Abstract
Background
The basic helix-loop-helix (bHLH) proteins are a large superfamily of transcription factors, and play a central role in a wide range of metabolic, physiological, and developmental processes in higher organisms. Tomato is an important vegetable crop, and its genome sequence has been published recently. However, the bHLH gene family of tomato has not been systematically identified and characterized yet.
Results
In this study, we identified 159 bHLH protein-encoding genes (SlbHLH) in tomato genome and analyzed their structures. Although bHLH domains were conserved among the bHLH proteins between tomato and Arabidopsis, the intron sequences and distribution of tomato bHLH genes were extremely different compared with Arabidopsis. The gene duplication analysis showed that 58.5% and 6.3% of SlbHLH genes belonged to low-stringency and high-stringency duplication, respectively, indicating that the SlbHLH genes are mainly generated via short low-stringency region duplication in tomato. Subsequently, we classified the SlbHLH genes into 21 subfamilies by phylogenetic tree analysis, and predicted their possible functions by comparison with their homologous genes of Arabidopsis. Moreover, the expression profile analysis of SlbHLH genes from 10 different tissues showed that 21 SlbHLH genes exhibited tissue-specific expression. Further, we identified that 11 SlbHLH genes were associated with fruit development and ripening (eight of them associated with young fruit development and three with fruit ripening). The evolutionary analysis revealed that 92% SlbHLH genes might be evolved from ancestor(s) originated from early land plant, and 8% from algae.
Conclusions
In this work, we systematically identified SlbHLHs by analyzing the tomato genome sequence using a set of bioinformatics approaches, and characterized their chromosomal distribution, gene structures, duplication, phylogenetic relationship and expression profiles, as well predicted their possible biological functions via comparative analysis with bHLHs of Arabidopsis. The results and information provide a good basis for further investigation of the biological functions and evolution of tomato bHLH genes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-014-1209-2) contains supplementary material, which is available to authorized users.
Keywords: Tomato, Solanum lycopersicum, bHLH gene family, transcription factor, fruit development
Background
The basic helix–loop–helix (bHLH) proteins are a large superfamily of eukaryotic transcription factors, and play a central role in a wide range of metabolic, physiological, and developmental processes [1-3]. Their bHLH domain contains approximately 60 amino acids, including a basic region and a HLH region [4]. The basic region, which consists of approximately 17 amino acids and is located at the N-terminus of the domain, is a DNA-binding region that allows HLH proteins to bind to a consensus hexanucleotide E-box (CANNTG) [5,6]. The HLH region is composed of two amphipathic helices consisting of hydrophobic residues linked by a divergent (both in length and primary sequence) loop, and functions as a dimerization domain [4,7]. The HLH domain promotes protein–protein interactions and allows for the formation of homodimeric or heterodimeric complexes [6]. Excluding the conserved bHLH domain, the proteins showed considerable sequence divergence [5]. Furthermore, groups of evolutionary and/or functionally related bHLH proteins shared additional motifs. Some of these motifs have been characterized in animals regarding specificity in the DNA-binding sequence recognition and dimerization activities responsible for the activation or repression of target genes or for binding to small molecules [1].
Previous classifications of animal bHLHs have led to the definition of six major functional and evolutionary lineages (groups A–F) [1,8], which have been further subdivided into several smaller orthologous subfamilies [9]. Group A of the bHLH proteins can bind to the E-box sequence. In group B, several proteins such as Max, Myc, MITF, SREBP and USF have diverse functions and bind to the G-box sequence CACGTG [10-12]. Members of Group C contain an additional protein–protein interaction region and bind to sequences (NACGTG or NGCGTG) that do not resemble the E-box. Proteins in group D contain only the HLH region and can form heterodimers with bHLH proteins and thus are functionally related to typical bHLH proteins [13]. Group E proteins contain Pro or Gly residues within the basic region and can bind preferentially to the CACGNG sequence [14,15]. Group F proteins contain divergent sequences compared with other groups and another domain for dimerization and DNA binding [16].
Studies on the bHLH gene family in various species will increase our understanding of their evolution and functions. However, systematic identification of the bHLH genes has been performed only in a few plants, such as Arabidopsis, rice, poplar, and moss [2,17-19]. Tomato (Solanum lycopersicum) is one of the most important vegetables in the world and is also a model plant for studying fruit development [20]. Tomato genome sequencing was recently completed and published [21]. But the bHLH gene family of tomato has still not been reported. In this study, we systematically identified and characterized the bHLH genes of tomato (SlbHLH) and compared them with the bHLHs of Arabidopsis thaliana. In addition, we also analyzed the expression profiles of SlbHLH genes in different tissues and at different stages of fruit development as well in response to iron-deficiency stress. Finally, we detected several genes associated with fruit development and ripening, and with iron-deficiency responses. In addition, we did the evolutionary analysis of SlbHLH genes by comparison of tomato bHLH genes with that of angiosperm, early land plants and algaes.
Results and discussion
Identification and classification of tomato bHLH genes
For genome-wide identification of bHLH genes in tomato, we initially identified the proteins using two approaches described in the Methods and filtered the results based on the criteria developed by Atchley et al. [5] and Toledo-Ortiz et al. [2]. The bHLH domain contained 19 conserved amino acid residues, including five in the basic region, five in the first helix, one in the loop, and eight in the second helix [5] (Table 1). Based on the criterion for identifying Arabidopsis bHLH proteins (AtbHLHs) [2], we allowed nine mismatches from the 19 conserved amino acid residues for the identification of tomato bHLHs. To further confirm and filter uncertain bHLH proteins, we performed SMART analysis and retained proteins with e-values less than 1 (the e-value setting based on testing AtbHLH family). Finally, we identified 159 genes encoding bHLH proteins (SlbHLHs) in the tomato genome (Table 1 and Additional file 1) and named them according to the system proposed for Arabidopsis [17] (Additional file 1). Compared with the recent report by The Tomato Genome Consortium [21], 57 bHLH proteins were absent in this study because they did not conform to our minimal criteria for inclusion. These proteins contained an incomplete bHLH domain, has a low similarity (e-value ≥ 0.01) with the bHLH domain profile, or more than nine mismatches within the 19 conserved amino acid residues.
