Abstract
Background
Rho GTPases function as molecular switches in many different signaling pathways and control a wide range of cellular processes. Rho GDP-dissociation inhibitors (RhoGDIs) regulate Rho GTPase signaling and can function as both negative and positive regulators. The role of RhoGDIs as negative regulators of Rho GTPase signaling has been extensively investigated; however, little is known about how RhoGDIs act as positive regulators. Furthermore, it is unclear how this opposing role of GDIs influences the Rho GTPase cycle. We constructed ordinary differential equation models of the Rho GTPase cycle in which RhoGDIs inhibit the regulatory activities of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) by interacting with them directly as well as by sequestering the Rho GTPases. Using this model, we analyzed the role of RhoGDIs in Rho GTPase signaling.
Results
The model constructed in this study showed that the functions of GEFs and GAPs are integrated into Rho GTPase signaling through the interactions of these regulators with GDIs, and that the negative role of GDIs is to suppress the overall Rho activity by inhibiting GEFs. Furthermore, the positive role of GDIs is to sustain Rho activation by inhibiting GAPs under certain conditions. The interconversion between transient and sustained Rho activation occurs mainly through changes in the affinities of GDIs to GAPs and the concentrations of GAPs.
Conclusions
RhoGDIs positively regulate Rho GTPase signaling primarily by interacting with GAPs and may participate in the switching between transient and sustained signals of the Rho GTPases. These findings enhance our understanding of the physiological roles of RhoGDIs and Rho GTPase signaling.
Electronic supplementary material
The online version of this article (doi:10.1186/s12918-015-0143-5) contains supplementary material, which is available to authorized users.
Keywords: RhoGDI, Rho GTPases, Ordinary differential equation, GAPs, GEFs
Background
Rho family GTPases are members of the Ras GTPase superfamily and act as molecular switches in numerous signaling pathways that control a variety of cellular processes, including actin cytoskeletal organization, microtubule dynamics, vesicle trafficking, cell cycle progression, and cell polarization [1]. Most Rho GTPases cycle between active GTP-bound and inactive GDP-bound states. There are three classes of regulators of Rho GTPases, namely, guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and GDP-dissociation inhibitors (GDIs).
GEFs activate GTPases by promoting the exchange of GDP for GTP. GAPs inactivate GTPases by stimulating their intrinsic GTP-hydrolyzing activity. GDIs are known to regulate only members of the Rho and Rab families and not other families of the Ras superfamily, although a GDI-like protein for Ras GTPases has been reported [2]. Unlike GEFs and GAPs, GDIs play several roles in the regulation of the Rho family GTPases [3-6]. First, GDIs bind GDP-bound GTPases and inhibit the dissociation of GDP from GTPases, thereby preventing the promotion of GDP/GTP exchange by GEFs and maintaining the GTPases in an inactive state [7]. Second, although the binding affinity of GDIs to GTP-bound GTPases remains controversial [8-15], it is possible that GDIs bind GTP-bound GTPases and inhibit both intrinsic and GAP-promoted GTP hydrolyzing activity [8,16,17], thereby maintaining GTPases in an active state. Third, GDIs mediate the cycling of GTPases between cytosolic and target sites [7].
GDIs for the Rho family GTPases can therefore act to inhibit both the activation and inactivation of GTPases by interacting with GDP- and GTP-bound GTPases, respectively. This dual function of GDIs is noteworthy, and adds to our understanding of the regulatory mechanisms of the Rho GTPase cycle, because GDIs for Rab family GTPases show a marked preference for the GDP-bound form [18]. Furthermore, it has also been suggested that Rho GTPases are regulated by a fine balance between GEF and GAP activities, and that the inactivation of GAP activity is a physiologically important regulatory mechanism for activating Rho GTPases [19]. Nonetheless, little is known about the significance of the inhibition of GAP-promoted GTP hydrolyzing activity by GDIs in the regulation of Rho signaling. How the opposing roles of GDIs influence the Rho GTPase cycle is also unclear.
Several ordinary differential equation models and process models of the Rho GTPase cycle have been constructed and analyzed [20-24]. In these models, GDIs inhibit the functions of GEFs and GAPs by sequestering GDP-bound and GTP-bound GTPases, respectively. However, the actual mechanisms involved in GDI inhibition of GEF and GAP activity are not fully understood. A previous report suggested that RhoGDIs can physically interact directly with both GEFs [25] and GAPs [26]. Based on these observations, we constructed a model of the Rho GTPase cycle in which GDIs inhibit the activities of GEFs and GAPs not only by sequestering GTPases, but also by direct physical interaction.
