Colombia. (A) Representative immunohistochemical staining for p53 protein of gastric biopsies collected from Helicobacter pylori-infected individuals from two areas: Túquerres where gastric cancer incidence is high (right) and Tumaco where gastric cancer incidence is low (centre). Left panel (Un) shows p53 staining of gastric biopsy collected from an uninfected patient. White arrows show epithelial cells expressing p53 protein. (B) Comparison of p53 nuclear expression in gastric biopsies collected from high (HR) and low (LR) gastric cancer risk areas. p53 expression was consistently lower in patients from the high gastric cancer risk area (p=0.05). The expression scoring is described in the Materials and Methods section.
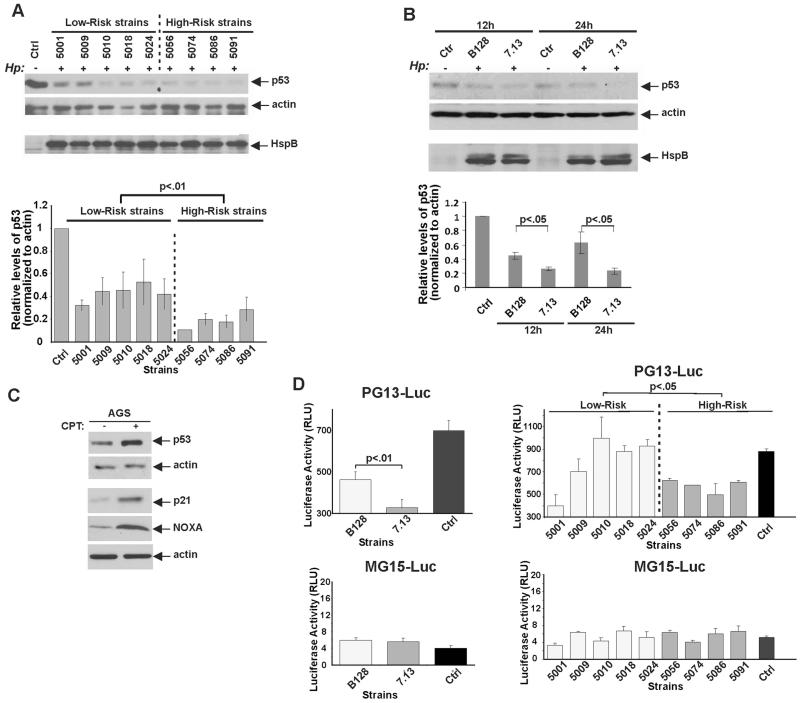
To further explore this phenomenon in a more controlled environment, five cagA-positive H. pylori strains isolated from patients residing in the low gastric cancer incidence area (PZ5001, PZ5009, PZ5010, PZ5018, PZ5024) and four strains from the area with high gastric cancer incidence (PZ5056, PZ5074, PZ5086, PZ5091) were co-cultured with AGS gastric cells, which harbour wild type p53 protein, for 24 h and analysed for expression of p53 in three independent experiments (figure 2A). We found that although the analysed strains varied in their ability to regulate p53, the high-risk strains were significantly more potent in decreasing expression of p53 than were the analysed low-risk strains (figure 2A; p=0.008, n=3). As a control for equal bacterial loading, cell lysates were analysed using an antiserum raised against the abundant H. pylori protein HspB, a bacterial chaperonin belonging to the Hsp60 class of proteins (GroEL) heat shock protein homologue (figure 2A).
Figure 2.
p53 expression is differentially regulated by high-risk and low-risk Helicobacter pylori. (A) Protein lysates were prepared from control AGS cells (−) or from those co-cultured with five low-risk and four high-risk Colombian H. pylori strains (+) for 24 h and analysed for expression of p53 protein by western blotting (upper panel). Bacterial load was assessed using HspB protein (bottom panel). Graphs show the results of densitometric analysis of immunoblots and depict expression of p53 protein normalised to actin (mean±SEM, n=3). p53 levels in uninfected control cells were arbitrarily set at 1. (B) The same as (A), but AGS cells were co-cultured with H. pylori strains B128 or 7.13 for 12 h or 24 h. Expression of the p53 protein was quantitated by densitometry and normalised to actin expression. (C) AGS cells were treated with 5 μM camptothecin (CPT) for 12 h and analysed for expression of p53 protein and its transcriptional targets p21/WAF1 and NOXA by western blotting. (D) AGS cells, stably expressing p53 reporter PG13-Luc, were co-cultured with H. pylori strains B128 or 7.13 for 8 h or left uninfected (Ctrl) and analysed for p53 activity. The luciferase data were normalised to total protein (right panel). Left panel: The same as above but AGS cells were co-cultured with Colombian low-risk and high-risk H. pylori strains. Bottom panels show data from mutant MG15-Luc reporter.