Table 1.
bHLH domain consensus motif
| Atchley et al. [ 5 ] | Toledo-Ortiz et al. [ 2 ] | This study | ||||
|---|---|---|---|---|---|---|
| Position in the alignment | Consensus motif amino acid frequency within the bHLH domain | Position in the alignment | Amino acid frequency within the Arabidopsis bHLH domains | Position in the alignment | Amino acid frequency within the tomato bHLH domains | |
| Basic | 1 | R (61%), K (27%) | 1 | R (24%), K (22%) | 1 | K(28%), R(25%), N(11%) |
| 2 | R (77%), K (16%) | 2 | R (35%) | 2 | R(32%), K(11%) | |
| 9 | E (93%) | 13 | E (76%), A (10%) | 13 | E(75%), A(11%) | |
| 10 | R (81%), K (14%) | 14 | R (74%), K (14%) | 14 | R(76%), K(18%) | |
| 12 | R (91%) | 16 | R (91%) | 16 | R(94%) | |
| Helix | 16 | I (35%), L (33%), V (23%) | 20 | I (52%), L (27%), M (12%) | 20 | I(53%), L(28%), M(17%) |
| 17 | N (74%) | 21 | N (51%), S (19%) | 21 | N(45%), S(26%) | |
| 20 | F (72%), L (14%), I (9%) | 24 | F (26%), L (26%), M (20%), I (14%) | 24 | L(28%), F(26%), M(19%), I(16%) | |
| 23 | L (98%) | 27 | L (100%) | 27 | L(99%) | |
| 24 | R (44%), K (35%) | 28 | Q (42%), R (35%) | 28 | Q(41%), R(37%) | |
| Loop | 47 | K (58%), R (24%) | 39 | K (66%) | 36 | K(68%) |
| Helix | 50 | K (93%) | 42 | K (45%), T (13%) | 47 | K(45%), T(21%) |
| 53 | I (74%), T (15%), V(7%) | 45 | M (33%), I (27%), V (16%), L (14%) | 50 | M(33%), I(28%), V(15%), L(14%) | |
| 54 | L (98%) | 46 | L (76%), V (14%) | 51 | L(78%), I(11%) | |
| 57 | A (76%) | 49 | A (60%), I (16%), V (12%) | 54 | A(60%), I(18%), V(11%), T(10%) | |
| 58 | I (31%), V (27%), T (23%) | 50 | I (63%), V (22%) | 55 | I(60%), V(25%) | |
| 60 | Y (77%) | 52 | Y (78%) | 57 | Y(74%), H(13%) | |
| 61 | I (69%), L (16%), V (8%) | 53 | I (40%), V (33%), L (13%) | 58 | I(43%), V(38%), L(13%) | |
| 64 | L (80%), M (7%) | 56 | L (93%) | 61 | L(97%) | |
Based on information of gene annotation [21], 158 of the predicted 159 SlbHLH genes were localized on tomato chromosomes, while SlbHLH073 was failed to locate on the 12 chromosomes [21]. As shown in Figure 1, the congregate region and the number of SlbHLHs are irregular although the SlbHLH genes distributed on all of the 12 tomato chromosomes. Chromosome 1 contained the largest number (22) of bHLH genes, whereas only four bHLH genes were present on chromosome 11. Based on the gene number of each chromosome, the percentage of bHLH genes per chromosome varied from 0.17% on chromosome 11 to 0.6% on chromosomes 6 and 9 with an average ratio of 0.41% (Additional file 2). According to similarity of SlbHLH proteins, we speculated that 103 genes were generated by gene duplication. Of them, 93 SlbHLHs belonged to the low-stringency duplication, and 10 to the high-stringency duplication (Additional file 3). Based on chromosome localization, 14 of 159 SlbHLHs were tandem array genes (TAGs) (Figure 1). The segment duplication analysis showed that SlbHLH149 and Solyc01g014960 were generated via a segment duplication (Additional file 4, Additional file 5A, B). However, Solyc01g014960 did not contain a complete bHLH domain, suggesting it was a pseudogene. Based on our analysis, we propose that the majority of SlbHLHs may have been generated via gene duplication rather than segment duplication in the genome of tomato.
Figure 1.
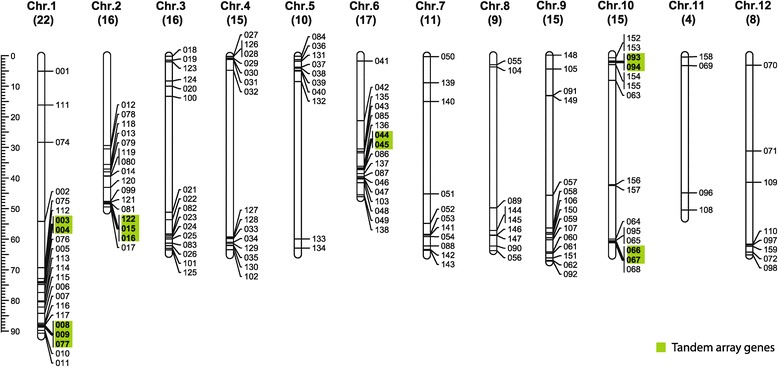
Chromosomal localizations and tandem array of SlbHLH genes. The numbers below the chromosome names show the total number of SlbHLH genes located on each chromosome. The green block indicates tandem array genes.
