Abstract
Current research targeting filtered macrobial environmental DNA (eDNA) often relies upon cold ambient temperatures at various stages, including the transport of water samples from the field to the laboratory and the storage of water and/or filtered samples in the laboratory. This poses practical limitations for field collections in locations where refrigeration and frozen storage is difficult or where samples must be transported long distances for further processing and screening. This study demonstrates the successful preservation of eDNA at room temperature (20 °C) in two lysis buffers, CTAB and Longmire's, over a 2-week period of time. Moreover, the preserved eDNA samples were seamlessly integrated into a phenol–chloroform–isoamyl alcohol (PCI) DNA extraction protocol. The successful application of the eDNA extraction to multiple filter membrane types suggests the methods evaluated here may be broadly applied in future eDNA research. Our results also suggest that for many kinds of studies recently reported on macrobial eDNA, detection probabilities could have been increased, and at a lower cost, by utilizing the Longmire's preservation buffer with a PCI DNA extraction.
Keywords: eDNA extraction, eDNA preservation, environmental DNA, Lepomis macrochirus
Introduction
The detection of macrobial DNA in environmental water samples, hereafter referred to as ‘eDNA’, is a burgeoning field of research often involving the detection of rare species, including invasive species (Dejean et al. 2012; Goldberg et al. 2013; Jerde et al. 2013; Takahara et al. 2013; Piaggio et al. 2014) and endangered species (Olson et al. 2012; Thomsen et al. 2012a). Three important considerations for eDNA research are the capture of the eDNA, the preservation of the eDNA and the successful extraction of the eDNA (Pilliod et al. 2013). The filtration of water samples is a routine capture mechanism of eDNA present in aquatic environments as it is scalable to the environment and allows for the concentration of rare eDNA fragments from large volumes of water. Various filter membrane types and filter pore sizes have been utilized for the filtration of water samples in eDNA studies (Minamoto et al. 2012; Thomsen et al. 2012b; Goldberg et al. 2013; Jerde et al. 2013; Piaggio et al. 2014). These choices can impact the efficiency of the eDNA capture (Liang & Keeley 2013), but are currently difficult to quantify between studies as other aspects (i.e. eDNA preservation and extraction) are not held constant (Turner et al. 2014).
For studies utilizing filtration of water samples, the preservation of eDNA is often reliant on cold ambient temperatures (Mahon et al. 2010; Takahara et al. 2012, 2013; Jerde et al. 2013; Wilcox et al. 2013). This is the case at multiple points of the sample collection process, including the use of ice in the transport of water samples from collection sites to laboratories, storage of water samples in freezers for filtering at later time points and freezing of filters following the processing of water samples (Mahon et al. 2010; Thomsen et al. 2012b; Pilliod et al. 2013; Takahara et al. 2013; Wilcox et al. 2013; Piaggio et al. 2014). This reliance on cold ambient temperatures poses practical limitations for field collections in locations where refrigeration and frozen storage is difficult (i.e. backcountry locations accessible only by foot) or where samples must be transported long distances for further processing and screening (i.e. international travel). The justification for such temperature control is to limit DNA degradation in eDNA samples, a concern of paramount importance in eDNA studies as the targeted fragments are often degraded at the time of collection and/or are present in vanishing amounts, due to the rarity of the organisms producing the eDNA. While in situ field filtration can be overcome with portable pumps (Pilliod et al. 2013; Wilcox et al. 2013), having a method that preserves filtered eDNA and prevents further degradation would benefit field scientists tasked with collecting samples under transport or temperature limitations.
The storage of filters in ethanol is a current room temperature preservation option (Goldberg et al. 2013; Pilliod et al. 2013), but the ethanol is not itself incorporated into the DNA extraction process. A number of preservation buffers, in addition to ethanol, have shown to be effective with tissue samples at room temperature (Seutin et al. 1991; Longmire et al. 1997; Kilpatrick 2002; Rhodes et al. 2003) and should serve in a similar capacity with eDNA captured on filters. Many of these room temperature preservation buffers simultaneously facilitate the lysis of cellular membranes, releasing intracellular components, such as DNA, into the preservation buffer. For eDNA studies, the assimilation of a room temperature preservation buffer into a DNA extraction protocol would increase the efficiency of the extraction (i.e. include eDNA that washes off the filter prior to extraction or eDNA released through lysis of cellular membranes) while avoiding hassles related to the storage of samples at a cold ambient temperature (Seutin et al. 1991; Kilpatrick 2002).
The extraction of filtered eDNA is often accomplished with a commercial kit, such as MoBio's PowerWater® DNA Isolation kit (Olson et al. 2012; Jerde et al. 2013; Wilcox et al. 2013; Piaggio et al. 2014) or Qiagen's DNeasy® Blood and Tissue kit (Minamoto et al. 2012; Goldberg et al. 2013; Pilliod et al. 2013; Kelly et al. 2014). Phenol–chloroform–isoamyl alcohol DNA extractions (Sambrook et al. 1989), hereafter ‘PCI’, are a popular extraction technique utilized in conjunction with room temperature preservation buffers for tissue samples in various applications of conservation genetics (Miller 2006; Smith & Hughes 2008; Wirgin et al. 2010). Additionally, this protocol has been recently employed for the extraction of macrobial eDNA captured on polycarbonate track-etch filters, nylon filters and glass fibre filters (Barnes et al. 2014; Deiner & Altermatt 2014; Turner et al. 2014). The PCI extraction protocol has the potential to drastically reduce per sample costs currently associated with eDNA research and assimilate a room temperature preservation buffer into the extraction process, integrating a transport and storage mechanism that does not rely on cold ambient temperatures.
