SUMMARY
Decreased human immunodeficiency virus (HIV)-specific CD8+ T cell proliferation is a hallmark of chronic infection, but the mechanisms of decline are unclear. We analyzed gene expression profiles from antigen-stimulated HIV-specific CD8+ T cells from patients with controlled and uncontrolled infection and identified caspase-8 as a correlate of dysfunctional CD8+ T cell proliferation. Caspase-8 activity was upregulated in HIV-specific CD8+ T cells from progressors and correlated positively with disease progression and programmed cell death-1 (PD-1) expression, but negatively with proliferation. In addition, progressor cells displayed a decreased ability to upregulate membrane-associated caspase-8 activity and increased necrotic cell death following antigenic stimulation, implicating the programmed cell death pathway necroptosis. In vitro necroptosis blockade rescued HIV-specific CD8+ T cell proliferation in progressors, as did silencing of necroptosis mediator RIPK3. Thus, chronic stimulation leading to upregulated caspase-8 activity contributes to dysfunctional HIV-specific CD8+ T cell proliferation through activation of necroptosis and increased cell death.
INTRODUCTION
HIV-specific CD8+ T cells have been strongly implicated in viral control, particularly in individuals who naturally suppress viremia to undetectable levels known as ‘elite controllers’ (EC) (Walker et al., 2013). While numerous qualitative features of CD8+ T cells have been identified in controllers, such as increased polyfunctionality and elevated expression of cytotoxic effector molecules, their robust proliferation in response to antigenic stimulation is among the most important correlates of immune control and delayed disease progression (Migueles et al., 2002; Lichterfeld et al., 2004; Betts et al, 2006; Day et al., 2007; Migueles et al., 2008; McKinnon et al., 2012). Recent data utilizing a high-dimensional immune monitoring model demonstrated that HIV-specific CD8+ T cell proliferation was the strongest single determinant of spontaneous viral control (Ndhlovu et al., 2013), while previous work illustrated that proliferative capacity is directly linked to the enhanced cytotoxicity observed in ex vivo inhibition assays (Migueles et al., 2002; Migueles et al., 2008).
In the vast majority of individuals however, chronic infection leads to a decline in HIV-specific CD8+ T cell proliferative capacity due to the development of intrinsic CD8+ T cell defects (Migueles et al., 2002; Day et al., 2007; Wherry et al., 2011). HIV-specific CD8+ T cells from chronic progressors (CP) exhibit elevated expression of inhibitory immunoregulatory receptors, such as programmed cell death-1 (PD-1), 2B4 and T-cell immunoglobulin domain and mucin domain-3 (Tim-3) (Day et al., 2006; Trautmann et al., 2006; Yamamoto et al., 2011; Pacheco et al., 2013), resulting in T cell exhaustion and reduced proliferative capacity. Recent work has also highlighted the role of transcription factors such as basic leucine transcription factor, ATF-like (BATF) and nuclear factor of activated T cells (NFAT) in mediating impaired proliferative states (Migueles et al., 2008; Quigley et al., 2010). While proliferation can be partially restored through blockade of PD-1 (Day et al., 2006) or by increased nuclear translocation of NFAT (Migueles et al., 2008), the molecular mechanisms that govern this diminished response are poorly understood. Thus, we sought to further characterize the molecular pathways that differentiate the highly proliferative responses in ECs from the dysfunctional HIV-specific CD8+ T cell responses that develop in CPs.