The tomato ‘Heinz 1706’ genome, with a genome size of approximately 900 Mb, was 7.2 times larger than the A. thaliana genome. However, the number of SlbHLH genes was similar to Arabidopsis (162). Based on the total genes, the ratio to SlbHLH genes in the tomato genome was about 0.46%, which was similar to rice (0.44%) and poplar (0.40%) [18,19], but was less than Arabidopsis (0.59%) [2,17].
Conserved amino acid residues in the bHLH domains and DNA-binding ability
The bHLH domain alignment of 159 SlbHLHs showed that 22 amino acid residues in their bHLH domains were conserved with more than 50% consensus ratio (Figure 2). Compared with bHLHs of animals that described by Atchley et al. [5], we found that the residues Ile-20, Leu-24, Gln-28, Lys-36, Met-50, Ile-55, Val-58 and Leu-61 in the bHLH domains of plants were more conserved than in animals, suggesting that the seven amino acid residues may play an important role in plants (Table 1). Further, the residues Arg-16, Leu-27, and Leu-61 showed extreme conservation among the 159 bHLH proteins of tomato. Previous reports revealed that Glu-13 and Arg-16/Arg-17 in the basic region of bHLH domain play an important role in DNA binding [8], and Leu-27 and Leu-61 in the helix regions function in dimerization activity [19].
Figure 2.
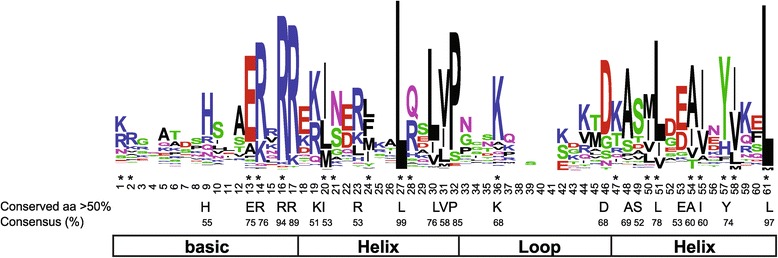
The bHLH domain is highly conserved across all SlbHLH proteins. The overall height of each stack represents the conservation of the sequence at that position. The asterisk indicates the position of the 19 conserved amino acids previously identified by Atchley et al. [5], and capital letters indicate over 50% conservation of amino acids among the 159 SlbHLH domains.
The basic region of the bHLH proteins contained 17 amino acids in tomato, which is four amino acids longer than that described by Atchley et al. [5] (Figure 2 and Table 1). The DNA-binding activity of target genes is determined in the basic region of the bHLH domain [6]. Based on the criteria developed by Toledo-Ortiz et al. [2], we defined proteins with more than five basic amino acid residues in the basic region as DNA-binding proteins. Furthermore, the DNA-binding proteins were subdivided into two subcategories, E-box (including G-box binder) and non-E-box binders (Table 2 and Additional file 1). Glu-13 and Arg-16 are known to be essential for recognition of the E-box, and His/Lys-9, Glu-13, and Arg-17 are required for binding of G-box [7,22]. According to the conservation of the residues, we predicted 98 SlbHLHs as putative E-box-binding proteins, and 12 as non-E-box-binding proteins (due to lacking Glu-13/Arg-16 residues). Among the 98 E-box-binding proteins, 72 belonged to G-box-binding proteins, while 26 proteins lacked the G-box-binding site (Table 2). Additionally, 49 of 159 SlbHLHs contain less than six amino acid residues in the basic region, and were classified as non-DNA-binding proteins (Table 2).
Table 2.
Predicted DNA-binding categories based on the bHLH domain
| Predicted activity | Predicted motif | Number of AtbHLHs | Number of SlbHLHs |
|---|---|---|---|
| (Toledo-Ortiz et al. [ 2 ]) | (This study) | ||
| DNA binding | |||
| E-box | |||
| G-box | bHLH | 89 (60.54%) | 72 (45.28%) |
| Non-G-box | bHLH | 20 (13.61%) | 26 (16.35%) |
| Non-E-box | bHLH | 11 (7.48%) | 12 (7.55%) |
| Total | 120 (81.63%) | 110 (69.18%) | |
| Non-DNA binding | HLH | 27 (18.37%) | 49 (30.82%) |
Intron distribution and phylogenetic analysis of SlbHLH genes
To analyze intron distribution within the coding sequence of the bHLH domain in all SlbHLH genes, we did alignment analysis with the coding and genome sequences using Blat software, and found 11 different intron distribution patterns (designated as I to XI). The intron number ranged from 0 to 3 within the coding sequence of the bHLH domain (Figure 3 and Additional file 1). As shown in Additional file 6, 85% of SlbHLHs contained introns in their coding sequence of the bHLH domain, and 74% had a conserved intron position, but the sequence length and similarity of the introns were quite different, even at the same position. In contrast, 15% of the SlbHLHs did not contain intron in their bHLH domain region (pattern XI). Pattern I (including three introns) was the most common pattern in Arabidopsis [2], but it was only the second common one in tomato. These results showed that intron sequences and distribution between tomato and Arabidopsis were different although their bHLH domains were conserved.
Figure 3.
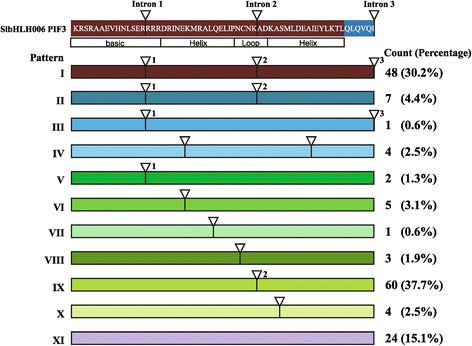
Intron distribution patterns in the coding sequence of the bHLH domain of SlbHLHs. Scheme of the intron distribution patterns (color coded and designated I to XI) within the bHLH domain of SlbHLHs. Position of introns is indicated by triangles and numbered (1 to 3) based on the bHLH region of PIF3, which is shown at the top. When the position of the intron coincides with that found in PIF3, the intron number is given above the triangle. The count and percentage of genes in each pattern are given on the right.