As eDNA projects currently employ a variety of capture, preservation and extraction protocols (Lodge et al. 2012; Pilliod et al. 2013), some standardization could help with comparisons of other aspects of the research that may be heavily influenced by environmental conditions, such as the use of various filter membrane types and pore sizes in the capture of targeted eDNA fragments (Barnes et al. 2014; Turner et al. 2014). The room temperature preservation would additionally allow for application in conditions not suited for cold storage of samples. With these considerations in mind, we conducted a set of four experiments to compare (i) preservation with CTAB and Longmire's buffers among fresh samples, and samples stored for 1 and 2 weeks at −20, 20 and 45 °C, (ii) the application of the PCI protocol for eDNA extraction from cellulose nitrate filters, polyethersulfone filters, polycarbonate track-etch filters and glass microfibre filters (iii) the PCI DNA extraction protocol with two commercial DNA extraction kits currently featured in eDNA research and (iv) different approaches to the PCI DNA extraction protocol.
Materials and methods
Separate rooms were used for fish husbandry, pre-PCR laboratory work and post-PCR laboratory work. The 70-gallon mesocosm, with approximately 100 juvenile bluegill (Lepomis macrochirus), was monitored throughout the experiment following institutional animal care and use protocols. For all the four experiments, 250 mL water samples were collected and filtered immediately through a single filter; unless otherwise specified, DNA extractions immediately followed the terminus of sample filtration for each experiment. Samples were filtered with 47-mm magnetic filter funnels (Pall), and each filter funnel was completely immersed in 10% bleach for a minimum of 10 min and thoroughly rinsed with DI water prior to any subsequent filtration. A single sample from each experimental treatment was filtered before filtering a second sample from each experimental treatment and then a third and so on, randomly spreading the filtering effort across all experimental treatments and minimizing the potential impact of contamination on any one experimental treatment. Unless otherwise specified, the filters were placed in 2-mL microcentrifuge tubes and completely immersed in 900 μL of CTAB buffer (1.4 M NaCl, 2% (w/v) cetyltrimethyl ammonium bromide, 100 mM Tris, 20 mM EDTA and 0.25 mM polyvinylpyrrolidone; Coyne et al. 2005). The CTAB buffer was chosen as it has been used successfully for ongoing research (Barnes et al. 2014; Turner et al. 2014). Unless otherwise specified, DNA extractions followed a modified PCI extraction and ethanol precipitation (Sambrook et al. 1989): (i) the 2-mL microcentrifuge tubes (filters and preservation buffer) were incubated in a 65 °C water bath for 10 min; (ii) 900 μL of PCI (one phase, 25:24:1, Amresco) was added to each tube, and samples were vortexed for 5 seconds. The addition of chloroform disintegrates polycarbonate track-etch (hereafter ‘PCTE’) and polyethersulfone (hereafter ‘PES’) filter membranes (Stark et al. 1998; Turner et al. 2014), and as such, extra care was given to mix the liquid layers for the cellulose nitrate (hereafter ‘CN’) and glass microfibre (hereafter ‘GMF’) filter membrane types, which remained intact; (iii) tubes were centrifuged at 15 000 g for 5 min, and 700 μL of the aqueous layer was transferred to a fresh set of 2-mL microcentrifuge tubes; (iv) 700 μL of chloroform–isoamyl alcohol, hereafter ‘CI’ (24:1, Amresco), was added to each tube and samples were vortexed for 5 seconds; (v) tubes were centrifuged at 15 000 g for 5 min, and 500 μL of the aqueous layer was transferred to a fresh set of 2 mL tubes; (vi) 1.25 mL of 100% ice-cold ethanol and 20 μL of 5 M NaCl were added to each tube, and samples were precipitated at −20 °C overnight; (vii) the precipitate was pelleted by centrifugation at 15 000 g for 10 min, and the liquid was decanted; (viii) pellets were dried in a vacuufuge at 45 °C for 15 min, followed by air drying until no visible liquid remained; and finally, (ix) pellets were rehydrated with 200 μL of 1× TE Buffer, low EDTA (USB).
All DNA extractions were assayed with qPCR TaqMan® primers and probe targeting a 100-bp fragment of the bluegill cytochrome b gene (Takahara et al. 2013) in the following 20 μL mixes: 10 μL of TaqMan® Environmental Master Mix 2.0 (Life Technologies), 1.8 μL of each primer (10 μM stock concentration), 0.25 μL of the TaqMan® probe (10 μM stock concentration), 4 μL of eDNA extract and 2.15 μL of sterile water. The cycling parameters were as follows: a single step at 50 °C for 2 min, a single step at 95 °C for 10 min and 55 cycles at 95 °C for 15 seconds followed by 60 °C for 1 min. To quantify the DNA copy number in each eDNA extract, a standard was created (as follows) and included on each qPCR plate along with the eDNA extracts. A DNA fragment was synthesized by Integrated DNA Technologies based on the sequence from GenBank Accession no. JN389795 starting at location 14 298 and ending at location 14 797. The 500-bp fragment included the 100-bp region of the bluegill cytochrome b gene targeted by the assay flanked by an additional 200-bp on either side. The copy number of the synthesized standard was determined by multiplying the number of moles by Avogadro's number. A serial dilution of the standard was run on each qPCR plate and provided a regression line from which the unknown copy numbers of the eDNA extracts could be estimated. All qPCR assays were run on a Mastercycler ep realplex real-time PCR system (Eppendorf) and analysed with the accompanying realplex 2.2 software. Two negative controls were included on each qPCR plate, both containing the aforementioned 20 μL mix except for additional sterile water in place of eDNA extract.