To further explore the H. pylori strains with varied tumorigenic potential, we next employed two previously characterised H. pylori strains, which have different abilities to induce gastric tumour: H. pylori clinical isolate B128 and its oncogenic derivative 7.13. The latter strain (but not the former) strongly activates cellular oncogenes, resulting in reproducible induction of premalignant and malignant gastric lesions in different rodent models.3 11 AGS gastric epithelial cells were cocultured with H. pylori strain 7.13 or B128 for 24 h and analysed for expression of p53. Similar to the Colombian high-risk strains shown above, we found that oncogenic H. pylori strain 7.13 was significantly more potent in downregulation of p53 than strain B128 (figure 2B; p=0.01, n=3).
To assess the functionality of the p53 pathway in the AGS cell line, p53 response was examined by treating cells with 5 μM camptothecin, a commonly used DNA-damaging drug. Treatment with camptothecin or other DNA-damaging drugs (cisplatin and etoposide) led to strong upregulation of the p53 protein in these cells (figure 2C). Drug treatment also induced transcriptional targets of p53: p21/WAF1 and NOXA (figure 2C, lower panels), showing that H. pylori downregulates the functionally active p53 protein. We next generated AGS cell lines stably expressing either p53 reporter (PG13-Luc) controlled by the promoter containing multiple repeats of the p53 binding site or its mutant version (MG15-Luc), which carries mutant p53 binding sites. PG13-Luc and MG15-Luc expressing cells were then co-cultured with the aforementioned H. pylori strains. Consistent with the analysis of p53 protein expression, oncogenic H. pylori strain 7.13 was significantly more potent in inhibition of p53 activity than B128 (figure 2D, left panel; p=0.01; n=5). Likewise, a statistically significant difference was found between Colombian high-risk and low-risk strains, where high-risk strains were significantly weaker inducers of PG13-Luc reporter activity than low-risk strains (figure 2D, right panel; p=0.05; n=3). No statistically significant differences between high-risk and low-risk strains were detected with the mutant MG15-Luc reporter (figure 2D; bottom panels) confirming the specificity of the analysis.
Regulation of p53 by H. pylori is mediated by p14ARF protein
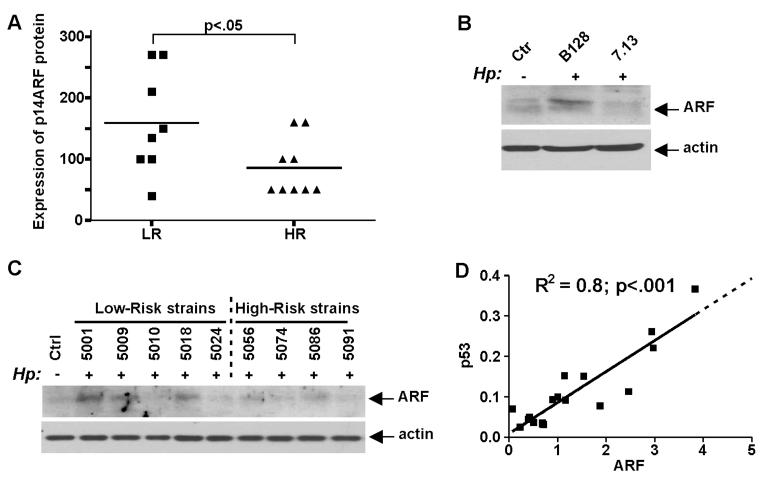
Previous studies have established that ARF is an upstream regulator of p53 that induces and activates p53 following oncogenic stress.12 To explore this mechanism in the settings of H. pylori infection, expression of p14ARF protein was assessed by immunohistochemistry in gastric biopsies (n=19) obtained from patients residing in Túquerres (high-risk) and Tumaco (low-risk) areas. We found that p14ARF staining was significantly lower in patients from the high gastric cancer risk area compared with the lower risk counterparts (figure 3A). As multiple factors may affect mucosal expression of ARF, we next analysed the regulation of p14ARF by various H. pylori strains. Cell lysates generated from AGS cells co-cultured with H. pylori strains 7.13 and B128 for 24 h were analysed for p14ARF protein by western blotting. We found that whereas co-culture of AGS cells with H. pylori strain B128 led to an increase in levels of p14ARF protein, weaker changes were found in cells co-cultured with the strain 7.13 (figure 3B) demonstrating that alterations in the p53 protein levels followed by infections with H. pylori strain B128 and 7.13 (shown in figure 2B) concur with the ARF protein levels.
