Retrotransposons have a way of staying under the radar. These mobile DNA elements are well-known agents of genome instability and evolution but are mostly thought of as oddities occasionally associated with an interesting phenotype or worse, because of their high copy number, confound the analysis of genome sequences. However, as much as two-thirds or more of the human genome is derived from repetitive sequences (1). Most of these are retrotransposon sequences that were active in the distant evolutionary past and are now present as fossils that litter our genomes. But this image belies the damage that can be wreaked by evolutionarily recent transposable elements.
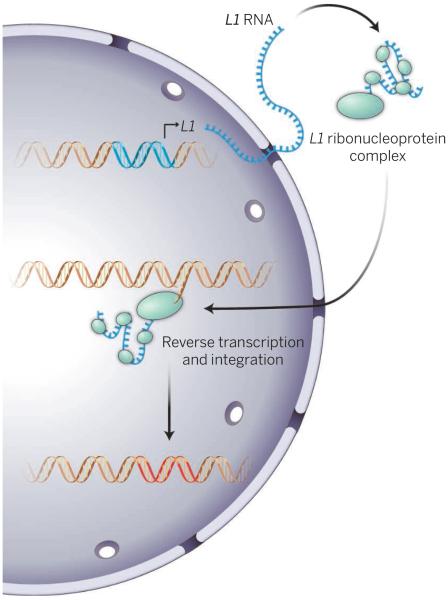
Retrotransposons can be copied into mRNA and then back to DNA that can integrate into the genome. The human genome harbors ~100 copies of an autonomously active retrotransposon called long interspersed nuclear element–1 (LINE-1) Homo sapiens (L1Hs). The L1Hs-encoded proteins, including a reverse transcriptase, are essential for the retrotransposition process and form a ribonucleoprotein particle with the mRNA. A second abundant human retrotransposon, the Alu element, depends on L1Hs for its movement as it does not encode any proteins, and its transcripts are assembled with L1Hs-encoded proteins. Both L1Hs and Alu ribonucleoprotein particles then enter the nucleus and by a process called target primed reverse transcription, insert new DNA elements at quasi-random sites throughout the genome (see the second figure).
Figure 2. Integration.
L1-encoded RNA and proteins assemble into ribonucleoprotein particles. Reverse transcription of L1 RNA is coupled to insertion into DNA at random sites throughout the genome.
As the “selfish gene” theory predicts (2), for a transposable element to succeed in the evolutionary race, the critical battleground is the germ line. There are numerous examples of insertions that affect human health (3). Because of this constant threat of genetic mayhem, cells have an impressive armamentarium to combat mobile elements. Where does this battle stand today? On one hand, the human retrotransposon load has been reduced to perhaps as few as 100 active elements; yet, two thirds of the human genome is scarred by the evidence of millions of years of warfare against mobile DNA elements, and new insertions occur at a frequency of 1 per 95 to 270 live births for L1Hs, and 1 in 20 for Alu (3).
What happens outside the germ line, in somatic tissues? Historically, little attention has been given to this question, because somatic retrotransposition is evolutionarily a dead-end process. However, rampant genomic instability and mutagenesis are deleterious to all cells, and somatic cells mount the same spectrum of defenses as those of the germ line. These include epigenetic chromatin modification, interfering RNAs, repression systems based on specific DNA-binding proteins (with zinc finger motifs), and autophagy. But these surveillance mechanisms are not foolproof. Derepression of L1 loci, L1 transcription, L1 proteins, and de novo L1 insertions have been detected in a variety of somatic contexts, including early embryos, adult brain, and certain stem cells (4). In the context of cancer, new L1 insertions have been found in a variety of tumor types, including colorectal, prostate, and ovarian tumors (5). Intriguingly, the incidence of retrotransposition in tumors appears to increase with age (6).
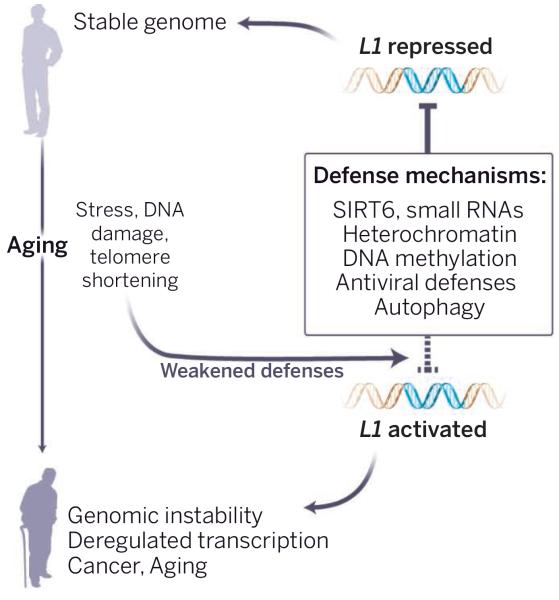
Aging presents an especially complex situation. Retrotransposon surveillance needs to be high in reproductively active individuals. However, because natural selection drops in the postreproductive period of life, host defense mechanisms may begin to fail with age. Could this allow the activation of retrotransposons (see the first figure)? Indeed, derepression of retrotransposons was documented during replicative senescence of human cells (7) and aging in yeast (8). It was also observed in the fly nervous system (9) and several mouse tissues, including liver, muscle, and brain (10, 11). In several contexts, derepression was associated with increased retrotransposon copy number and genome instability. Mutations in flies that derepress retrotransposons exacerbate age-dependent memory impairment and shorten life span (9). Calorie restriction delays age-related disorders and extends life span in most species, and also opposes the derepression of retrotransposons in mouse liver and skeletal muscle (10). A genetic intervention that increases surveillance by small interfering RNA reduces retrotransposon expression and extends life span in flies (12).