To explore whether the intron distribution pattern and DNA-binding activity correlate with their phylogenetic classification, a neighbor-joining phylogenetic tree was built using alignments of bHLH domains. The phylogenetic tree showed that 159 bHLH domains were classified into 21 subfamilies (Figure 4). The genes with E-box binding region was mostly clustered within the subfamilies 1–5, 12–14, and 16–20, whereas the genes with non-DNA-binding region were grouped in the subfamilies 6–11 and 21. Moreover, intron distribution pattern I (corresponding to the subfamily 6) and pattern IX (corresponding to the subfamilies 8, 12, 13, and 17–21) mainly belonged to the E-box-binding type, and pattern XI (corresponding to the subfamilies 7, 9, 11 and 15) to non-DNA-binding type. These results suggest that some rules and dependencies exist between the intron distribution pattern and DNA-binding activity among the SlbHLHs.
Figure 4.
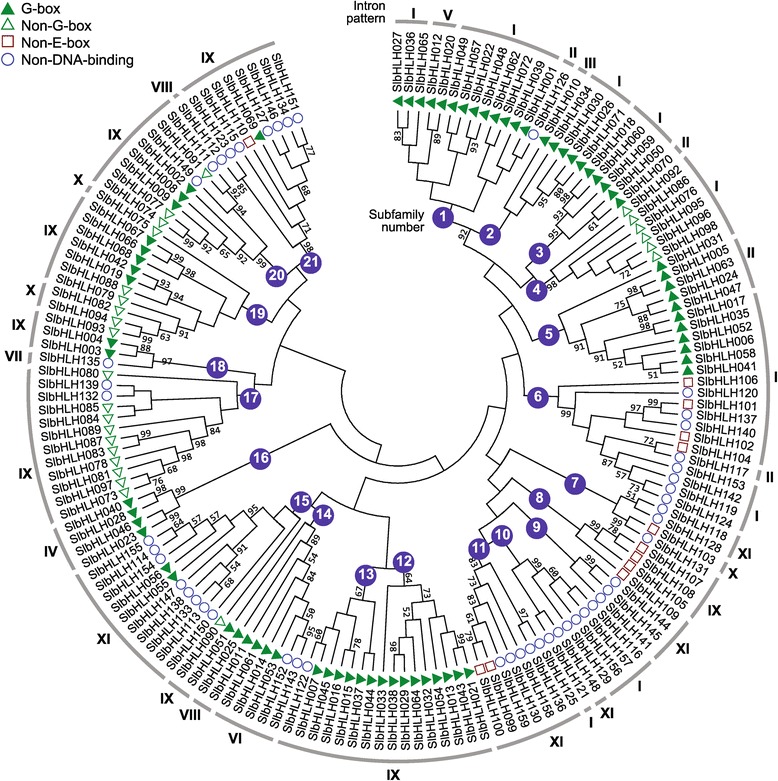
Relationship of neighbor-joining phylogenetic tree of the SlbHLH domains with the predicted DNA-binding activities and the intron distribution patterns. The tree shows the 21 phylogenetic subfamilies marked with white font on a colored background. Roman numerals correspond to the intron patterns shown in Figure 3. The different shape on the left side of SlbHLH represents the predicted DNA-binding activity of each protein.
Up to now, the biological functions of the most SlbHLHs remain unclear. However, approximately 40% of Arabidopsis bHLH proteins have been functionally characterized. Hence, the clustering and comparison of tomato bHLH proteins with AtbHLHs can help to predict their functions via ortholog analysis. The phylogenetic analysis revealed that 43 SlbHLHs were tightly grouped with the AtbHLHs, in which their functions are known. These suggest that the 43 SlbHLH proteins may have the similar functions as their Arabidopsis ortholog (Additional file 7 and Additional file 8).
Expression pattern of SlbHLH genes among different tissues
To analyze the expression pattern of SlbHLH genes among ten different tissues, we used the Reads Per Kilobase per Million (RPKM) normalized data from RNA-seq [21]. Additional file 9 showed the expression profiles of SlbHLH genes in the ten tomato tissues. Among 159 SlbHLHs genes, 122 expressed at least in one of the ten tissues with an RPKM value greater than 1.4, while the rest 37 exhibited a low expression (RPKM ≤ 1.4) in all ten tissues. Moreover, we found that 21 SlbHLHs displayed the tissue-specific expression preference (the expression intensity was more than 2 times higher in a particular tissue than that of other tissues). They include eleven genes (SlbHLH034, SlbHLH038, SlbHLH050, SlbHLH053, SlbHLH057, SlbHLH085, SlbHLH092, SlbHLH105, SlbHLH120, SlbHLH128, SlbHLH137) in root, three genes (SlbHLH024, SlbHLH041, SlbHLH094) in leaf, three genes (SlbHLH001, SlbHLH022, SlHLH059) in flower, one gene (SlbHLH089) in bud, and three genes (SlbHLH006, SlbHLH078, SlbHLH095) in fruit (Additional file 9), implying that they may have some functions in these tissues, respectively.