Figure 3.
Regulation of p14ARF protein by Helicobacter pylori. (A) Comparison of p14ARF expression in gastric biopsies collected from high (HR) and low (LR) gastric cancer risk areas. ARF expression was significantly higher in patients from the low gastric cancer risk area (p<0.05). (B) Western blot analysis of p14ARF protein in AGS cells co-cultured with H. pylori strain B128 or 7.13. H. pylori strain B128 is a stronger inducer of ARF than its oncogenic derivative 7.13. (C) The same as (B), but Colombian low-risk and high-risk H. pylori strains were analysed. (D) Correlation analysis of p14ARF and p53 protein expression in AGS cells co-cultured with Colombian low-risk and high-risk H. pylori strains. p53 and ARF protein expression was analysed by densitometry and normalised to actin. p53 and ARF levels significantly correlate (R2=0.8, p<0.001).
Next, expression of p14ARF protein was assessed in AGS cells co-cultured with Colombian H. pylori high-risk and low-risk strains. p14ARF was found to be strongly induced by some low-risk strains while high-risk strains had generally lower levels of ARF induction (figure 3C). When p14ARF protein levels measured by densitometry were plotted against levels of p53, we found that expression of ARF and p53 levels strongly correlate (R2=0.8; p<0.001) (figure 3D).
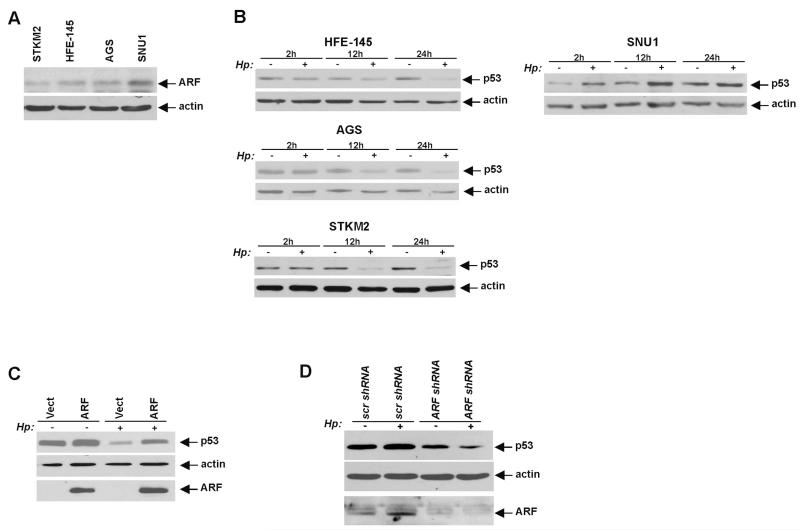
To explore the p14ARF regulatory mechanism in more detail, we analysed the ARF protein in a panel of gastric epithelial cell lines harbouring wild type p53 (HFE-145, AGS, STKM2 and SNU1). Cell lines were assessed for the p14ARF expression by western blotting and qPCR. Although expression of p14ARF protein and mRNA were low in AGS, HFE-145 and STKM2 cells and generally require more starting material for western blotting (100 μg) and enhanced PCR techniques, the ARF expression was significantly higher in SNU1 cells (figure 4A and see online supplementary figure S1). Given that the ARF promoter is frequently methylated in gastric cancer and gastric cancer cell lines, we also assessed the methylation of the p14ARF gene promoter by pyrosequencing and found that the levels of p14ARF mRNA cor-relate with methylation levels of the p14ARF gene promoter (see online supplementary figure S1).