Using this model, we showed that the functions of GEFs and GAPs are integrated into Rho GTPase signaling through the interactions of these regulators with GDIs and that the negative role of GDIs is to suppress the overall Rho activity by inhibiting GEFs. Additionally, the positive role of GDIs is to sustain Rho activation by inhibiting GAPs. These observations illustrate the more detailed roles RhoGDIs and further enhance our understanding of the physiological functions of Rho GTPase signaling.
Results
Interaction of GDI with GAP sustains Rho activation
A Rho GTPase switch can be regulated by three classes of regulators: GEFs, GAPs, and GDIs. In the canonical model of the Rho GTPase cycle (Figure 1A, left), GEFs promote GDP/GTP exchange, thereby activating Rho GTPases. In contrast, GAPs promote GTP hydrolysis, thereby inactivating Rho GTPases. GDIs sequester GDP-bound GTPases from GEFs and keep them inactive; however, GDIs can also sequester GTP-bound GTPases from GAPs and keep them active. In this model, the Rho GTPase cycle functions as a simple ON/OFF switch and Rho activation is transiently elevated upon stimulation (Figure 1A, right).
Figure 1.
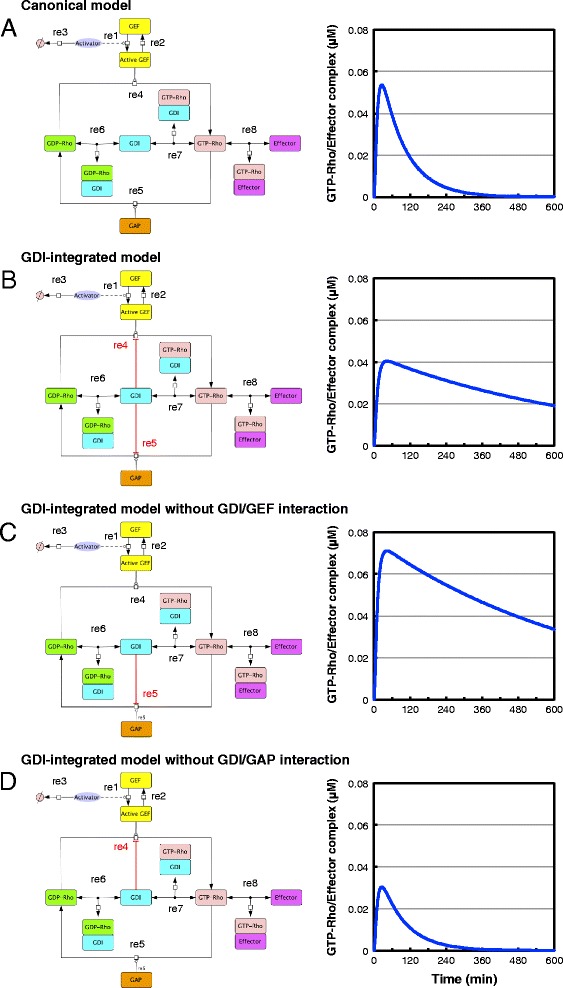
Representation of the models of Rho GTPase cycle regulation (left) and simulations of their Rho activation dynamics (right). The activation levels of GTPases were defined as the concentration of the GTP-Rho/Effector complex. A) The canonical model of the Rho GTPase cycle in which GDIs inhibit the activities of GEFs and GAPs by sequestering GTPase. B) The GDI-integrated model of the Rho GTPase cycle in which GDIs inhibit the activities of GEFs and GAPs not only by sequestering GTPase but also by interacting with GEFs and GAPs. C) GDI/GEF interaction was removed from the GDI-integrated model. D) GDI/GAP interaction was removed from the GDI-integrated model. All parameters and reactions in the models are shown in Additional file 1: Tables S1 and S2. Reaction numbers (re#) correspond to the reaction numbers in Additional file 1: Table S2.
The majority of Rho GTPases exist in biologically inactive cytosolic complexes with GDIs, and the dissociation of GTPases from GDIs is hypothesized to be a prerequisite for activation by GEFs. However, it has been suggested that GDI and Rho GTPase can simultaneously bind GEF or GAP and form a ternary complex (GEF/GDI/Rho GTPase or GAP/GDI/Rho GTPase) [25-27]. According to these observations, we constructed a model of the Rho GTPase cycle (Figure 1B, left) in which GDIs inhibit the activities of GEFs and GAPs by physically interacting with them as well as by sequestering Rho GTPases (see Methods). We designated this model the ‘GDI-integrated model’ because the activation dynamics and ultimate output of GEFs and GAPs are integrated by GDIs to regulate Rho activity. Rho activation is sustained for a longer period of time in this model (Figure 1B, right), compared with the canonical model (Figure 1A, right).