Fruit-development-related SlbHLH genes and their cis-element analysis
Tomato is a model plant for studying fruit development and ripening. Based on gene expression data, we identified 11 SlbHLH genes which showed a gradually increased or decreased expression with fruit development and ripening. Of them, 6 genes (SlbHLH022, SlbHLH065, SlbHLH069, SlbHLH073, SlbHLH078, and SlbHLH127) exhibited a high expression in 1-cm large fruit, and two (SlbHLH006 and SlbHLH108) in 3-cm large fruit in comparison with other tissues (Figure 5A). These data indicate that the eight SlbHLH genes may be involved in fruit development. The expression of the rest three genes (SlbHLH025, SlbHLH095, and SlbHLH113) was gradually upregulated during the ripening process, suggesting that they may function in fruit ripening. Indeed, it was reported recently that SlbHLH095 (Solyc10g079050) was associated with fruit ripening [23]. Therefore, further characterization of the 11 SlbHLHs is highly important and will provide a new insight to understand the molecular mechanism of fruit development and ripening.
Figure 5.
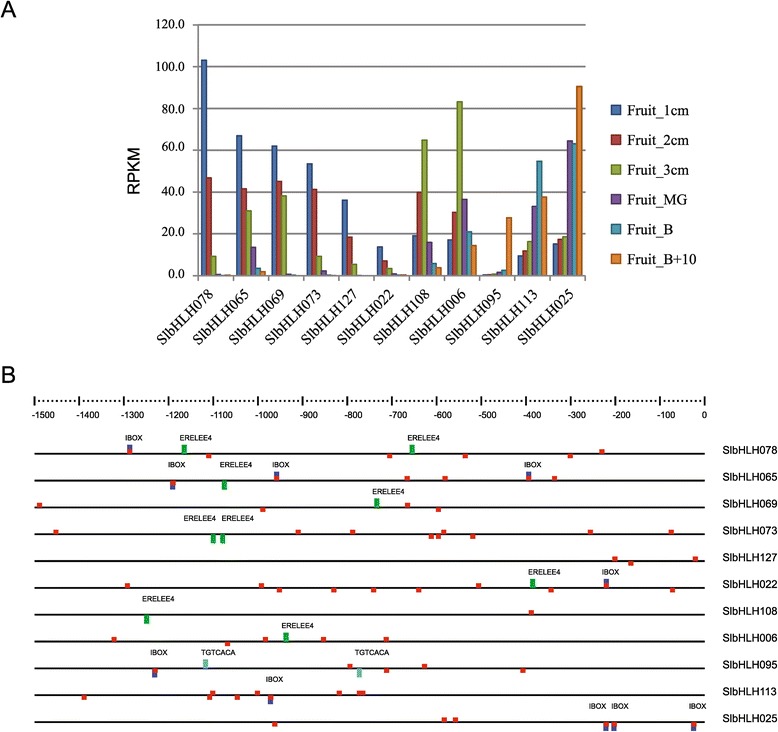
Expression patterns of the SlbHLH genes related fruit development and their cis -element analysis. (A) The expression profiles of the indicated genes at different stages of fruit development and ripening. The expression data of the SlbHLH genes were obtained from the RNA-seq data of Solanaceae Genomics Network (http://solgenomics.net; ITAG Release 2.3). Fruit_1cm, Fruit_2cm and Fruit_3cm mean the RNAs extracted from 1 cm, 2 cm and 3 cm large fruits, respectively. Fruit_MG indicates mature green (MG), Fruit_B is breaker (early ripening), and Fruit_B + 10 means 10 days post-B (red ripe). (B) Cis-element analysis of the indicated genes from upstream 1500 bp sequence to the transcription start site. The upside represents the forward sequence element and downside represents the matching element with the reverse. Red-, blue-, green-, and light blue-colored blocks mean I-box core-, I-box-, Erelee4-, and TGTCACA-elements, respectively.
To investigate the regulation mechanisms of the 11 fruit-related SlbHLH genes, we analyzed their cis-elements from the transcriptional start site to the –1500-bp upstream region (Figure 5B). In the 11 SlbHLHs, 172 elements were detected using PLACE (http://www.dna.affrc.go.jp/PLACE/), of which 22 were mutual elements. Ethylene is known to be important for fruit development and ripening [20]. Thus, we examined ethylene-regulated elements using bioinformatics tool. Ethylene-responsive elements (ERELEE4, AWTTCAAA) were found in seven genes (SlbHLH006, SlbHLH022, SlbHLH065, SlbHLH069, SlbHLH073, SlbHLH078, and SlbHLH108), which were involved in young fruit development, but were not detected in the three SlbHLH genes related to fruits ripening (SlbHLH025, SlbHLH095, and SlbHLH113). In previous study, the TGTCACA motif was shown to be an enhancer element required for fruit-specific expression of the cucumisin gene from melon, and the I-box-like sequence AGATATGATAAAA functions as a negative regulatory element [24]. Among the fruit-specific bHLH genes of tomato, only SlbHLH095 contained the TGTCACA element in promoter. Although the I-box-like motif was not detected in the promoter region of the 11 SlbHLHs, the I-box core element (GATAA) and I-box (GATAAG) presented commonly. Considering the distribution of cis-elements in the promoter of these genes, we speculate that they may play some roles in regulating the expression of the corresponding genes for the fruit development and ripening.