To accomplish this, we performed whole genome transcriptional profiling on HIV tetramer+ CD8+ T cells from patients with controlled and uncontrolled infection following stimulation with cognate antigen. We sorted HIV-specific CD8+ T cells from 4 HLA-B*2705+ ECs and 4 HLA-B*2705+ CPs, in the presence and absence of the immunodominant B*2705-restricted Gag p24 KK10 (amino acids 263–272) epitope. Gene expression analysis revealed a unique set of genes that were differentially expressed in KK10-specific EC and CP CD8+ T cells following 6-day peptide stimulation. Further refinement of this list using an in silico approach (DAPPLE) (Rossin et al., 2011) revealed genes directly involved in protein-protein network connectivity, which included the aspartate-specific cysteine protease caspase-8. While caspase-8 has primarily been associated with activation-induced cell death (AICD) mediated by Fas/CD95 (Green et al., 2003; Wilson et al., 2009), it also plays a non-redundant role in CD8+ T cell proliferation by suppressing the endogenous necrotic cell death pathway necroptosis following TCR activation (Salmena et al., 2003; Leverrier et al., 2011; Ch’en et al., 2011). These distinct roles for caspase-8 are regulated by cellular compartmentalization, with membrane-associated caspase-8 (in association with c-FLIPL) playing a key role in inhibiting the formation of the receptor-interacting protein kinase-1/3 complex (RIPK1, RIPK3) that mediates necroptosis (Misra et al., 2007, Cho et al., 2009; Oberst et al., 2011; Koenig et al., 2014).
In our study, CP HIV-specific CD8+ T cells had elevated levels of caspase-8 activity, which was predominantly localized in the cytoplasm and correlated strongly with impaired proliferation. CP HIV-specific CD8+ T cells also displayed decreased upregulation of membrane-associated caspase-8 activity following T cell receptor (TCR) stimulation, and consequently an increase in necrotic cell death. Blockade of necroptosis by chemical inhibition reversed the proliferative defect, which was further validated by the improvement in proliferation following shRNA-mediated silencing of RIPK3. Collectively, these data suggest that necroptosis plays a previously undefined role in mediating dysfunctional HIV-specific CD8+ T cell proliferation in chronic HIV infection.
RESULTS
Transcriptional profiling differentiates peptide-stimulated HIV-specific CD8+ T cells from ECs and CPs
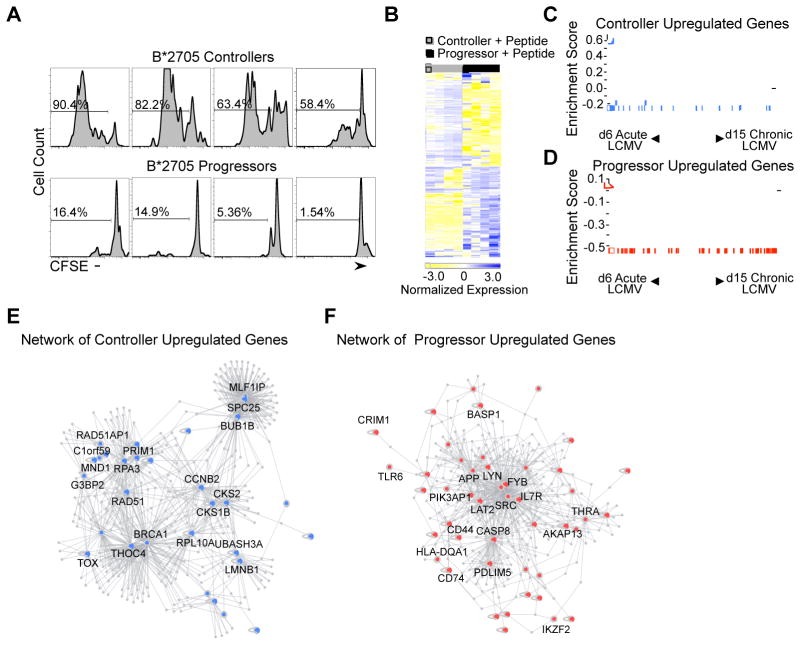
To elucidate the mechanisms underlying functional and dysfunctional HIV-specific CD8+ T cell proliferation, we focused on CD8+ T cells specific for the HLA-B*2705-restricted Gag p24 KK10 epitope within a group of B*2705 ECs (n = 4) and CPs (n = 4; Table S1). We selected this epitope because of its strong association with the protective features of HLA-B*2705 (Goulder et al., 1997; Kelleher et al., 2001), its high immunodominance (Streeck et al., 2009), and the marked difference in KK10 tetramer+ CD8+ T cell proliferation following 6d peptide stimulation within our two patient groups (P = 0.029; Figure 1A). Given these striking differences, we chose to further analyze these patients’ responses by performing transcriptional profiling following 6d KK10 peptide stimulation on three sorted populations: 1) peptide-stimulated KK10 tetramer+ CD8+ T cells, 2) non-peptide stimulated KK10 tetramer+ CD8+ T cells and 3) peptide-stimulated bulk CD8+ T cells (Figure S1B).