Filter preservation experiment
A pilot experiment evaluated the use of four storage buffers: a 20% DMSO buffer (20% DMSO, 0.25M EDTA, saturated with NaCl; Seutin et al. 1991), RNAlater (Qiagen #76106), CTAB buffer and Longmire's buffer (0.1 M Tris, 0.1 M EDTA, 10 mM NaCl, 0.5% (w/v) SDS; Longmire et al. 1997). The 20% DMSO and RNAlater were not compatible with the PCI DNA extraction protocol as they both yielded a substantial precipitate that appeared to completely inhibit qPCR amplification. The removal of the 20% DMSO and RNAlater immediately prior to the DNA extraction and replacement with CTAB reduced the resulting precipitate, but the qPCR assays again failed to amplify. As such, only the CTAB buffer and Longmire's buffer were evaluated in the filter preservation experiment.
For each experimental treatment, five 250 mL water samples were each immediately filtered through a single PCTE filter (1.2 μm; Millipore). Filters were placed in 2-mL microcentrifuge tubes and completely immersed in 900 μL of either CTAB buffer (35 samples total) or Longmire's buffer (25 samples total). For each storage buffer, a set of five filters was extracted immediately, hereafter ‘fresh’. For the CTAB, an additional 10 filters were kept at each of the three temperature regimes: −20, 20 and 45 °C. For each temperature regime, 5 of the filters were extracted after a 1-week storage period, while the remaining 5 were extracted after a 2-week storage period. For the Longmire's buffer, the same protocol was applied for only two of the temperature regimes, 20 and 45 °C. The −20 °C regime was only used with the CTAB storage buffer as this has been used with success in the past (Barnes et al. 2014; Turner et al. 2014) and served as a benchmark against which the other temperature regimes and storage buffer could be compared. All DNA extracts from a given storage buffer were assayed once simultaneously on the same qPCR plate with a serial dilution of the standard for the quantification of DNA copy number. Each plate was then run a second time to produce two qPCR replicates for each sample.
Filter membrane type experiment
The feasibility of DNA extraction from different filter membrane types with the PCI extraction protocol was evaluated for four different filter membrane types: 0.8 μm CN (Whatman), 0.8 μm PES (Pall), 1.0 μm PCTE (GE Osmonics) and 1.5 μm GMF (Whatman). For each filter membrane type, ten 250 mL water samples were each filtered through a single filter and completely immersed with 900 μL of CTAB in a 2-mL microcentrifuge tube. DNA was extracted and ethanol precipitated with the previously described PCI protocol. All of the samples (in duplicate) were run simultaneously on a single qPCR plate with a serial dilution of the standard for the quantification of DNA copy number.
PCI kit comparison experiment
Two historically popular combinations of macrobial eDNA filtration membrane types with commercial kit eDNA extractions have utilized 0.45-μm CN filters with Qiagen's DNeasy® Blood and Tissue kit (Ahmed et al. 2010, 2013; Goldberg et al. 2011, 2013; Pilliod et al. 2013, 2014) and 1.5-μm GMF filters with MoBio's PowerWater® DNA Isolation kit (Olson et al. 2012; Jerde et al. 2013; Wilcox et al. 2013; Piaggio et al. 2014). As such, comparisons between the PCI DNA extraction protocol and these two commercial DNA extraction kits focused on the same pairings of filters and extraction kits. A total of forty 250 mL water samples were collected, and twenty of them were each filtered immediately through a single 0.45-μm CN filter (Spectrum). Ten of the CN filters were completely immersed in 900 μL of CTAB, incubated in a 65 °C water bath for 1 h and then put through the PCI extraction and ethanol precipitation protocol as outlined previously. DNA was extracted from the remaining 10 CN filters following Qiagen's recommendations for the DNeasy® Blood and Tissue kit, with some modifications. Filters were completely immersed in 567 μL buffer ATL and 63 μL Proteinase-K (rather than the recommended 180 and 20 μL, respectively) and incubated in a 65 °C water bath for 1 h. Following the incubation time, 630 μL buffer AL and 630 μL 100% ethanol were added to the 2-mL tube, instead of the recommended 200 μL of each solution. A total of three centrifugation iterations were required to load the entire contents of the 2-mL tube, minus the CN filter, onto the silica membrane, as compared to the single centrifugation step normally required. The remainder of the protocol followed the manufacturer's recommendations. The other 20 samples were each filtered immediately through a single 1.5 μm GMF filter (Whatman). Ten of the GMF filters were completely immersed in 900 μL of CTAB, incubated in a 65 °C water bath for 1 h and then put through the PCI extraction and ethanol precipitation protocol as outlined previously. DNA was extracted from the remaining 10 GMF filters following MoBio's recommendations for the PowerWater® DNA Isolation kit, with the exception that the bead-beating step was performed until the filters appeared to be completely liquefied rather than the 5 min recommended maximum. DNA extractions from both kits were eluted from their respective spin columns with the addition of 200 μL of 1× TE Buffer, low EDTA (USB), rather than the recommended Buffer AE (Qiagen) and Solution PW6 (MoBio). All of the samples (in duplicate) were run simultaneously on a single qPCR plate with a serial dilution of the standard for the quantification of DNA copy number.