Prediction of SlbHLHs involved in the regulation of iron deficiency responses and homeostasis
Iron deficiency is one of mostly limiting factors for plant growth and development. Its uptake and homeostasis are tightly regulated because excess and deficiency of iron are pernicious for plants. Several bHLH transcription factors involved in the regulation of iron uptake and homeostasis have been cloned and characterized in Arabidopsis. They are the FER-like iron-deficiency-induced transcription factor (FIT), bHLH38, bHLH39, bHLH100, bHLH101, POPEYE (PYE), and IAA–Leu Resistant3 (ILR3) [25-28]. In tomato, only one bHLH transcriptional regulator (FER), which is involved in regulating iron-deficiency responses and uptake, has been characterized [29]. In Arabidopsis, FIT is an ortholog of tomato FER [25]. Under iron deficiency, FIT interacts with bHLH38, bHLH39, bHLH100 or bHLH101 to form heterodimer(s) and activate the expression of their downstream genes, such as FRO2 and IRT1 [26,28]. PYE interacts with ILR3, functioning in the negative regulation of iron deficiency responses [27]. To predict SlbHLHs which may function in the responses to iron deficiency and homeostasis, we compared the tomato bHLHs with AtbHLHs. As shown in Additional file 7, six SlbHLHs (SlbHLH025, SlbHLH066, SlbHLH067, SlbHLH068, SlbHLH085, SlbHLH143) were correspondingly clustered with the 7 AtbHLHs (FIT, AtbHLH38, AtbHLH39, AtbHLH100, AtbHLH101, PYE, ILR3), which are involved in iron deficiency responses and homeostasis. SlbHLH085 is FER and the ortholog of FIT (AtbHLH29) [25]. SlbHLH025 is corresponded to AtbHLH047 (PYE), SlbHLH143 to AtbHLH105 (ILR3), whereas SlbHLH066, SlbHLH067 and SlbHLH068 were grouped with AtbHLH38, AtbHLH39, AtbHLH100 and AtbHLH101 together. Further, we analyzed the expression profiles of the six SlbHLHs in shoots and roots using qRT-PCR. The genes SlbHLH066, SlbHLH067 and SlbHLH068, which are corresponding to AtbHLH38, AtbHLH39, AtbHLH100 and AtbHLH101 of Arabidopsis, expressed specifically in roots under iron-deficient conditions (Figure 6), whereas the four Arabidopsis genes expressed in roots and also in leaves [26,28]. These results imply that the transcriptional regulation of these bHLH genes has some differences between tomato and Arabidopsis. In addition, SlbHLH025 (ortholog of PYE) and SlbHLH085 (FER) were also specifically expressed in root, and were highly expressed under iron deficiency. However, the expression of SlbHLH143 (corresponding to Arabidopsis ILR3) was not distinct between iron deficiency and sufficiency in shoots while its expression was upregulated only in roots under iron deficiency (Figure 4). All these suggest that SlbHLH066, SlbHLH067 and SlbHLH068 may have similar function as AtbHLH38, AtbHLH39, AtbHLH100 and AtbHLH101 in the responses to iron deficiency and uptake, and that SlbHLH025, like PYE of Arabidopsis, may function in the negative regulation of iron deficiency responses. However, more experiments are needed to confirm it.
Figure 6.
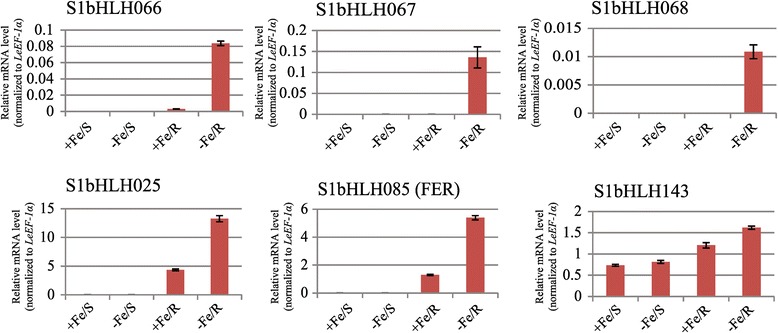
Expression analyses of the six SlbHLH genes under iron-deficient stress by qRT-PCR. For qRT-PCR, the relative amount of mRNA (y-axis) was calculated by according to the description in Methods. The x-axis indicates the shoot (S) and root (R) of tomato under iron-sufficient (+Fe) and iron deficient (-Fe) conditions.
Evolutionary relationship of SlbHLH genes
Plant kingdom is divided into algae (including red algae, chlorophyta) and land plant (mosses, lycophytes, and angiosperms) [30]. Its evolution is from algae to land plant. Recently, Pires and Dolan [31] defined the relationships of bHLH proteins in plant kingdom using the whole-genome sequences of nine species from algae and land plants. They showed that only few (less than 5) bHLH proteins were detected in the genomes of chlorophytes and red algae. In contrast, many bHLH proteins (100–170) are identified in the genomes of land plants (embryophytes). Phylogenetic analyses suggest that plant bHLH proteins are monophyletic and much of the bHLH protein diversity in plant kingdom was established in early land plants, over 440 million years ago [31].
To observe the evolutionary relationship of tomato bHLH genes, SlbHLHs were compared with bHLH protein data of other plants described by Pires and Dolan [31]. The phylogenetic tree was constructed with the bHLH domain regions of bHLH proteins of 8 species (1 monocot, 2 eudicots, 1 lycophyte, 1 moss, 2 chlorophyceaes, and 1 red algae). As shown in Figure 7 and Additional file 10, total 696 bHLH proteins of the 8 species were classified into 18 subfamilies based on clade and evolution of species in the topology of the trees. The 159 bHLHs of tomato were distributed into 16 of 18 subfamilies except of the subfamily 1 and 11. Based on the subfamily classification, 13 SlbHLHs were clustered together with algae bHLHs in the subfamily 2 and 3, 127 with moss bHLHs in 12 subfamilies (subfamily 4-9, 12-17), and 19 with lycophyte bHLHs in the subfamily 10 and 18. These results suggest that the 13, 127 and 19 SlbHLHs may evolve from ancestors of algae, moss and lycophyte, respectively. The oldest evidence for the existence of vascular plants comes from trilete spores found in Upper Ordovician sediments, over 443 Ma [32], and green algae chlorophyceae from 1 billion years ago [33]. Hence, we speculate that 92% of SlbHLH proteins should be evolved from ancestor(s) originated in land plants over 443 Ma, and 8% from the ancestor(s) appeared in algae over 1 billion years ago. These are consistent with the report by Pires and Dolan [31].
Figure 7.