Figure 4.
p14ARF regulates p53 protein in Helicobacter pylori-infected cells. (A) Protein lysates were prepared from SNU1, AGS, STKM2 and HFE-145 cells and analysed for p14ARF protein expression by western blotting. (B) Protein lysates were prepared from control uninfected (−) HFE-145, AGS, SNU1 and STKM2 cells or those co-cultured with H. pylori strain J166 (+) for the indicated time and analysed using western blotting with p53 antibody. (C) p53 level was assessed in AGS cells (which have low levels of endogenous ARF protein) transfected with either p14ARF expression vector (ARF) or empty control vector (Vect) and co-cultured with H. pylori (Hp) strain J166 (+) for 12 h or left uninfected (−). p14ARF transfection inhibited degradation of p53 by H. pylori. (D) Protein level of p53 was analysed in SNU1 cells (high levels of endogenous ARF protein), stably transfected with either p14ARF shRNA or scrambled control shRNA for 48 h and co-cultured with H. pylori strain J166 for an additional 12 h. Inhibition of p14ARF by shRNA suppressed H. pylori-mediated upregulation of p53.
Based on these data, we next examined the regulation of p53 by H. pylori strain 7.13 in these cell lines. A notable decrease of endogenous p53 protein was observed in all cell lines expressing low levels of ARF (HFE-145, AGS and STKM2) 12 h after co-culture with H. pylori strain 7.13 followed by a further decrease after 24 h. In contrast, the regulation of p53 in SNU1 cells, which express higher levels of p14ARF, was different from that of other cells lines. Following the co-culture with H. pylori strain 7.13, p53 protein was induced implying that ARF may protect the p53 protein against H. pylori-induced degradation (figure 4B).
To confirm these findings, we next asked whether H. pylori-induced degradation of p53 is affected by the alteration of ARF expression. AGS cells, which have low ARF expression, were transiently transfected with a vector expressing p14ARF and co-cultured with H. pylori strain 7.13 for 24 h. We found that downregulation of p53 was hindered in AGS cells transfected with p14ARF (figure 4C). A similar effect on p53 was observed in HFE-145 cells, which also have a low ARF expression (data not shown). Conversely, when ARF was down-regulated by a stable transfection of specific shRNA against p14ARF in SNU1 cells, which express high levels of ARF, p53 protein was not induced and its level remained low after co-culture with H. pylori strain 7.13, compared with control cells expressing scrambled shRNA (figure 4D). Thus, p14ARF is a critical regulator of p53 in H. pylori-infected cells.
HDM2 and ARF-BP1/Mule regulate levels of p53 protein in H. pylori-infected cells
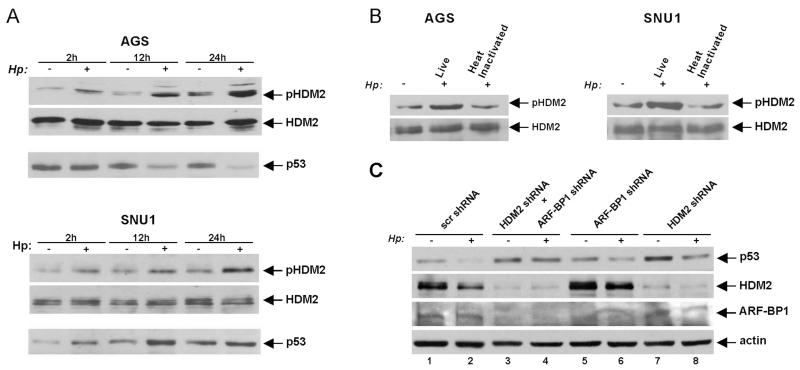
Previously, it has been demonstrated that p14ARF regulates p53 by inhibiting HDM2 protein.5 To examine the HDM2 protein in H. pylori-infected cells, we assessed the activity of HDM2 in cells that express low levels of ARF. AGS cells were co-cultured with H. pylori strain 7.13 and analysed for phosphorylation of HDM2 protein at serine 166, which is known to activate HDM2.13 Consistent with previous observations,8 HDM2 (Ser166) phosphorylation was strongly increased by H. pylori in a time-dependent manner (figure 5A). This increase coincided with a downregulation of p53 (figure 5A). We next repeated this experiment in SNU1 cells, which express high levels of ARF. Similar to AGS cells, H. pylori induced strong phosphorylation of HDM2 at Ser166. However, bacteria were unable to degrade p53 in SNU1 cells despite activation of HDM2 (figure 5A, bottom panel). Notably, heat-inactivated bacteria were unable to phosphorylate HDM2, showing that phosphorylation of HDM2 requires live bacteria (figure 5B; bottom panel).