Figure 1. Transcriptional profiling analysis of peptide-stimulated HIV-specific CD8+ T cells from ECs and CPs.
(A) Histograms of gated viable CD8+ KK10-specific tetramer+ lymphocytes in B*2705 controllers and CPs stimulated with KK10 peptide (10 ng/mL). Numbers over bracketed lines indicate the percentages of the gated population (KK10 Tetramer+ CD8+ T cells) that have undergone at least one cell division after 6 days in culture. (B) The top differentially expressed genes in peptide-stimulated controller and CP KK10-specific CD8+ T cells are shown. Each column represents an individual sample and each row an individual gene, colored to indicate normalized expression (blue = increased expression, yellow = decreased expression). (C, D) Gene set enrichment analysis of upregulated gene sets in ECs and CPs with Day 6 LCMV Arm-specific CD8+ T cells (Acute LCMV) and Day 15 LCMV Cl13-specific CD8+ T cells (Chronic LCMV). The upregulated genes in HIV-specific CD8+ T cells from ECs were strongly enriched in the Acute LCMV Armstrong gene set (FDR q-value < 0.25). The upregulated genes in HIV-specific CD8+ T cells from CPs were strongly enriched in the Chronic LCMV Clone 13 gene set (FDR q-value < 0.25). The vertical blue and red bars indicate the individual genes that are enriched in both gene sets. (E, F) Direct and indirect DAPPLE networks built from upregulated gene expression sets from peptide-stimulated ECs (blue) and CPs (red) using known high-confidence pairwise protein-protein interactions (Rossin et al., 2011; Lage et al., 2008). See also Supplemental Table 2 and 3.
Analysis of differentially expressed genes between peptide-stimulated and non-peptide stimulated KK10 tetramer+ CD8+ T cells revealed a strong effector phenotype in controllers (n = 284 genes; Table S2; Figure S2A), while progressors were enriched for interferon-related genes and zinc finger transcription factors, which is characteristic of a persistently activated state (n = 92 genes; Table S2) (Wherry et al., 2007). However, to preferentially identify those genes involved in functional and dysfunctional CD8+ T cell proliferation, we focused on transcripts that differentiated EC and CP peptide-stimulated KK10 tetramer+ CD8+ T cells (n = 136 genes; Table S2; Figure 1B). This comparison revealed that proliferating EC KK10 tetramer+ CD8+ T cells were enriched for genes involved in mitosis, cell cycle and DNA double strand break repair, while CP cells were enriched for genes involved in T lymphocyte homeostasis and constitutive T cell activation (Table S2, S3).
We next compared these results in humans to the well-defined murine lymphocytic choriomeningitis virus (LCMV) model (Doering et al., 2012) during infection with two strains of LCMV: LCMV Armstrong (Arm), which results in an acute infection, and LCMV Clone 13 (Cl13), which results in a persistent infection. Transcriptional profiles of CD8+ T cells specific for the LCMV epitope DbGP33 during LCMV Arm infection were noted to be highly similar to the genetic signature of EC cells. These included genes involved in cell cycle, mitosis, DNA replication and DNA repair. In addition, genes upregulated in GP33-specific CD8+ T cells isolated during LCMV Cl13 infection were enriched in the CP gene set. These included markers of activation (CD44, CD74, LAT2), regulators of T cell homeostasis (CASP8, IL7R) and transcription factors (IKZF2). Furthermore, the CP gene signature contained genes involved in constitutive T cell activation (SRC, LYN and FYB), which are rarely expressed in T cells except during pathogenic states such as leukemia and lymphoma (Weil et al., 1999). This was similar to the transcriptional phenotype of LCMV Cl13 GP33-specific CD8+ T cells, which lacked a subset of quiescence genes allowing them to disarm activation and return to homeostasis (Doering et al., 2012).