DNA extraction experiment
Variations on the PCI extraction protocol were evaluated, utilizing a total of forty 250 mL water samples each filtered immediately through a single 1.2-μm PCTE filter (Millipore). Twenty of the filters were placed in 2-mL tubes with 900 μL of CTAB, and DNA was extracted with the PCI protocol. An alternate precipitation protocol was evaluated using isopropyl alcohol with 10 of the samples in conjunction with the previously mentioned ethanol precipitation on the remaining 10 samples. The isopropyl alcohol precipitation was conducted as follows: (i) 500 μL of ice-cold isopropyl alcohol and 250 μL of 5M NaCl were added to the 500 μL recovered from the aqueous layer (step 5 in the PCI protocol), and tubes were precipitated at −20 °C overnight; (ii) the precipitate was pelleted by centrifugation at 15 000 g for 10 min, and the liquid was decanted; (iii) 150 μL of room temperature 70% ethanol was added to each tube; (iv) tubes were centrifuged at 15 000 g for 5 min, and the liquid was decanted; (v) 150 μL of room temperature 70% ethanol was added to each tube a second time; (vi) tubes were centrifuged at 15 000 g for 5 min, and the liquid was decanted; (vii) pellets were dried in a vacuufuge at 45 °C for 15 min, followed by air drying until no visible liquid remained; and finally, (viii) pellets were rehydrated with 200 μL of 1× TE Buffer, low EDTA (USB).
An alternate DNA extraction protocol eliminated the use of phenol (step 2 in the PCI protocol). The remaining 20 filters were completely immersed with 700 μL of CTAB in 2-mL microcentrifuge tubes and incubated in a 65 °C water bath for 10 min. These samples were then extracted starting with the addition of 700 μL of CI (step 4 in the PCI protocol). The ethanol and isopropanol precipitations were again both evaluated, each on 10 of the samples. All of the samples (in duplicate) were run simultaneously on a single qPCR plate with a serial dilution of the standard for the quantification of DNA copy number.
Statistical analyses
ANOVA statistical tests were conducted individually for each of the four experiments to test for differences between mean DNA copy numbers. A two-sided t-test was used to test differences in the average amount of DNA recovered from fresh CTAB and Longmire's extractions within the ‘filter preservation experiment’. Technical replicates were averaged for the analysis, residuals from the ANOVAs and t-test were checked for normality using normal Q–Q plots, and pairwise comparisons in the ANOVA were performed using Tukey's post hoc test. All statistics and plots were conducted and created in Mathematica 9.0.1.0 (Wolfram Research, Inc., Version 9.0.1.0, Champaign, IL 2013). All tests conformed to the normality assumptions unless otherwise indicated.
Results
For all of the qPCR plates run for this study, the qPCR standard curve slope ranged from −3.342 to −3.498, the y-intercept ranged from 38.43 to 39.26, the efficiency ranged from 0.93 to 0.99, and the R2 values ranged from 0.994 to 1.000. All of the negative controls failed to amplify throughout the entire experiment.
Filter preservation experiment
All replicates amplified and were incorporated into the statistical analyses for all twelve of the experimental treatments (Table1). For the CTAB preservation buffer, relative to fresh samples, Tukey's post hoc comparisons of the ANOVA results revealed a significantly higher DNA copy number in samples stored at all the three temperatures (−20, 20 and 45 °C) following the 2-week time interval (Fig.1a–c). For the Longmire's preservation buffer, the same result was observed for the 45 °C temperature (Fig.1e), but no significant difference in copy number existed between fresh samples and those stored at 20 °C (Fig.1d). A two-sided t-test of the fresh extractions revealed a significantly higher yield in DNA copy number for the Longmire's preservation buffer as compared to the CTAB preservation buffer (P-value < 0.001; Fig.1f).
Table 1.
Outline of all four experiments, with the treatments evaluated in each experiment (Treatment) and the number of samples analysed per experimental treatment (N)
| Experiment | Treatment | N |
|---|---|---|
| Filter preservation | CTAB; fresh | 5 |
| CTAB; −20 °C; 1 week | 5 | |
| CTAB; −20 °C; 2 weeks | 5 | |
| CTAB; 20 °C; 1 week | 5 | |
| CTAB; 20 °C; 2 weeks | 5 | |
| CTAB; 45 °C; 1 week | 5 | |
| CTAB; 45 °C; 2 weeks | 5 | |
| Longmire's; fresh | 5 | |
| Longmire's; 20 °C; 1 week | 5 | |
| Longmire's; 20 °C; 2 weeks | 5 | |
| Longmire's; 45 °C; 1 week | 5 | |
| Longmire's; 45 °C; 2 weeks | 5 | |
| Filter membrane type | 0.8 μm; cellulose nitrate (CN) | 10 |
| 0.8 μm; polyethersulfone (PES) | 10 | |
| 1 μm; polycarbonate track-etch (PCTE) | 10 | |
| 1.5 μm; glass microfibre (GMF) | 9 | |
| PCI kit comparison | 0.45 μm; cellulose nitrate (CN); PCI | 10 |
| 0.45 μm; cellulose nitrate (CN); Qiagen | 10 | |
| 1.5 μm; glass microfibre (GMF); PCI | 10 | |
| 1.5 μm; glass microfibre (GMF); MoBio | 10 | |
| DNA extraction | PCI start; ethanol precipitation | 10 |
| PCI start; isopropanol precipitation | 10 | |
| CI start; ethanol precipitation | 10 | |
| CI start; isopropanol precipitation | 10 |
Fig 1.