Figure 5.
Regulation of p53 protein stability by HDM2 and ARF-BP1 E3 ubiquitin ligases in Helicobacter pylori-infected cells. (A) Protein lysates were prepared from control uninfected (−) AGS and SNU1 cells or those co-cultured with H. pylori strain 7.13 (+) for the indicated time and analysed by western blotting with pHDM2 antibody, which recognises HDM2 phosphorylation at position Ser166. (B) Analysis of HDM2 phosphorylation in AGS and SNU1 cells co-cultured with H. pylori strain 7.13 or heat-inactivated bacteria for 12 h. (C) AGS cells were transfected with either HDM2 siRNA or ARF-BP1 siRNA alone or both for 48 h. Cells were then co-cultured with H. pylori 7.13 for 12 h and assessed for p53 protein expression by western blotting.
To investigate the role of HDM2 in more detail, expression of HDM2 was downregulated with specific siRNA in AGS cells, which were then co-cultured with H. pylori strain 7.13 for 12 h. We found a strong decrease of p53 degradation mediated by H. pylori. However, even after inhibition of HDM2, H. pylori retained residual degrading activity, suggesting that additional factors contribute to p53 degradation (figure 5C; compare lanes 7 and 8 with 1 and 2). This led us to investigate additional E3 ubiquitin ligases. Since ARF-BP1/Mule protein regulates p53 in an ARF-dependent manner, expression of ARF-BP1 was inhibited with a specific siRNA.14 Similar to HDM2, inhibition of ARF-BP1 had only a partial effect on p53 degradation, implicating both E3 ligases in the degradation of p53 (figure 5C; compare lanes 5 and 6 with 1 and 2). To explore this possibility, HDM2 and ARF-BP1 were concurrently downregulated with siRNAs as described above. This led to significant suppression of p53 degradation, demonstrating that both E3 ligases, HDM2 and ARF-BP1, contribute to the degradation of p53 mediated by H. pylori (figure 5C; compare lanes 3 and 4 with 1 and 2).
p14ARF protects p53 protein against CagA-induced degradation
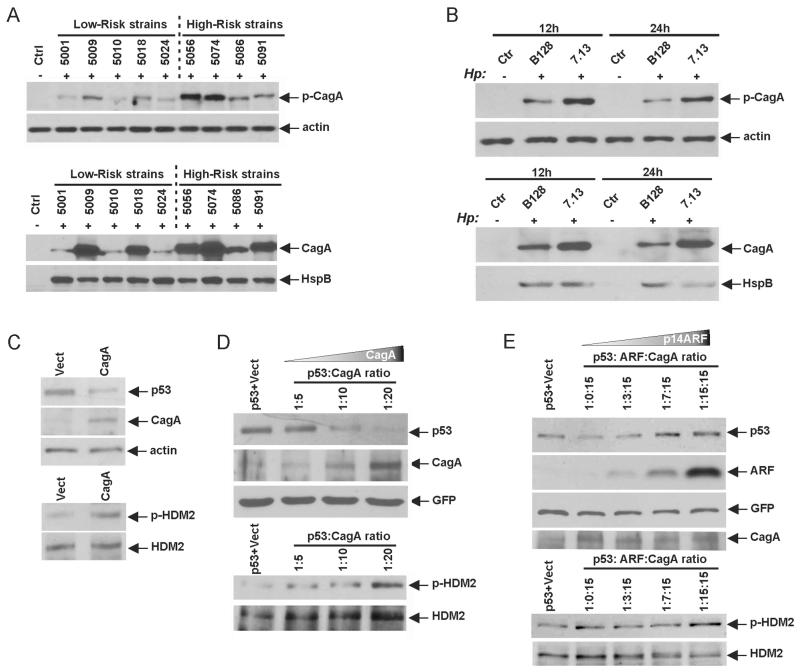
To explore how bacterial factors affect the ARF-p53 regulatory pathway, we next investigated the CagA protein, a bacterial oncogene, which is injected into gastric cells by H. pylori and then becomes phosphorylated on tyrosine residues by intracellular kinases.15 Using levels of tyrosine phosphorylation of CagA as a readout for CagA intracellular delivery, we determined the protein phosphorylation of CagA following the co-culture of AGS cells with the aforementioned high-risk and low-risk strains (figure 6A, upper panel). Our analyses revealed that CagA proteins injected by the Colombian high-risk H. pylori strains have higher levels of tyrosine phosphorylation than low-risk strains. Moreover, total protein levels of CagA were also higher in high-risk strains (figure 6A, bottom panel). Similar differences were found between H. pylori strains 7.13 and B128, where expression and phosphorylation of CagA protein were significantly higher after co-culture of AGS cells with the oncogenic H. pylori strain 7.13 (figure 6B). These data suggest that H. pylori strain 7.13 delivers more intracellular CagA protein causing stronger inhibition of p53.