To clarify which interaction of GDIs with GEFs or GAPs participates in this sustained Rho activation, we further modified our GDI-integrated model. When the interaction of GDIs with GEFs was removed (Figure 1C, left), similar Rho activation dynamics, with a two-fold increase in the overall level, were obtained (Figure 1C, right). In contrast, when the interaction of GDIs with GAPs was removed (Figure 1D, left), Rho activation level decreased and was not sustained (Figure 1D, right). These results therefore suggest that GDIs sustain Rho activation through interaction with GAPs.
Influence of free (non-GTPase-complexed) GDI levels on Rho activation dynamics
To confirm the contribution of GDIs in sustaining Rho activation, we simulated Rho activation dynamics in the presence of various cellular concentrations of free GDIs, i.e., GDIs not complexed with GTPases. Based on the literature [28], we calculated the concentration of free RhoGDIα to be 0.7 μM (Additional file 1: Table S1). We used a range of concentrations of free GDIs close to this value to simulate the Rho activation dynamics. The canonical model predicted that an increase in free GDIs would simply lead to an overall decrease in Rho activation (Figure 2A). However, in our GDI-integrated model, while the increase of free GDIs also led to an overall decrease in Rho activation, this did not negate the sustained Rho activation (Figure 2B). Unexpectedly, the presence of free GDIs sustained the Rho activation level beyond 1,800 min after stimulation, in contrast to the cessation observed at this time point in the absence of free GDIs (Figure 2C).
Figure 2.
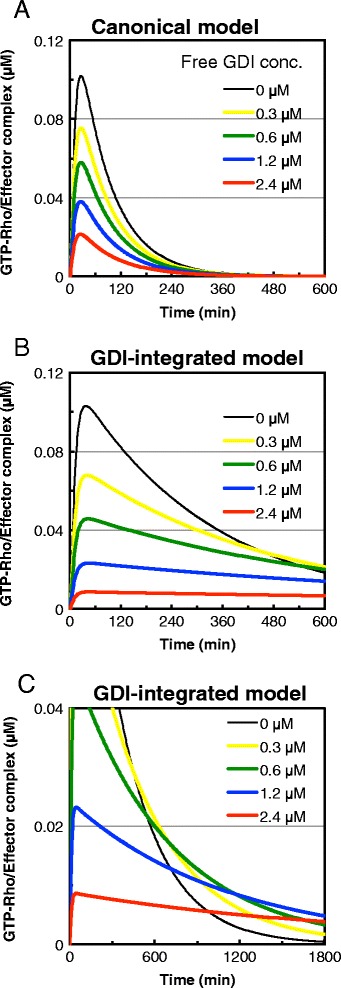
Free (non-GTPase-complexed) GDI concentration affects the prolongation of Rho activation in the GDI-integrated model. Rho activation dynamics were simulated at various concentration of free GDI. A) 600 min after stimulation in the canonical model. B) 600 min after stimulation in the GDI-integrated model. C) 1,800 min after stimulation in the GDI-integrated model. The activation levels of GTPases were expressed as the concentration of GTP-Rho/Effector complex.
Influence of GDI affinity for GEF and GAP and the concentration of GEF and GAP on Rho activation dynamics
Phosphorylation affects the affinity of GDIs for various Rho GTPases [29-33] and affects the function of GEFs [34,35] and GAPs [36-38]. Therefore, phosphorylation may modify the regulation of Rho signaling by GDIs, GEFs and GAPs. To examine how the affinity of GDIs for GEFs (KmGEF/GDI) and GAPs (KmGAP/GDI) affects the ability of GDIs to sustain Rho activation, we simulated the Rho activation dynamics at 0.01, 0.1, and 1.0 μM of KmGEF/GDI and KmGAP/GDI in our model. The decrease of KmGEF/GDI resulted in overall decrease of Rho activation at all the tested concentrations of KmGAP/GDI (Figure 3A). The Rho activation was markedly sustained at 0.01 and 0.1 μM of KmGAP/GDI and decreasing KmGEF/GDI did not negate the sustained Rho activation (Figure 3A). Conversely, as the KmGAP/GDI value became smaller, the Rho activation was sustained to a greater degree at all KmGEF/GDI (Figure 3B). These results indicate that the sustained Rho activation can primarily be attributed to the interaction between GAPs and GDIs, and the higher affinity of GDIs for GAPs promotes sustained Rho activation.
Figure 3.