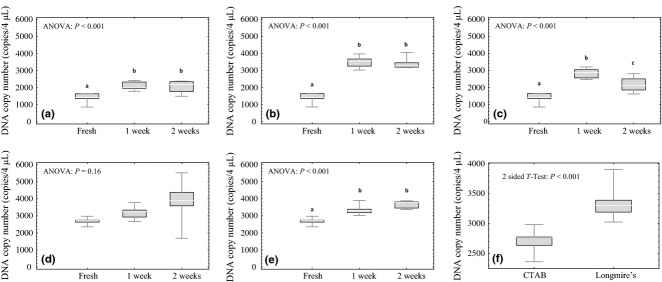
Box and whisker plots for the filter preservation experiment. The top and bottom of the whiskers represent the maximum and minimum values, the top and bottom of the boxes represent the 75% and 25% quartiles, and the lines inside the boxes represent the median values. Significance in pairwise comparisons of treatments is noted by letters a, b and c where different letters represent statistically significant differences. Two preservation buffers, CTAB and Longmire's, were evaluated over a 2-week interval of time. (a) CTAB with −20 °C storage, (b) CTAB with 20 °C storage, (c) CTAB with 45 °C storage, (d) Longmire's with 20 °C storage, (e) Longmire's with 45 °C storage and (f) comparison between CTAB and Longmire's for fresh extractions.
Filter membrane type experiment
For three of the four filter membrane types (CN, PES and PCTE), all 10 samples amplified; one of the 10 samples for the GMF membrane type failed to amplify, and as such, only nine of the samples were used in the statistical analyses (Table1). Tukey's post hoc comparisons of the ANOVA results revealed that the 0.8-μm CN and 0.8-μm PES filters did not differ significantly and both yielded significantly more copies of DNA than the 1.0-μm PCTE and 1.5-μm GMF filters; the 1.0-μm PCTE filters yielded significantly more copies of DNA than the 1.5-μm GMF filters (Fig.2).
Fig 2.
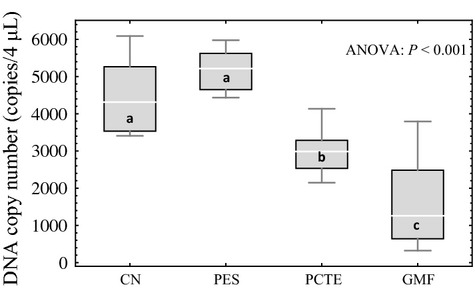
Box and whisker plots for the filter membrane type experiment. The top and bottom of the whiskers represent the maximum and minimum values, the top and bottom of the boxes represent the 75% and 25% quartiles, and the lines inside the boxes represent the median values. Significance in pairwise comparisons of treatments is noted by letters a, b and c where different letters represent statistically significant differences between experimental treatments. The four treatments were 0.8-μm cellulose nitrate filters (CN), 0.8-μm polyethersulfone filters (PES), 1.0-μm polycarbonate track-etch filters (PCTE), and 1.5-μm glass microfibre filters (GMF).
PCI kit comparison experiment
All 10 of the samples amplified and were incorporated into the statistical analyses for each of the four experimental treatments (Table1). Tukey's post hoc comparisons of the ANOVA results revealed that the CN filter with PCI extraction yielded significantly more copies of DNA than the other three experimental treatments; the GMF filter with the MoBio extraction yielded significantly more copies of DNA than both the GMF filter with PCI extraction and the CN filter with Qiagen extraction, which were not significantly different from one another (Fig.3).
Fig 3.
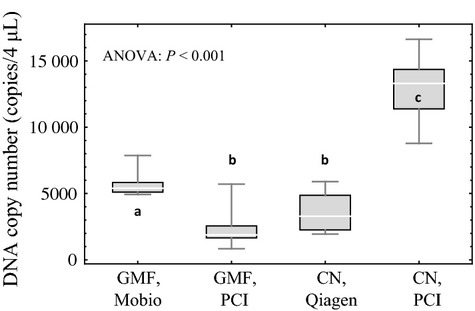
Box and whisker plots for the PCI-kit comparison experiment. The top and bottom of the whiskers represent the maximum and minimum values, the top and bottom of the boxes represent the 75% and 25% quartiles, and the lines inside the boxes represent the median values. Significance in pairwise comparisons of treatments is noted by letters a, b and c where different letters represent statistically significant differences between experimental treatments. The four treatments were 1.5-μm glass microfibre filters (GMF) with MoBio extraction, 1.5-μm glass microfibre filters (GMF) with PCI extraction, 0.45-μm cellulose nitrate filters (CN) with Qiagen extraction and 0.45-μm cellulose nitrate filters (CN) with PCI extraction.
DNA extraction experiment
All 10 of the samples amplified and were incorporated into the statistical analyses for each of the four experimental treatments (Table1). Tukey's post hoc comparisons of the ANOVA results revealed no statistically significant differences among the four experimental treatments (Fig.4).
Fig 4.
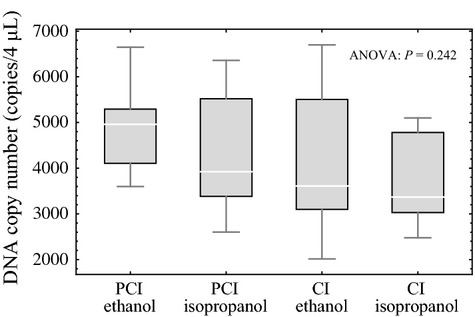
Box and whisker plots for the DNA extraction experiment. The top and bottom of the whiskers represent the maximum and minimum values, the top and bottom of the boxes represent the 75% and 25% quartiles, and the lines inside the boxes represent the median values. There was no statistical significance in pairwise comparisons between the four experimental treatments: PCI extraction with ethanol precipitation, PCI extraction with isopropanol precipitation, CI extraction with ethanol precipitation and CI extraction with isopropanol precipitation.