Figure 6.
p14ARF prevents degradation of p53 induced by bacterial cytotoxin-associated gene A (CagA) protein. (A) Analyses of levels (bottom panel) and global tyrosine phosphorylation of CagA protein (upper panel) after co-culture of AGS cells with the indicated high-risk and low-risk Helicobacter pylori strains for 24 h. Phosphorylation of CagA protein and its level were assessed using pTyr and CagA antibodies, respectively. Gel loading was normalised for expression of actin and HspB proteins. (B) The same as (A) but H. pylori strain B128 and 7.13 were analysed. The 12 h and 24 h time points are shown. (C) AGS cells were transfected with CagA-expressing plasmid for 24 h and analysed for the expression of endogenous p53, pHDM2(Ser166) and CagA proteins using western blotting. (D) p53-null Kato III cells were cotransfected with p53 and CagA-expressing plasmids at the indicated ratios for 48 h and analysed for p53, pHDM2(Ser166) and CagA. To normalise for transfection efficiency cells were cotransfected with an equal amount of green fluorescent protein (GFP)-expressing plasmid. (E) The same as (D) but Kato III cells were cotransfected with p53, CagA and an increasing amount of p14ARF-expressing plasmid at the indicated ratios for 48 h and analysed for p53, ARF, pHDM2(Ser166) and CagA proteins using western blotting.
To recapitulate bacterial injection of CagA protein, we trans-fected AGS cells with CagA-expressing plasmid and analysed the expression of p53 and phosphorylation of HDM2 (Ser166). We found that CagA transfection induced a strong phosphorylation of HDM2 and downregulation of endogenous p53 (figure 6C). We next cotransfected p53 plasmid with an increased amount of CagA plasmid into Kato III gastric cells, which lack the expression of both p53 and p14ARF.16 To normalise for differences in transfection efficiency, cells were also cotransfected with an equal amount of GFP-expressing plasmid. Similar to AGS cells, CagA decreased levels of the p53 protein in Kato III cells (figure 6D). Notably, downregulation of p53 and phosphorylation of HDM2 (Ser166) were determined by relative expression levels of the CagA protein. To assess the role of p14ARF in the CagA-dependent downregulation of p53, we next cotransfected p14ARF into Kato III cells and found that p14ARF completely blocked downregulation of p53 induced by CagA (figure 6E). Importantly, a suppressive effect of p14ARF on CagA-induced degradation was dependent on its relative expression. Interestingly, p14ARF did not prevent phosphorylation of the HDM2 protein induced by CagA similar to what was seen in SNU1 cells. Taken together, our data demonstrate that p14ARF protects p53 protein against CagA-induced degradation.