To rigorously test these associations, we performed gene set enrichment analyses (GSEA) (Subramanian et al., 2005) between our EC and CP gene sets and the transcriptional profiles of LCMV Arm and Cl13 GP33-specific CD8+ T cells. Gene expression signatures of GP33-specific CD8+ T cells were available at a number of time points (d6, d8, d15 and d30). We therefore performed GSEAs between all permutations of d6-d30 LCMV Arm and Cl13 gene sets and our peptide-stimulated EC and CP KK10 tetramer+ CD8+ T cell transcriptional profiles. We found the strongest enrichments between the d6 LCMV Arm and our EC signature, and between the d15 LCMV Cl13 and our CP signature (FDR q-value < 0.25; Figures 1C,D). These data showed a likeness between the transcriptional profiles of HIV-specific and LCMV-specific CD8+ T cells during chronic infection (Quigley et al., 2010), demonstrating the similarity in the genetic signatures of LCMV Arm-specific and antigen-stimulated EC HIV-specific CD8+ T cells, while also confirming the transcriptional differences between peptide-stimulated EC and CP cells.
Network analysis identifies key upregulated genes in HIV-specific CD8+ T cells
To further evaluate our differential gene list, we made use of an in silico network analysis platform (DAPPLE) (Rossin et al., 2011) that examines the protein-protein connectivity of a gene list as defined by InWeb (Lage et al., 2007). We performed distinct analyses on the upregulated genes in the EC and CP signatures, which revealed that each gene set exhibited significantly greater direct connectivity than would be expected by chance (EC upregulated genes, P < 0.001; CP upregulated genes, P < 0.05; Figure 1E,F). Genes involved in each direct network, which were identified as primary drivers of overall network connectivity, revealed 16 candidate genes in the EC network (SPC25, MLF1IP, BUB1B, RAD51, RAD51AP1, C1orf59, BRCA1, RPA3, PRIM1, THOC4, MND1, CKS2, CKS1B, CCNB2, LMNB1 and UBASH3A) and 11 candidate genes in the CP network (APP, CASP8, SRC, FYB, LYN, IL-7R, AKAP13, THRA, CD44, CD74 and HLA-DQA1) (Table S3). For our analysis, we focused on genes potentially involved in dysfunctional HIV-specific CD8+ T cell proliferation. We therefore selected caspase-8 (Casp8) for further evaluation given the aforementioned DAPPLE analysis, its known role in regulating T cell homeostasis (Salmena et al., 2003) and also its involvement in 7 of the 10 most significant CP gene ontology pathways (Figure S2B).
Caspase-8 activity is associated with HIV disease progression
Caspase-8, an aspartate-specific cysteine protease, has a well-established role in apoptosis following activation of death receptors, such as Fas (Wilson et al., 2009). However, it also plays an essential role in antigen-induced T cell proliferation by suppressing necroptosis at the cell membrane (Salmena et al., 2003; Leverrier et al., 2011; Ch’en et al., 2011). To define the role of caspase-8 in dysfunctional HIV-specific CD8+ T cell proliferation, we first confirmed differences in caspase-8 gene expression by qRT-PCR within sorted peptide-stimulated HIV tetramer+ CD8+ T cells from a group of ECs (n = 7) and CPs (n = 7) distinct from those used in our transcriptional profiling studies. Measurements of caspase-8 transcript expression were carried out relative to GAPDH expression and revealed significantly higher Casp8/GAPDH expression in peptide-stimulated CP tetramer+ CD8+ T cells in comparison to peptide-stimulated EC cells (P = 0.027; Figure 2A), which was consistent with our transcriptional profiling data (Table S2).