Discussion
Both preservation buffers, CTAB and Longmire's, successfully preserved filtered eDNA at 20 °C over a 2-week period of time. The DNA copy number increased from the initial time point to the 2-week time point, assuming the results from the fresh treatment indicate the initial copy number in all corresponding experimental treatments. This trend was observed for the filters in the −20 and 45 °C regimes as well. One possible explanation for the observed trend is increased cell lysis efficiency for both buffers with an increase in time. This explanation indicates it is possibly beneficial to leave the filters in the lysis buffer for an extended period of time prior to the DNA extraction. Alternatively, longer incubation times in the 65 °C water bath could significantly increase the yield of extracted DNA as heat facilitates cell lysis and the release of intracellular components, such as DNA (Lu et al. 2005). In a comparison between the two buffers, the Longmire's buffer produced a significantly higher DNA copy number than the CTAB buffer for the fresh extractions. In the 45 °C regime, the Longmire's buffer demonstrated a copy number increase from the initial time point to the 1-week time point to the 2-week time point; in comparison, a significant reduction in copy number was observed in the CTAB buffer from the 1-week time point to the 2-week time point, possibly representative of DNA degradation in the elevated storage temperature. These results suggest that while both preservation buffers are adequate for the room temperature preservation of filtered macrobial eDNA, the Longmire's buffer outperformed the CTAB buffer.
The reliance of the PCI extraction approach on chemicals that are harmful to humans is a disadvantage associated with the handling and proper disposal of the phenol and chloroform. When handling these chemicals, certain measures should be considered to reduce the risk of skin contact (i.e. laboratory coat and gloves), eye contact (i.e. safety glasses) or inhalation (i.e. fume hood). It is also recommended to equip laboratories with eyewash stations and safety showers in the event of exposure. In addition to safety measures for handling, the hazardous nature of both chemicals requires them to be disposed of in a manner that conforms to the safety regulations as governed by the organization with which any laboratory is affiliated. One viable alternative for researchers that are interested in using either the CTAB or Longmire's preservation buffers without relying on phenol and chloroform for the DNA extraction is the integration of the room temperature preservation buffer with Qiagen's DNeasy® Blood and Tissue kit (Herath et al. 2010; Miller et al. 2010).
On the other hand, the PCI approach provides several advantages. First, the PCI extraction protocol potentially yields significantly more copies of targeted eDNA fragments, as demonstrated by the use of 0.45-μm CN filters with both Qiagen's DNeasy® Blood and Tissue kit and the PCI protocol. It is noteworthy that a number of recommended modifications of the DNeasy® protocol exist for increasing the detection probability of targeted species, including the addition of Qiagen's QIAshredder and increasing incubation times (Goldberg et al. 2011). These modifications could potentially provide more comparable results to those achieved in the current study with the CTAB preservation buffer and PCI DNA extraction. In the comparison between MoBio's PowerWater® DNA Isolation Kit and the PCI protocol with the 1.5-μm GMF filters, however, the commercial kit yielded significantly more eDNA than the PCI protocol. The MoBio kit, as implemented in the current study, shreds apart the GMF filters through a bead-beating step, exposing the internally captured material (as accomplished by GMF filters through a tortuous path) and possibly increasing lysis efficiency. These results suggest that the addition of a bead-beating step should be considered for the use of the GMF filters with the preservation buffer and PCI extraction.
Second, the PCI approach is substantially cheaper than the commercial kits. Potential per sample costs for eDNA extraction with MoBio's PowerWater® DNA Isolation kit are over $8 (USD), for Qiagen's DNeasy® Blood and Tissue kit are over $2 (USD) and for the PCI extraction protocol are <$0.20 (USD). These values are rough estimates as a number of factors can impact their calculation, but the relative difference in costs between methods is well represented. And finally, the reagents used in the process are well understood, in the public domain, and easily tested and adapted to individual research projects. The lack of statistical significance between the approaches employed in the current study (DNA extraction with and without the use of phenol and DNA precipitation with both ethanol and isopropanol) suggests that researchers have flexibility with the extraction protocols while producing comparable results.
The PCI extraction protocol was successful for all four evaluated filter membrane types: cellulose nitrate (CN), polyethersulfone (PES), polycarbonate track-etch (PCTE) and glass microfibre (GMF). The discrepancy between membrane types in the amount of DNA extracted can, in part, be attributed to differences in pore size. A consistent relationship between pore size and amount of DNA present in the extract has been previously demonstrated (Liang & Keeley 2013), with a measurable decrease in DNA recovery with an increase in pore size (see Fig. S1, Supporting information for independent confirmation of this relationship). The one comparison between membrane types with an equal stated pore size, 0.8 μm CN and 0.8 μm PES, produced statistically comparable copies of targeted eDNA. This relationship differs from previous observations by Liang & Keeley (2013), where mixed cellulose ester filters recovered between 2.6 and 3.9 times more copies of plasmid DNA than PES filters. In addition to potential differences between the type of cellulose filter utilized (mixed cellulose esters comprises both cellulose nitrate and cellulose acetate), the current study may also highlight the complexity of an eDNA sample. Only a singular contributor to eDNA yields (free-floating, extracellular DNA) was evaluated by Liang & Keeley (2013). The increase in relative capture efficiency of the PES filters in the current study may highlight differences between filter types in the potential capture efficiencies of other sources of eDNA, such as those that are intracellular and/or extracellular but bound to other material in the environment (Siuda & Gude 1996; Converse et al. 2012; Thomsen et al. 2012a), and more closely reflect macrobial eDNA capture potentials in aquatic systems where free-floating DNA is a minority contributor to total eDNA (Turner et al. 2014).