DISCUSSION
H. pylori represents a fine example of a microbial pathogen that has successfully adapted to its host environment by modulating intracellular signalling pathways in ways beneficial to the bacteria. Unfortunately, these alterations carry serious risks for the human hosts, increasing the likelihood of cancer development. Our current studies have characterised a novel mechanism of p53 tumour suppressor regulation in gastric epithelial cells interacting with H. pylori. We found that degradation of p53 protein induced by bacterial CagA protein is controlled by the p14ARF tumour suppressor and that two host E3 ubiquitin ligases, ARF-BP1 and HDM2, are involved in this process. The extent of p53 degradation is linked to the efficiency of the T4SS that delivers CagA protein into host cells. We found that in our model system, where we studied the degradation of p53 with clinical isolate B128 and its oncogenic derivate 7.13, the latter strain was more efficient in CagA delivery. Providing that the CagA sequence of H. pylori strain 7.13 is identical to that of strain B128,3 a higher concentration of intracellular CagA may explain the stronger effect of H. pylori strain 7.13 on p53. However, CagA protein variability may also contribute to the extent of the p53 degradation for Colombian high-risk and low-risk strains. Further studies are needed to investigate this mechanism.
Our data suggest that p53 degradation is linked to tumorigenic potential of a particular H. pylori strain. We found that high-risk H. pylori strains were generally more potent in down-regulation of p53 than were low-risk counterparts. This effect was found using Colombian H. pylori strains and the clinical isolate B128 and its oncogenic derivative 7.13. The latter two strains have been previously characterised using gerbil and mouse animal models in vivo, where H. pylori strain 7.13 has been found to rapidly induce gastric dysplasia and adenocarcinoma while the B128 strain was non-tumorigenic.3 11 It was also supported by our findings that Colombian patients in the areas of high gastric cancer incidence and infected with high-risk H. pylori strains had generally lower expression of p53 than patients with H. pylori infection from the coastal areas where gastric cancer incidence is low.
We propose a model in which the fate of the p53 protein in H. pylori-infected cells is determined by relative levels of p14ARF. In gastric cells expressing p14ARF protein at levels that are not sufficient to neutralise activation of HDM2 and ARF-BP1, bacterial CagA protein causes degradation of p53 protein. Of note, degradation of p53 was recapitulated by transfection of CagA implying that CagA protein alone is sufficient to decrease levels of p53. In contrast, H. pylori-induced degradation of p53 is inhibited and levels of p53 are increased in cells with high p14ARF levels. Indeed, our data show that levels of p14ARF strongly correlate with p53 in gastric cells co-cultured with H. pylori. p53 degradation was observed only in gastric cells in which the p14ARF gene promoter is partially methylated and, as a result, expression of ARF is decreased. These findings were further confirmed by manipulating ARF expres-sion by either overexpression of ectopic protein or downregulation of endogenous ARF with shRNA. We found that when levels of p14ARF were sufficiently increased, it completely blocked the CagA-induced degradation of p53.
It has been reported that methylation of the p14ARF promoter is increased during tumour progression in the stomach. Compared with normal gastric mucosa, a twofold to threefold increase in ARF gene methylation has been found in patients with chronic gastritis and intestinal metaplasia.6 These early epigenetic alterations are further exacerbated in gastric cancer affecting approximately 30% of patients with tumour.6 Notably, the frequency of p14ARF gene methylation is increased as a function of aging, suggesting that old people with gastric precancerous lesions may be vulnerable to degradation of p53 by H. pylori.17
In summary, we found that ARF tumour suppressor protects p53 proteins against degradation induced by injection of bacterial CagA protein into gastric epithelial cells. H. pylori degrades p53 in a strain-specific manner; strains that are able to deliver more intracellular CagA have higher degrading ability and tumorigenic potential.
Supplementary Material
Significance of this study.
What is already known on this subject?
-
▶
Helicobacter pylori infection is strongly associated with the development of gastric cancer. Using the type IV secretion system (T4SS), H. pylori delivers the bacterial cytotoxin-associated gene A (CagA) protein into gastric epithelial cells causing activation of multiple oncogenic pathways. CagA has been characterised as a bacterial oncoprotein that plays an important role in H. pylori-induced tumorigenesis.