Figure 1. Aging guards invite a jailbreak.
With aging, increased stress, DNA damage, and telomere shortening weaken the multiple systems that keep retrotransposons in check. Aged cells lose repressive heterochromatin, SIRT6 relocalizes away from L1 promoters, and autophagy becomes less efficient. Other defense pathways (see box) may also lose their effectiveness. The consequent unleashing of L1 elements could lead to profound somatic damage, driving age-associated cell and tissue dysfunction.
What could be the processes that fail and awaken these “sleeping dogs?” One prime candidate is the maintenance of repressive heterochromatin. Repetitive regions lose DNA methylation during aging, and widespread changes in chromatin modifications are increasingly being documented. Recently, a connection between aging, chromatin, and L1 has been established through SIRT6 (11). SIRT6 is a prototypical longevity gene–mice without Sirt6 age prematurely, and mice overexpressing Sirt6 exhibit life-span extension (13). SIRT6 is a protein deacetylase and mono-ADP ribosyltransferase that promotes chromatin silencing and facilitates DNA repair. SIRT6-deficient cells show a marked derepression of L1 transcription. SIRT6 silences L1 by binding to its promoter and recruiting additional silencing factors. Interestingly, upon DNA damage, SIRT6 leaves L1 promoters and relocalizes to the sites of DNA breaks. It is likely that a similar process plays out during aging: Chronic DNA damage and short telomeres accumulate, SIRT6 is redeployed, and the dormant retrotransposons are left unguarded.
Beyond generating new insertions, retrotransposons can result in aberrant expression of nearby genes through the promoters and cryptic splice sites that they harbor. Ribonucleoprotein particles that they form in the cytoplasm could trigger immune responses (antiviral defenses) or overwhelm the capacity of homeostatic mechanisms such as autophagy (14), and contribute to neurodegeneration or autoimmune disorders. Further, abortive retrotransposition can cause DNA damage and genotoxic stress.
Although aging is perhaps the most basic aspect of life, the mechanisms that explain it remain a puzzle. Retrotransposon activation brings in yet another dimension: We may be bogged down in a complex host–parasitelike struggle (with evolution acting on both parties), leaving open the possibility for profound collateral damage on our soma.
What can be done against this broad attack? Clearly, letting sleeping dogs lie by keeping them mired in heterochromatin is a compelling strategy. For this we need drugs targeted at chromatin regulators that could maintain distinct euchromatin and heterochromatin characteristics of the youthful state. Shoring up other processes that may decrease in effectiveness during aging, like small RNA pathways (12) or autophagy, should also help to rein in retrotransposons. In addition, reverse transcriptase inhibitors can prevent the spread of new elements throughout the genome. These drugs have been highly successful in treating HIV/AIDS but have side effects. However, experiments in mice are a feasible way to explore the merit of the overall strategy, and may warrant the development of new drugs highly specific to L1Hs.
Many questions remain. For example, the landscape of somatic retrotransposition across our tissues is unclear. It is also uncertain how the mechanisms that oppose retrotransposons change with age. Most importantly, investigating the impact of somatic retrotransposon activation on cellular physiology, disease, and aging should be a high research priority. Could controlling retrotransposons have beneficial therapeutic effects?
ACKNOWLEDGEMENTS
We gratefully acknowledge the National Institute on Aging program staff (M. Guo, R. Kohansky, F. Sierra) for organizing a workshop in August 2014 where many of these ideas were discussed and consolidated. We acknowledge the following funding sources: National Institutes of Health (NIH) grants R01AG027237 and P01AG047200 (V.G.); NIH grant P50GM107632 (J.D.B.); NIH grants R37AG016667 and R01AG024353 (S.L.H.); NIH grants R37AG016694, P30GM103410, and T32AG041688 and Samsung GRO program grant (J.M.S.); and Ellison Medical Foundation (V.G., S.L.H., J.M.S.), Life Extension Foundation (V.G.), Glenn Foundation for Medical Research (V.G., S.L.H., J.M.S.), and Glenn/American Federation for Aging Research BIG award (S.L.H. and J.M.S.).
REFERENCES AND NOTES
- 1.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawkins R. The Selfish Gene. Oxford Univ. Press; New Your: 1976. [Google Scholar]
- 3.Solyom S, Kazazian HH., Jr. Genome Med. 2012;4:12. doi: 10.1186/gm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faulkner GJ. FEBS Lett. 2011;585:1589. doi: 10.1016/j.febslet.2011.03.061. [DOI] [PubMed] [Google Scholar]
- 5.Lee E, et al. Science. 2012;337:967. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solyom S, et al. Genome Res. 2012;22:2328. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Cecco M, et al. Aging Cell. 2013;12:247. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell PH, Burhans WC, Curcio MJ. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20376. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, et al. Nat. Neurosci. 2013;16:529. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Cecco M, et al. Aging. 2013;5:867. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Meter M, et al. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savva YA, et al. Nat. Commun. 2013;4:2745. doi: 10.1038/ncomms3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kugel S, Mostoslavsky R. Trends Biochem Sci. 2014;39:72. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H, et al. Nat. Commun. 2014;5:5276. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]