The burgeoning field of macrobial eDNA research has already produced noteworthy results in both freshwater and marine environments, including the detection of fish (Thomsen et al. 2012a,b; Jerde et al. 2013; Kelly et al. 2014), amphibians (Dejean et al. 2012; Olson et al. 2012; Thomsen et al. 2012a; Pilliod et al. 2013, 2014), reptiles (Piaggio et al. 2014), insect larvae and crustaceans (Thomsen et al. 2012a; Deiner & Altermatt 2014), mammals (Foote et al. 2012; Thomsen et al. 2012a) and molluscs (Goldberg et al. 2013; Deiner & Altermatt 2014). And as the field continues to grow, the application of next-generation sequencing platforms opens avenues into biodiversity estimates on a large scale (Thomsen et al. 2012a) and further integration into the conservation and management of natural resources (Lodge et al. 2012). The main goal of this study was to evaluate eDNA preservation and extraction for filtered macrobial environmental DNA, with the potential broad application for studies in a variety of aquatic environments. The Longmire's preservation buffer provides researchers with a room temperature storage buffer that adequately handles elevated temperatures (up to 45 °C in the current study), and the assimilation of the Longmire's preservation buffer into a PCI DNA extraction protocol has the potential to simultaneously reduce per sample costs and increase the recovery of targeted eDNA fragments. The resulting increase in detection probabilities for rare species will benefit future eDNA research for both species-specific assays and large-scale biodiversity estimates.
Acknowledgments
We would like to thank K. Uy and C. Gantz for assistance in the laboratory; C. Gantz for assistance in the ongoing care of the bluegill; L. de Souza and J. Godwin for stimulating ideas for this study. We would also like to thank three anonymous reviewers who provided excellent insight and feedback from which the manuscript benefitted greatly. EPA Great Lakes Restoration Initiative, Great Lakes Fisheries Trust and DoD SERDP provided funding. This is a publication of the Notre Dame Environmental Change Initiative.
Author contributions
M.A.R., B.P.O. and C.L.J. participated in experimental design. M.M.M. and M.A.R. conducted laboratory work. C.L.J. performed data analysis. B.P.O. and M.A.R. cared for captive animal test subjects and performed laboratory experiments. M.A.R. wrote the manuscript with help from B.P.O., C.L.J., M.M.M. and D.M.L. D.M.L. was responsible for providing funding and laboratory infrastructure. All authors read and approved the final manuscript.
Data Accessibility
DNA copy numbers from qPCR plates are available as Data S1 (Supporting information).
Supporting Information
Data S1 DNA copy numbers from qPCRs for all four experiments.
Fig. S1 A total of 40-250 mL water samples were each filtered through single PCTE filters, 10 each for four different pore sizes: 1, 3, 8 and 20 µm.
References
- Ahmed W, Goonetilleke A, Gardner T. Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Research. 2010;44:4662–4673. doi: 10.1016/j.watres.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Yusuf R, Hasan I, et al. Fecal indicators and bacterial pathogens in bottled water from Dhaka, Bangladesh. Brazilian Journal of Microbiology. 2013;44:97–103. doi: 10.1590/S1517-83822013005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science and Technology. 2014;48:1819–1827. doi: 10.1021/es404734p. [DOI] [PubMed] [Google Scholar]
- Converse RR, Griffith JF, Noble RT, Haugland RA, Schiff KC, Weisberg SB. Correlation between quantitative PCR and culture-based methods for measuring Enterococcus spp. over various temporal scales at three California marine beaches. Applied and Environmental Microbiology. 2012;78:1237–1242. doi: 10.1128/AEM.07136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne KJ, Handy SM, Demir E, et al. Improved quantitative real-time PCR assays for enumeration of harmful algal species in field samples using an exogenous DNA reference standard. Limnology and Oceanography: Methods. 2005;3:381–391. [Google Scholar]
- Deiner K, Altermatt F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE. 2014;9:e88786. doi: 10.1371/journal.pone.0088786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C. Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. Journal of Applied Ecology. 2012;49:953–959. [Google Scholar]
- Foote AD, Thomsen PF, Sveegaard S, et al. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE. 2012;7:e41781. doi: 10.1371/journal.pone.0041781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg CS, Pilliod DS, Arkle RS, Waits LP. Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE. 2011;6:e22746. doi: 10.1371/journal.pone.0022746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg CS, Sepulveda A, Ray A, et al. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum. Freshwater Science. 2013;32:792–800. [Google Scholar]
- Herath P, Hoover GA, Angelini E, Moorman GW. Detection of elm yellows phytoplasma in elms and insects using real-time PCR. Plant Disease. 2010;94:1355–1360. doi: 10.1094/PDIS-12-09-0783. [DOI] [PubMed] [Google Scholar]
- Jerde CL, Chadderton WL, Mahon AR, et al. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Canadian Journal of Fisheries and Aquatic Sciences. 2013;70:522–526. [Google Scholar]
- Kelly RP, Port JA, Yamahara KM, Crowder LB. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE. 2014;9:e86175. doi: 10.1371/journal.pone.0086175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick CW. Noncryogenic preservation of mammalian tissues for DNA extraction: an assessment of storage methods. Biochemical Genetics. 2002;40:53–62. doi: 10.1023/a:1014541222816. [DOI] [PubMed] [Google Scholar]
- Liang Z, Keeley A. Filtration recovery of extracellular DNA from environmental water samples. Environmental Science and Technology. 2013;47:9324–9331. doi: 10.1021/es401342b. [DOI] [PubMed] [Google Scholar]
- Lodge DM, Turner CR, Jerde CL, et al. Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Molecular Ecology. 2012;21:2555–2558. doi: 10.1111/j.1365-294X.2012.05600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire JL, Maltbie M, Baker RJ. Use of ‘lysis buffer’ in DNA isolation and its implications for museum collections. Museum of Texas Tech University. 1997;163:1–3. [Google Scholar]
- Lu H, Schmidt MA, Jensen KF. A microfluidic electroporation device for cell lysis. Lab on a Chip. 2005;5:23–29. doi: 10.1039/b406205a. [DOI] [PubMed] [Google Scholar]
- Mahon AR, Rohly A, Budny M, et al. 2010. Environmental DNA monitoring and surveillance: standard operation procedures. Report to the United States Army Corps of Engineers, Environmental Laboratories, Cooperative Environmental Studies Unit, Vicksburg, Mississippi.