-
▶
p53 and p14ARF proteins are tumour suppressors that prevent gastric tumorigenesis. As a result of gastric tumour development, both proteins are frequently inactivated.
-
▶
p14ARF and p53 proteins functionally interact; p14ARF activates p53 under conditions of oncogenic stress limiting cellular proliferation and eliminating transformed cells that otherwise may cause tumour development.
-
▶
H. pylori affects the expression of p53 protein following the direct interaction of gastric epithelial cells with the bacteria.
What are the new findings?
-
▶
This study provides the first evidence that the ability of H. pylori to affect p53 is associated with its tumorigenic potential. High-risk H. pylori strains were found to be more potent in downregulation of p53 than low-risk counterparts.
-
▶
Degradation of p53 protein induced by H. pylori is linked to the efficiency of the T4SS that delivers CagA protein into host cells.
-
▶
This study characterised a novel mechanism of p53 tumour suppressor regulation in gastric epithelial cells interacting with H. pylori. Degradation of p53 protein induced by bacterial CagA protein is controlled by the p14ARF tumour suppressor. Two host E3 ubiquitin ligases, ARF-BP1 and HDM2, are involved in this process.
-
▶
p14ARF protects the p53 protein against H. pylori-induced degradation. Hypermethylation of the p14ARF promoter facilitates degradation of the p53 protein by H. pylori.
How might it impact on clinical practice in the foreseeable future?
-
▶
Taken together, our studies show that the ability of H. pylori to affect the protein degradation of p53 may serve as a prediction maker of the tumorigenic potential of a particular H. pylori strain. This may help to develop novel strategies for prevention of gastric cancer.
Acknowledgements
The authors thank Dr T Cover (Vanderbilt University, Nashville, Tennessee, USA) for providing help and HspB antibody.
Funding This work was supported by the National Cancer Institute grants NIH CA138833, Vanderbilt Digestive Disease Research Center (P30DK058404), Vanderbilt Ingram Cancer Center (P30CA68485), P01 CA28842 and CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences.
Footnotes
Contributors All authors meet the criteria for authorship. JW, JMN, EZ, JR-G, MBP and BS conducted experimental and clinical work described in the manuscript. BS, WE-R, PC, RMP, AIZ contributed to analyses, planning and reporting results described in the manuscript.
Competing interests None.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Zhu Y, Zhong X, Zheng S, et al. Transformed immortalized gastric epithelial cells by virulence factor CagA of Helicobacter pylori through Erk mitogen-activated protein kinase pathway. Oncogene. 2005;24:3886–95. doi: 10.1038/sj.onc.1208551. [DOI] [PubMed] [Google Scholar]
- 2.Ohnishi N, Yuasa H, Tanaka S, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003–8. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco AT, Israel DA, Washington MK, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:10646–51. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodaman N, Pazos A, Schneider BG, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA. 2014;111:1455–60. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol. 2012;1:731–41. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106:483–93. doi: 10.1002/cncr.21657. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto Y, Marusawa H, Kinoshita K, et al. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–6. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 8.Wei J, Nagy TA, Vilgelm A, et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology. 2010;139:1333–43. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Sablet T, Piazuelo MB, Shaffer CL, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60:1189–95. doi: 10.1136/gut.2010.234468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei J, O’Brien D, Vilgelm A, et al. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology. 2008;134:1412–23. doi: 10.1053/j.gastro.2008.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox JG, Rogers AB, Ihrig M, et al. Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific. Cancer Res. 2003;63:942–50. [PubMed] [Google Scholar]
- 12.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–9. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhou BP, Liao Y, Xia W, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 14.Chen D, Shan J, Zhu WG, et al. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–7. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asahi M, Azuma T, Ito S, et al. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med. 2000;191:593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida S, Akiyama Y, Nakajima T, et al. Alterations and hypermethylation of the p14 (ARF) gene in gastric cancer. Int J Cancer. 2000;87:654–8. doi: 10.1002/1097-0215(20000901)87:5<654::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Kang GH, Lee HJ, Hwang KS, et al. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–6. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.