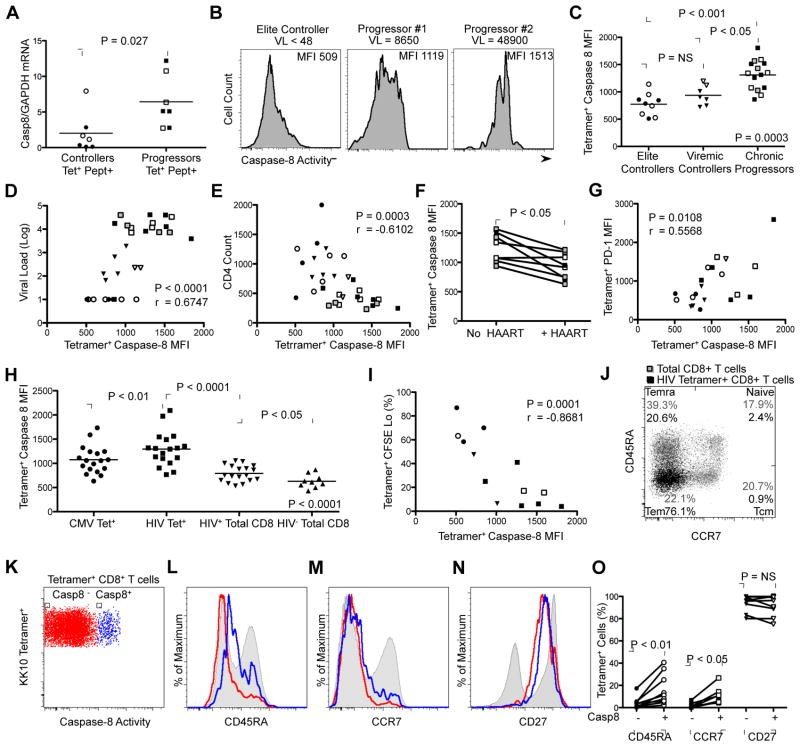
Figure 2. Caspase-8 activity is upregulated in HIV-specific CD8+ T cells and is associated with disease progression.
(A) Real-time qPCR measurement of caspase-8 expression relative to GAPDH expression from sorted peptide-stimulated HIV-specific Tetramer+ CD8+ T cells from ECs (n = 7) and CPs (n = 7). Filled symbols represent HLA-B*2705 KK10 tetramer+ responses and open symbols represent HLA-B*5701 KF11 tetramer+ responses. Relative expression was calculated using the 2−ΔΔ Ct method (Livak et al., 2001). Statistical analysis was made using the Mann-Whitney test. (B) Representative caspase-8 mean fluorescence intensity (MFI) values of KK10-specific CD8+ T cells in three HLA-B*2705 HIV+ patients with diverse viral loads. (C) Relative mean fluorescence intensity (MFI) of caspase-8 activity on HIV-specific CD8+ T cells from ECs (n = 9), VC (n = 7) and CPs (n = 15). Filled symbols represent HLA-B*2705 KK10-specific response, open symbols represent HLA-B*5701 KF11-specific responses and gray symbols represent non-HLA B*2705/non-HLA B*5701 responses. Horizontal bars indicate mean MFI of caspase-8 activity. Kruskal-Wallis test was used for comparison among all groups of subjects; the Dunns post-test was used for comparisons between groups. (D) Positive correlation between MFI of caspase-8 activity of HIV-specific CD8+ T cells and viral load (n = 31). (E) Negative correlation between MFI of caspase-8 activity of HIV-specific CD8+ T cells and CD4+ T cell count (n = 31). (F) Caspase-8 activity decreases in HIV-specific CD8+ T cells following initiation of HAART (n = 9). Statistical comparisons were made using the Wilcoxon matched pairs test. (G) Positive correlation between MFI of caspase-8 activity and MFI of PD-1 expression on HIV-specific CD8+ T cells (n = 21). (H) Mean fluorescence intensity of caspase-8 activity of A*0201 SL9-specific CD8+ T cells compared with A*0201 CMV pp65 NV9-specific CD8+ T cells and total CD8+ T cells in patients naïve from anti-retroviral therapy (n = 18), and total CD8+ T cells from A*0201 HIV-seronegative controls (n = 9). Repeated measures analysis of variance was used for comparison CMV Tet+, HIV Tet+ and HIV Total CD8+ given that these were paired observations. Mann-Whitney test was used for comparison of HIV+ and HIV− Total CD8+. (I) Negative correlation between MFI of caspase-8 activity of HIV-specific CD8+ T cells and percentage of CFSE lo Tetramer+ cells following peptide stimulation (n = 13). Correlation statistics for D, E, G and I were calculated using the Spearman correlation. (J) Representative CCR7 and CD45RA staining of total CD8+ T cells (gray) and