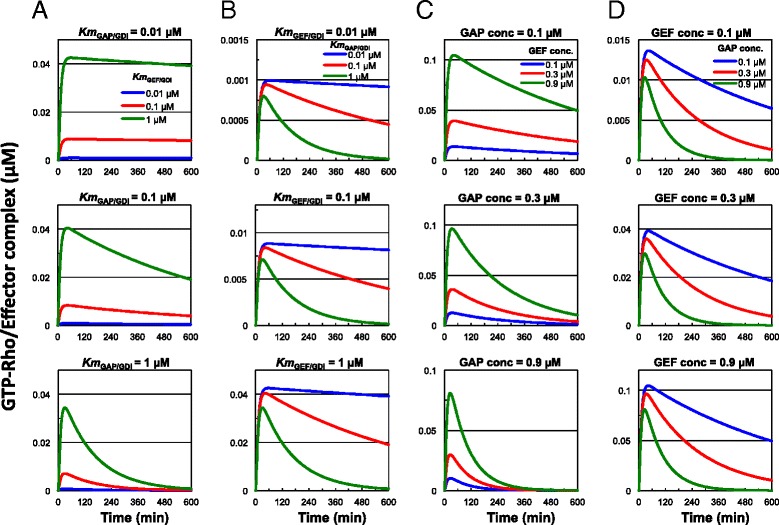
Prolongation of Rho activation in the GDI-integrated model is dependent on K mGAP/GDI and the GAP concentration. Rho activation dynamics were simulated at various K mGEF/GDI values (A), K mGAP/GDI values (B), GEF concentration (C), and GAP concentrations (D) in the GDI-integrated model. The activation levels of GTPases were expressed as the concentration of GTP-Rho/Effector complex.
It was also suggested that the local concentration of GEFs and GAPs defined the modes of Rho GTPase signaling [22]. We examined how the concentration of GEFs and GAPs affected the ability of GDIs to sustain Rho activation. We simulated the Rho activation dynamics at 0.1, 0.3, and 0.9 μM concentrations of GEFs and GAPs in our model. The decrease of GEF concentration resulted in overall decrease of Rho activation at all of the tested GAP concentrations (Figure 3C). The sustained Rho activation was apparent only at 0.1 μM of GAP and the decrease of GEF concentration did not negate this sustained Rho activation (Figure 3C). However, at all of the tested GEF concentrations, as the GAP concentrations became smaller, Rho activation was sustained to a higher degree, and increasing GAP concentration negated this sustained Rho activation (Figure 3D). These results indicate that the sustained Rho activation is dependent on the concentration of GAPs, and a lower GAP concentration sustains Rho activation. Finally, we compared the Rho activation dynamics at 0.01, 0.1, and 1.0 μM KmGAP/GDI under various concentrations of free GDI. A decrease in the KmGAP/GDI value enhanced the prolongation of Rho activation regardless of free GDI concentration (Figure 4A). Surprisingly, at 0.01 μM KmGAP/GDI, Rho activation was sustained for a significant period of time, longer than 12,000 min (8.3 days), after stimulation in the presence of free GDI (Figure 4B). However, the overall levels of Rho activation markedly decreased in association with an increase in free GDI. These results suggest that GDIs enable extremely long-term retention of the activated state of the Rho GTPases.
Figure 4.

GDI enables extremely long-term retention of the activation state of Rho GTPases. Simulation of Rho activation dynamics at K mGAP/GDI = 0.01, 01, and 1.0 μM in the presence of various free GDI concentrations (0–2.4 μM) in the GDI-integrated model. A) 600 min after stimulation. B) 12,000 min after stimulation. The activation levels of GTPases were expressed as the concentration of GTP-Rho/Effector complex.
Discussion
It is well established that the main function of RhoGDIs is to maintain Rho GTPases in inactive soluble complexes. In many canonical models of the Rho GTPase cycle, GDIs extract GTPases from the membrane and sequester them as inactive cytosolic complexes. RhoGDIs are therefore predominantly thought to act as negative regulators; however, they inhibit both activation [7] and inactivation [8,16,17] of GTPases. Little is known about how the opposing functions of GDIs influence the Rho GTPase cycle.
In the present study, we constructed a model of the Rho GTPase cycle, designated as the GDI-integrated model, in which GDIs inhibit the activities of GEFs and GAPs by interacting with them in addition to sequestering the Rho GTPases. This model indicated that GDIs sustain the activation of Rho GTPase by interacting with GAPs. Furthermore, as expected from the positive regulatory role of GDIs, (in other words, the inhibition of GAP activity by GDIs), an increase in the intracellular concentration of free GDIs enhanced the prolongation of Rho activation despite the overall decrease in the Rho activation level.