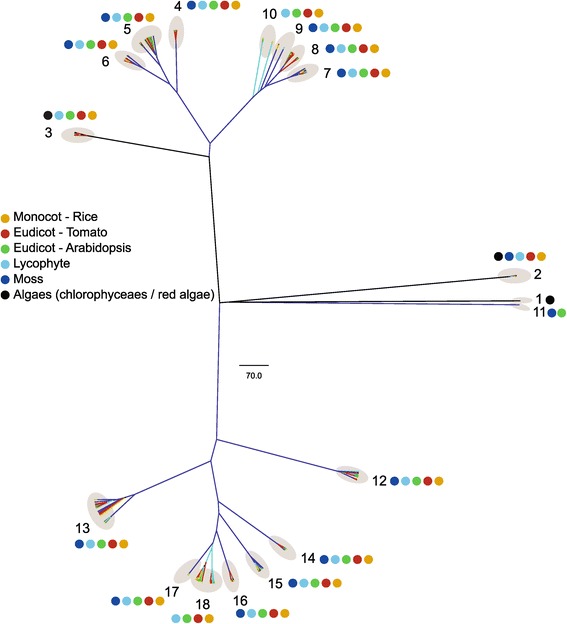
Phylogenetic relationships in the vascular plants, moss, and algaes. Maximum likelihood analysis of 696 plant bHLHs show as cladogram, and was rooted with a node of red algae protein. The gray balloons delineate the 18 subfamilies of bHLH proteins. Colored dots symbolize the species to which the bHLH proteins in each group belong (yellow: Oryza sativa [monocot]; red: Solanum lycopersicum [eudicot]; green: Arabidopsis thaliana [eudicot]; light-blue: Selaginella moellendorffii [lycophyte]; purple: Physcomitrella patens [moss]; black: Volvox carteri, Chlamydomonas reinhardtii, and Cyanidioschyzon merolae [chlorophytes and red algae]). A full tree with protein names is given in Additional file 10.
Conclusions
In this work, we systematically analyzed the sequence of tomato genome, identified 159 bHLH genes, and characterized their structure, duplication, chromosomal distribution, phylogenetic tree, and expression patterns. Among the 159 SlbHLHs, the expression of 11 and 6 SlbHLHs was related to fruit development and ripening, and to response of iron deficiency, respectively. Further, we annotated the possible biological functions of 43 SlbHLHs by comparative analysis with bHLHs of Arabidopsis. The evolution analysis showed that all of SlbHLHs are highly conserved in plant evolution and much of the diversity of SlbHLH proteins was established in early land plants. Taken together, the results and information described in this work provide a good basis for further investigation of the biological functions and evolution of tomato bHLH genes.
Methods
Data set collection and identification of bHLH genes
Tomato genome sequence data were obtained from the Solanaceae Genomics Network (SGN) in May 2012 (http://solgenomics.net; ITAG Release 2.3) [21]. The information and sequences of A. thaliana bHLHs (AtbHLHs) were retrieved from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/), Oryza sativa bHLHs (OsbHLHs) were obtained from Li et al. [18], and a data set of bHLH proteins from early land plant (Lycophyte - Selaginella moellendorffii, and Moss - Physcomitrella patens) and algaes (Chlorophyceae - Volvox carteri and Chlamydomonas reinhardtii, and Red algae - Cyanidioschyzon merolae) was retrieved from Pires and Dolan [31]. The bHLH proteins of tomato (SlbHLHs) were predicted using the HLH hidden Markov model (HMM) profile obtained from Pfam (http://pfam.xfam.org, PF00010) and used as queries to search the bHLH proteins from tomato sequences with HMMER software (http://hmmer.janelia.org). In addition, the previously known AtbHLH sequences were applied as input to build a bHLH consensus domain profile with MEME software (http://meme.nbcr.net/meme/). The profile was then used as queries to identify bHLH proteins using the MAST program (http://meme.nbcr.net/meme/) with tomato sequences. To further confirm and filter uncertain bHLH proteins, the predicted bHLH domains were examined with the SMART tool (http://smart.embl-heidelberg.de). The all of the bHLH protein sequences used in this study were showed in Additional file 11.
Alignment and phylogenetic analysis
Multiple domain alignments were performed using Multalin (http://multalin.toulouse.inra.fr) and Clustal-omega tool (v 1.2, http://www.clustal.org). To visualize the conserved motifs, the sequences were analyzed with WEBLOGO programs (http://weblogo.berkeley.edu). Phylogenetic tree was constructed using MEGA5 (http://www.megasoftware.net) with the neighbor-joining method and the following parameters: pairwise deletion option, 1000 replicates of bootstrap and Poisson correction distance. The consensus tree showed only branches with a bootstrap consensus > 50. The maximum likelihood (ML) analyses were done with the program PhyML version 3.0 (http://www.atgc-montpellier.fr/phyml) using the JTT model of amino acid substitution, the radial tree was drawn using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree).
Gene duplication pattern and location of SlbHLH genes on chromosomes
Gene duplication was classified into two groups: low-stringency duplication (protein pairs with ≥ 30% identity and covering ≥ 70% protein length) and high-stringency duplication (protein pairs with ≥ 50% identity and covering ≥ 90% protein length) [34]. TAGs were defined if they belonged to the same superfamily and were either physically adjacent or separated by a specific number of nonhomologous intervening “spacer” genes [34]. Segmental duplications (length ≥ 3 kb; identity ≥ 90%) were identified using MUMmer (http://sourceforge.net/projects/mummer) in the whole genome sequences of tomato. The SlbHLH genes were mapped onto the corresponding chromosomes by identifying their chromosomal positions provided in the SGN. The distribution of SlbHLH genes on chromosomes were drawn using MapChart software (http://www.wageningenur.nl/en/show/Mapchart.htm).