- Miller HC. Cloacal and buccal swabs are a reliable source of DNA for microsatellite genotyping of reptiles. Conservation Genetics. 2006;7:1001–1003. [Google Scholar]
- Miller BF, DeYoung RW, Campbell TA, Laseter BR, Ford WM, Miller KV. Fine-scale genetic and social structuring in a central Appalachian white-tailed deer herd. Journal of Mammalogy. 2010;91:681–689. [Google Scholar]
- Minamoto T, Yamanaka H, Takahara T, Honjo MN, Kawabata Z. Surveillance of fish species composition using environmental DNA. Limnology. 2012;13:193–197. [Google Scholar]
- Olson ZH, Briggler JT, Williams RN. An eDNA approach to detect eastern hellbenders (Cryptobranchus a. alleganiensis) using samples of water. Wildlife Research. 2012;39:629–636. [Google Scholar]
- Piaggio AJ, Engeman RM, Hopken MW, et al. Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Molecular Ecology Resources. 2014;14:374–380. doi: 10.1111/1755-0998.12180. [DOI] [PubMed] [Google Scholar]
- Pilliod DS, Goldberg CS, Arkle RS, Waits LP. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Canadian Journal of Fisheries and Aquatic Sciences. 2013;70:1123–1130. [Google Scholar]
- Pilliod DS, Goldberg CS, Arkle RS, Waits LP. Factors influencing detection of eDNA from a stream-dwelling amphibian. Molecular Ecology Resources. 2014;14:109–116. doi: 10.1111/1755-0998.12159. [DOI] [PubMed] [Google Scholar]
- Rhodes KL, Lewis RI, Chapman RW, Sadovy Y. Genetic structure of camouflage grouper, Epinephelus polyphekadion (Pisces: Serranidae), in the western central Pacific. Marine Biology. 2003;142:771–776. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Harbor Laboratory Press; 1989. [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology. 1991;69:82–90. [Google Scholar]
- Siuda W, Gude H. Determination of dissolved deoxyribonucleic acid concentration in lake water. Aquatic Microbial Ecology. 1996;11:193–202. [Google Scholar]
- Smith S, Hughes J. Microsatellite and mitochondrial DNA variation defines island genetic reservoirs for reintroductions of an endangered Australian marsupial, Perameles bougainville. Conservation Genetics. 2008;9:547–557. [Google Scholar]
- Stark KDC, Nicolet J, Frey J. Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Applied and Environmental Microbiology. 1998;64:543–548. doi: 10.1128/aem.64.2.543-548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z. Estimation of fish biomass using environmental DNA. PLoS ONE. 2012;7:e35868. doi: 10.1371/journal.pone.0035868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Minamoto T, Doi H. Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS ONE. 2013;8:e56584. doi: 10.1371/journal.pone.0056584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen PF, Kielgast J, Iversen LL, et al. Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology. 2012a;21:2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x. [DOI] [PubMed] [Google Scholar]
- Thomsen PF, Kielgast J, Iversen LL, Moller PR, Rasmussen M, Willerslev E. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE. 2012b;7:e41732. doi: 10.1371/journal.pone.0041732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CR, Barnes MA, Xu CCY, Jones SE, Jerde CL, Lodge DM. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods in Ecology and Evolution. 2014 , doi: 10.1111/2041-210X.12206, in press. [Google Scholar]
- Wilcox TM, McKelvey KS, Young MK, et al. Robust detection of rare species using environmental DNA: the importance of primer specificity. PLoS ONE. 2013;8:e59520. doi: 10.1371/journal.pone.0059520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Grunwald C, Stabile J, Waldman JR. Delineation of discrete population segments of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequence analysis. Conservation Genetics. 2010;11:689–708. doi: 10.1046/j.1365-294x.2002.01575.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 DNA copy numbers from qPCRs for all four experiments.
Fig. S1 A total of 40-250 mL water samples were each filtered through single PCTE filters, 10 each for four different pore sizes: 1, 3, 8 and 20 µm.
Data Availability Statement
DNA copy numbers from qPCR plates are available as Data S1 (Supporting information).