It was previously reported that the molar amount of RhoGDIα is roughly equal to the molar total of the RhoA, Rac1, and Cdc42 GTPases in several types of cultured cells [28]. RhoGDIβ is strongly expressed in hematopoietic cell lineages [39,40] and in other cell types [41-43], though it is not as ubiquitous as RhoGDIα. Specificities of RhoGDIs for Rho GTPases are largely unknown, but both RhoGDIα and RhoGDIβ can associate with RhoA, Cdc42, and Rac1 in some cell types [3]. Therefore, in RhoGDIβ-expressing cells, the total amount of RhoGDIα and RhoGDIβ may exceed the total amount of these Rho GTPases. In such cases, RhoGDIs may exist in a non-GTPase-complexed form in the cell and may function to sustain Rho activation for long periods.
RhoGDIβ is implicated in cancer progression, however, reports have presented contradictory evidence as to the nature of the correlation between cancer progression and RhoGDIβ expression level [44]. We have also reported that RhoGDIβ plays a positive [41,45,46] and negative [47] role in cancer progression. Several explanations for this contradictory behavior of RhoGDIβ have been proposed [44]. Our present study suggests that RhoGDI can act both as a positive and negative regulator of GTPases, and which role RhoGDI plays may depend on its expression level. This presents at least a partial explanation for the inconsistent correlation of RhoGDIβ with cancer progression.
It has been proposed that intracellular signals are transmitted through the dynamic activities of signaling molecules (defined as the temporal change in activity of a molecule) [48]. For example, in the case of ERK (extracellular signal-regulated kinases), transient and sustained activation states have been shown to result in different cellular responses [49]. It is well established that GEFs and GAPs function as positive and negative regulators of Rho GTPase cycles, respectively. We have shown that the functions of GEFs and GAPs are modulated by their interactions with GDIs, and that the interconversion between transient and sustained Rho activation occurs mainly through changes in the affinities of GDIs to GAPs and the concentrations of GAPs. The properties of GDIs and GAPs are regulated by posttranscriptional modifications [29-33,36-38] and the affinity between GDIs and GAPs may be altered by such modifications. Therefore, RhoGDIs and GAPs might participate in the switching between transient and sustained signals of the Rho GTPases. Although this mode seems not to be common in the regulation of Rho GTPases, certain sets of GTPases, GEFs, and GAPs may use this mode of regulation.
In the present study, we proposed a simplified model for positive regulation of Rho GTPases by GDIs. However, the model does not take into account GTPase cycling between membrane and cytosol. Cells contain membranous and cytoplasmic compartments, and typically, Rho GTPases function within the membranous compartments. It has been shown that the efficient cycling between inactive and active states of GTPases can occur entirely within protein complexes assembled on membrane surfaces [22]. RhoGDIs mediate the membrane-cytoplasmic shuttling of GTPases, and likely can alter the concentrations of GTPases and their RhoGDI-associated regulators at target sites in cells. Therefore, it is necessary to take into account the shuttling processes in developing a truly comprehensive model. Membrane-cytoplasmic shuttling has been considered in a simulation of the distribution of activated Cdc42 during the early phase of yeast bud formation [23]. Additionally, a modeling framework describing Rac cycling between membrane and cytosol has been reported [21]. Because our model for the Rho GTPase switch can be regarded as a basal signaling module, these studies that have taken into account the Rho GTPase shuttling processes should be incorporated into our model of the Rho GTPase switch for a more detailed and biologically-relevant model.
Conclusions
We constructed models of the Rho GTPase cycle in which RhoGDIs inhibit the activities of GEFs and GAPs by physically interacting with them, as well as by sequestering the Rho GTPases. This model showed that the functions of GEFs and GAPs are integrated into Rho GTPase signaling through the interactions of these regulators with GDIs, and thus, the interconversion between transient and sustained Rho activation occurs by changes mainly in the affinities of GDIs to GAPs and the concentrations of GAPs. These results provide new insights into the physiological roles of Rho GTPase signaling.
Methods
Construction of models
The pathway diagrams of the Rho GTPase cycle and their simulation programs were described using CellDesigner (Systems Biology Institute, Tokyo, Japan) [50], and were simulated by SOSLib in CellDesigner. All kinetic reactions in the pathway diagrams in Figure 1 were described by ordinary differential equations based on mass-action kinetics (reactions 1, 2, 3, 6, 7, and 8) or Michaelis–Menten kinetics (reactions 4 and 5) [51,52].
In the canonical model (Figure 1A), we used a typical Michaelis–Menten kinetic model to describe the promoting activities of GEFs (reaction 4 in Figure 1A and C) and GAPs (reaction 5 in Figure 1A and D) towards the Rho GTPases. GDIs inhibit the activities of GEFs and GAPs only by sequestering Rho GTPases.