Gene expression analysis and cis-element prediction
The expression pattern of the genes in different tissues was drawn using R script based on an average of normalized expression (RPKM mapped reads) of tomato bHLH genes from RNA-seq data [21]. The cis-element was predicted by PLACE (http://www.dna.affrc.go.jp/PLACE/).
qRT-PCR analysis
After germination for 5 days, the tomato seedlings (Heinz) were cultured in a modified half-strength Hoagland nutrient solution [35] for 4 days. The nutrient solution contained 3.0 mM KNO3, 2.0 mM Ca(NO3)2·4H2O, 1.0 mM NH4H2PO4, 0.5 mM MgSO4·7H2O, 1.0 μM KCl, 25.0 μM H3BO3, 2.0 μM MnSO4·4H2O, 2.0 μM ZnSO4·7H2O, 0.1 μM CuSO4·5H2O, 0.1 μM (NH4)6Mo7O24·4H2O, 2.0 mM MES, and 20.0 μM Fe(Na)–EDTA, and the pH of the solution was adjusted to 5.5 with KOH. The 5-days-old seedlings were transferred into the one-half strength modified Hoagland solution with or without iron supply, and cultivated for 3 days. Subsequently, the roots and shoots were separately harvested, and their total RNAs were extracted. After eliminating genomic DNA contamination by DNase I (Fermentas, Waltham, MA, USA), about 2.0 μg of total RNA were used for the synthesis of first-strand complementary DNA (cDNA) with the cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) and subjected to qRT-PCR analysis using the LightCycler (Roche Diagnostics, Indianapolis, IN, USA). The relative expression level for each candidate gene was calculated using the 2–ΔΔCT method with LeEF-1α as an internal reference gene. The primers used for qRT-PCR reactions are listed in the Additional file 12.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (grant No. 2011CB100304), the Ministry of Agriculture of China (Grant No. 2011ZX08009-003-005).
Additional files
List of the predicted SlbHLHs and their corresponding information.
The distribution ratio of SlbHLH genes on chromosome of tomato.
The duplication of SlbHLHs with low- and high-stringency.
Segmental duplication analysis for SlbHLH genes in whole tomato genome.
Segmental duplication and gene alignment.
Intron distribution in the coding sequence of the bHLH domain of SlbHLH genes.
Neighbor-joining phylogenetic tree based on the bHLH domains of AtbHLHs and SlbHLHs.
Summary of the predicted functions of SlbHLHs by comparative analysis with AtbHLHs.
RPKM values of expression of SlbHLHs in ten tissues of tomato.
Maximum likelihood tree of plant bHLH domains. This is a detailed representation of the tree shown in Figure 7.
The bHLH protein sequences of tomato and other species.
Primers used for qRT-PCR analysis.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HS designed the experiments, performed all the bioinformatics analysis, and drafted the manuscript. HF prepared the materials and did qRT-PCR analysis. HQL made suggestions and discussions for experiments and analysis, and revised the manuscript. All the authors have read and approved the final manuscript.
Contributor Information
Hua Sun, Email: sunhua229@genetics.ac.cn.
Hua-Jie Fan, Email: hjfan@sjziam.ac.cn.
Hong-Qing Ling, Email: hqling@genetics.ac.cn.
References
- 1.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–70. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–70. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenfeld MJ, Delvecchio C, Sun X. Analysis of the transcriptional activation domain of the Drosophila tango bHLH-PAS transcription factor. Dev Genes Evol. 2005;215:221–9. doi: 10.1007/s00427-004-0462-9. [DOI] [PubMed] [Google Scholar]
- 4.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–83. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 5.Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–16. doi: 10.1007/PL00006494. [DOI] [PubMed] [Google Scholar]
- 6.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–40. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferre-D’Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–9. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–6. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7(33):1–18. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–82. doi: 10.1016/S0065-230X(08)60353-X. [DOI] [PubMed] [Google Scholar]
- 11.Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–51. [PubMed] [Google Scholar]
- 12.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–28. [PubMed] [Google Scholar]
- 13.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–11. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Steidl C, Leimeister C, Klamt B, Maier M, Nanda I, Dixon M, et al. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000;66:195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- 16.Crozatier M, Valle D, Dubois L, Ibnsouda S, Vincent A. Collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr Biol. 1996;6:707–18. doi: 10.1016/S0960-9822(09)00452-7. [DOI] [PubMed] [Google Scholar]
- 17.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–47. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–84. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 21.Consortium TTG. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–41. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, et al. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–97. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujisawa M, Nakano T, Shima Y, Ito Y. A Large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–86. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagata H, Yonesu K, Hirata A, Aizono Y. TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of the cucumisin gene. J Biol Chem. 2002;277:11582–90. doi: 10.1074/jbc.M109946200. [DOI] [PubMed] [Google Scholar]
- 25.Yuan YX, Zhang J, Wang DW, Ling HQ. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005;15:613–21. doi: 10.1038/sj.cr.7290331. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18:385–97. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 27.Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–36. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant. 2013;6:503–13. doi: 10.1093/mp/sss089. [DOI] [PubMed] [Google Scholar]
- 29.Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci U S A. 2002;99:13938–43. doi: 10.1073/pnas.212448699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pires ND, Dolan L. Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond B Biol Sci. 2012;367:508–18. doi: 10.1098/rstb.2011.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–74. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–30. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 33.Steemans P, Herisse AL, Melvin J, Miller MA, Paris F, Verniers J, et al. Origin and radiation of the earliest vascular land plants. Science. 2009;324:353. doi: 10.1126/science.1169659. [DOI] [PubMed] [Google Scholar]
- 34.Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:989–1000. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schat H, Vooijs R, Kuiper E. Identical major gene loci for heavy metal tolerances that have independently evolved in different local populations and subspecies of Silene vulgaris. Evolution. 1996;50:1888–95. doi: 10.2307/2410747. [DOI] [PubMed] [Google Scholar]