The majority of Rho GTPases exist in biologically inactive cytosolic complexes with GDIs and the dissociation of GTPases from GDIs is hypothesized to be a prerequisite for activation by GEFs. However, it has been suggested that βPIX (GEF), Rac1, and RhoGDIα form a ternary complex [27] and that Bcr (GAP), Rac, and RhoGDIα also form a ternary complex [26]. Furthermore, several studies have shown that GDIs directly interact with both GEFs [25] and GAPs [26]. These observations suggest that GDIs and Rho GTPases can simultaneously bind GEFs or GAPs, and form ternary complexes. According to these observations, we constructed a model of the Rho GTPase cycle (Figure 1B, left) in which GDIs inhibit the activities of GEFs and GAPs by interacting with them as well as by sequestering Rho GTPases. We used the non-competitive inhibition model of Michaelis–Menten kinetics to describe the reactions in which GDIs inhibit the actions of GEFs (reaction 4 in Figure 1B and D) and GAPs (reaction 5 in Figure 1B and C), because in the non-competitive inhibition model the inhibitor and substrate can simultaneously bind the enzyme. The processes of GTPase cycling between membrane and cytosol are very important for understanding Rho activation dynamics. However, in the present study we focused on the interaction of GDIs and GEFs/GAPs and address how GDIs regulate GTPase activity through these interactions. Therefore, to simplify the model we did not consider the membrane localization of Rho GTPases and their regulators in our models.
Parameters and equations in the models are listed in Tables S1 and S2 (Additional file 1). The kinetic parameters and initial concentrations of molecules were determined based on previous studies [24,28,53-56] or arbitrary values. The activation levels of GTPases were defined as the concentrations of the GTP-Rho/Effector complex. Model files are provided as .xml files (Additional files 2 and 3) in the supplementary materials and can be viewed using CellDesigner [50].
Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (16590642 and 20591596 to TO and 13671354, 16591368, 18590706, and 20591596 to MM) and Grant for Assist KAKEN from Kanazawa Medical University (K2014-9).
Additional files
Tables S1-2.pdf. Table S1. lists the initial concentrations of molecules. Table S2. lists the kinetic reactions, ordinary differential equations, and parameters used in the models.
Canonical model.xml. The canonical model file is provided as an .xml file and can be viewed using CellDesigner.
GDI-integrated model.xml. The GDI-integrated model file is provided as an .xml file and can be viewed using CellDesigner.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TO conceived of the project. TO, MM, MO, and MT carried out the studies. TO drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Takahide Ota, Email: takahide@kanazawa-med.ac.jp.
Masayo Maeda, Email: nellie@kanazawa-med.ac.jp.
Mayumi Okamoto, Email: q231002cg@pu-hiroshima.ac.jp.
Masaaki Tatsuka, Email: tatsuka@pu-hiroshima.ac.jp.
References
- 1.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–35. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 2.Nancy V, Callebaut I, El Marjou A, de Gunzburg J. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J Biol Chem. 2002;277(17):15076–84. doi: 10.1074/jbc.M109983200. [DOI] [PubMed] [Google Scholar]
- 3.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15(7):356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 2005;390(Pt 1):1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dransart E, Olofsson B, Cherfils J. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 2005;6(11):957–66. doi: 10.1111/j.1600-0854.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Mata R, Boulter E, Burridge K. The 'invisible hand': regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12(8):493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki T, Takai Y. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem Biophys Res Commun. 1998;245(3):641–5. doi: 10.1006/bbrc.1998.8253. [DOI] [PubMed] [Google Scholar]
- 8.Hancock JF, Hall A. A novel role for RhoGDI as an inhibitor of GAP proteins. Embo J. 1993;12(5):1915–21. doi: 10.1002/j.1460-2075.1993.tb05840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JL, Erickson JW, Cerione RA. New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J Biol Chem. 2009;284(35):23860–71. doi: 10.1074/jbc.M109.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch G, Tanaka K, Masuda T, Yamochi W, Nonaka H, Takai Y. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene. 1997;15(4):417–22. doi: 10.1038/sj.onc.1201194. [DOI] [PubMed] [Google Scholar]
- 11.Nomanbhoy TK, Cerione R. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J Biol Chem. 1996;271(17):10004–9. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki T, Kato M, Takai Y. Consequences of weak interaction of rho GDI with the GTP-bound forms of rho p21 and rac p21. J Biol Chem. 1993;268(32):23959–63. [PubMed] [Google Scholar]
- 13.Tiedje C, Sakwa I, Just U, Hofken T. The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol Biol Cell. 2008;19(7):2885–96. doi: 10.1091/mbc.E07-11-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tnimov Z, Guo Z, Gambin Y, Nguyen UT, Wu YW, Abankwa D, et al. Quantitative analysis of prenylated RhoA interaction with its chaperone, RhoGDI. J Biol Chem. 2012;287(32):26549–62. doi: 10.1074/jbc.M112.371294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda T, Kikuchi A, Ohga N, Yamamoto J, Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265(16):9373–80. [PubMed] [Google Scholar]
- 16.Chuang TH, Xu X, Knaus UG, Hart MJ, Bokoch GM. GDP dissociation inhibitor prevents intrinsic and GTPase activating protein-stimulated GTP hydrolysis by the Rac GTP-binding protein. J Biol Chem. 1993;268(2):775–8. [PubMed] [Google Scholar]
- 17.Hart MJ, Maru Y, Leonard D, Witte ON, Evans T, Cerione RA. A GDP dissociation inhibitor that serves as a GTPase inhibitor for the Ras-like protein CDC42Hs. Science. 1992;258(5083):812–5. doi: 10.1126/science.1439791. [DOI] [PubMed] [Google Scholar]
- 18.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302(5645):646–50. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 19.Vincent S, Settleman J. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur J Cell Biol. 1999;78(8):539–48. doi: 10.1016/S0171-9335(99)80019-3. [DOI] [PubMed] [Google Scholar]
- 20.Cardelli L, Caron E, Gardner P, Kahramanogullari O, Phillips A. A Process Model of Rho GTP-binding Proteins. Theor Comput Sci. 2009;410(33–34):3166–85. doi: 10.1016/j.tcs.2009.04.029. [DOI] [Google Scholar]
- 21.Falkenberg CV, Loew LM. Computational analysis of Rho GTPase cycling. PLoS Comput Biol. 2013;9(1):e1002831. doi: 10.1371/journal.pcbi.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goryachev AB, Pokhilko AV. Computational model explains high activity and rapid cycling of Rho GTPases within protein complexes. PLoS Comput Biol. 2006;2(12):e172. doi: 10.1371/journal.pcbi.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582(10):1437–43. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 24.Lipshtat A, Jayaraman G, He JC, Iyengar R. Design of versatile biochemical switches that respond to amplitude, duration, and spatial cues. Proc Natl Acad Sci U S A. 2010;107(3):1247–52. doi: 10.1073/pnas.0908647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groysman M, Russek CS, Katzav S. Vav, a GDP/GTP nucleotide exchange factor, interacts with GDIs, proteins that inhibit GDP/GTP dissociation. FEBS Lett. 2000;467(1):75–80. doi: 10.1016/S0014-5793(00)01121-2. [DOI] [PubMed] [Google Scholar]
- 26.Kweon SM, Cho YJ, Minoo P, Groffen J, Heisterkamp N. Activity of the Bcr GTPase-activating domain is regulated through direct protein/protein interaction with the Rho guanine nucleotide dissociation inhibitor. J Biol Chem. 2008;283(6):3023–30. doi: 10.1074/jbc.M705513200. [DOI] [PubMed] [Google Scholar]
- 27.Shin EY, Shim ES, Lee CS, Kim HK, Kim EG. Phosphorylation of RhoGDI1 by p21-activated kinase 2 mediates basic fibroblast growth factor-stimulated neurite outgrowth in PC12 cells. Biochem Biophys Res Commun. 2009;379(2):384–9. doi: 10.1016/j.bbrc.2008.12.066. [DOI] [PubMed] [Google Scholar]
- 28.Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152(1):111–26. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DerMardirossian C, Rocklin G, Seo J-Y, Bokoch GM. Phosphorylation of RhoGDI by Src Regulates Rho GTPase Binding and Cytosol-Membrane Cycling. Mol Biol Cell. 2006;17(11):4760–8. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell. 2004;15(1):117–27. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Dovas A, Choi Y, Yoneda A, Multhaupt HA, Kwon SH, Kang D, et al. Serine 34 phosphorylation of rho guanine dissociation inhibitor (RhoGDIα) links signaling from conventional protein kinase C to RhoGTPase in cell adhesion. J Biol Chem. 2010;285(30):23296–308. doi: 10.1074/jbc.M109.098129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elfenbein A, Rhodes JM, Meller J, Schwartz MA, Matsuda M, Simons M. Suppression of RhoG activity is mediated by a syndecan 4-synectin-RhoGDI1 complex and is reversed by PKCα in a Rac1 activation pathway. J Cell Biol. 2009;186(1):75–83. doi: 10.1083/jcb.200810179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knezevic N, Roy A, Timblin B, Konstantoulaki M, Sharma T, Malik AB, et al. GDI-1 phosphorylation switch at serine 96 induces RhoA activation and increased endothelial permeability. Mol Cell Biol. 2007;27(18):6323–33. doi: 10.1128/MCB.00523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe K, Rossman KL, Liu B, Ritola KD, Chiang D, Campbell SL, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275(14):10141–9. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- 35.Schuebel KE, Movilla N, Rosa JL, Bustelo XR. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998;17(22):6608–21. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doki N, Kawashima T, Nomura Y, Tsuchiya A, Oneyama C, Akagi T, et al. Constitutive phosphorylation of a Rac GAP MgcRacGAP is implicated in v-Src-induced transformation of NIH3T3 cells. Cancer Sci. 2009;100(9):1675–9. doi: 10.1111/j.1349-7006.2009.01235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pullikuth AK, Catling AD. Extracellular signal-regulated kinase promotes Rho-dependent focal adhesion formation by suppressing p190A RhoGAP. Mol Cell Biol. 2010;30(13):3233–48. doi: 10.1128/MCB.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieillemard A, Prouzet-Mauleon V, Hugues M, Lefebvre F, Mitteau R, Claverol S, et al. The Saccharomyces cerevisiae RhoGAP Rgd1 is phosphorylated by the Aurora B like kinase Ipl1. Biochem Biophys Res Commun. 2013;433(1):1–5. doi: 10.1016/j.bbrc.2013.02.081. [DOI] [PubMed] [Google Scholar]
- 39.Lelias JM, Adra CN, Wulf GM, Guillemot JC, Khagad M, Caput D, et al. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP- binding proteins. Proc Natl Acad Sci U S A. 1993;90(4):1479–83. doi: 10.1073/pnas.90.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherle P, Behrens T, Staudt LM. Ly-GDI, a GDP-dissociation inhibitor of the RhoA GTP-binding protein, is expressed preferentially in lymphocytes. Proc Natl Acad Sci U S A. 1993;90(16):7568–72. doi: 10.1073/pnas.90.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang YS, Maeda M, Okamoto M, Fujii M, Fukutomi R, Hori M, et al. Centrosomal localization of RhoGDIβ and its relevance to mitotic processes in cancer cells. Int J Oncol. 2013;42(2):460–8. doi: 10.3892/ijo.2012.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leffers H, Nielsen MS, Andersen AH, Honore B, Madsen P, Vandekerckhove J, et al. Identification of two human Rho GDP dissociation inhibitor proteins whose overexpression leads to disruption of the actin cytoskeleton. Exp Cell Res. 1993;209(2):165–74. doi: 10.1006/excr.1993.1298. [DOI] [PubMed] [Google Scholar]
- 43.Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF., Jr Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10(11):3800–6. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 44.Griner EM, Theodorescu D. The faces and friends of RhoGDI2. Cancer Metastasis Rev. 2012;31(3–4):519–28. doi: 10.1007/s10555-012-9376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ota T, Maeda M, Murakami M, Takegami T, Suto S, Tatsuka M. Activation of Rac1 by Rho-guanine nucleotide dissociation inhibitor-β with defective isoprenyl-binding pocket. Cell Biol Int. 2007;31(1):92–6. doi: 10.1016/j.cellbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Ota T, Maeda M, Suto S, Tatsuka M. LyGDI functions in cancer metastasis by anchoring Rho proteins to the cell membrane. Mol Carcinog. 2004;39(4):206–20. doi: 10.1002/mc.20006. [DOI] [PubMed] [Google Scholar]
- 47.Ota T, Maeda M, Sakita-Suto S, Zhou X, Murakami M, Takegami T, et al. RhoGDIβ lacking the N-terminal regulatory domain suppresses metastasis by promoting anoikis in v-src-transformed cells. Clin Exp Metastasis. 2006;23(7–8):323–34. doi: 10.1007/s10585-006-9041-y. [DOI] [PubMed] [Google Scholar]
- 48.Behar M, Hoffmann A. Understanding the temporal codes of intra-cellular signals. Curr Opin Genet Dev. 2010;20(6):684–93. doi: 10.1016/j.gde.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80(2):179–85. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 50.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol. 2005;23(8):961–6. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 51.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8(11):1195–203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 52.Ota T, Maeda M, Odashima S. Mechanism of action of ginsenoside Rh2: uptake and metabolism of ginsenoside Rh2 by cultured B16 melanoma cells. J Pharm Sci. 1991;80(12):1141–6. doi: 10.1002/jps.2600801210. [DOI] [PubMed] [Google Scholar]
- 53.Aoki K, Nakamura T, Inoue T, Meyer T, Matsuda M. An essential role for the SHIP2-dependent negative feedback loop in neuritogenesis of nerve growth factor-stimulated PC12 cells. J Cell Biol. 2007;177(5):817–27. doi: 10.1083/jcb.200609017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang TH, Bohl BP, Bokoch GM. Biologically active lipids are regulators of Rac. GDI complexation. J Biol Chem. 1993;268(35):26206–11. [PubMed] [Google Scholar]
- 55.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435(7041):513–8. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 56.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem. 2000;275(33):25299–307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]