HIV tetramer+ CD8+ T cells (black) from an HIV+ CP to identify naïve, central memory (Tcm), effector memory (Tem) and terminal effector (Temra) cells. (K) Representative FACS plot of KK10 tetramer+ CD8+ T cells identifying caspase-8 negative (Casp8-; red) and positive (Casp8+; blue) cells. (L, M, N) Representative histograms of CD45RA, CCR7 and CD27 staining of KK10 tetramer+ Casp8− CD8+ T cells (red), KK10 tetramer+ Casp8+ CD8+ T cells (blue) and total CD8+ T cells (gray). (O) Summary of phenotypic data for HIV tetramer+ Casp8− and Casp8+ CD8+ T cells analyzed for CD45RA, CCR7 and CD27 (n = 10). Horizontal bars indicate mean percentage of HIV tetramer+ Casp8− and Casp8+ CD8+ T cells that were positive for the indicated marker. Statistical comparisons were made by the Wilcoxon matched pairs test.
We next measured the mean fluorescence intensity (MFI) of caspase-8 activity in HIV tetramer+ CD8+ T cells directed against four immunodominant epitopes: HLA-B*2705 KK10, HLA-B*5701-restricted Gag p24 KF11 (amino acids 162–172), HLA-A*0201-restricted Gag p17 SL9 (amino acids 77–85) and HLA-B*0801-restricted Nef FL8 (amino acids 90–97) using the cell-permeable, fluorescently-labeled caspase-8 probe FAM-LETD-FMK, which selectively binds only to active caspase-8 molecules (Figures 2B, 2C) (Ekert et al., 1999). Within a group of 31 treatment-naïve HIV+ individuals, which consisted of ECs, ‘viremic controllers’ (VC; with viral loads between 50 and 2,000 RNA copies/mL) and CPs, we found that caspase-8 activity in HIV tetramer+ CD8+ T cells was significantly higher in CPs in comparison to both ECs (P < 0.001) and VCs (P < 0.05). Examination of HIV tetramer+ CD8+ T cell caspase-8 activity and markers of disease progression revealed a positive correlation between caspase-8 MFI and viral load (P < 0.0001; Figure 2D) and a negative correlation with CD4+ T cell count (P = 0.0003; Figure 2E). Analysis of the effect of highly active anti-retroviral therapy (HAART) on caspase-8 activity in nine CPs, for whom peripheral blood mononuclear cells (PBMCs) were available before and after the initiation of treatment, revealed a significant decrease in caspase-8 MFI similar to that observed in ECs and VCs, coincident with the decline in plasma viral load (P < 0.05; Figure 2F). Caspase-8 MFI of HIV tetramer+ CD8+ T cells was also significantly correlated with the MFI of PD-1 expression (P = 0.0108; Figure 2G), a known marker of chronic activation and T cell exhaustion (Day et al., 2006; Trautman et al., 2006). Collectively, these data indicate that high amounts of antigenemia during chronic infection leads to increased caspase-8 activity within ex vivo HIV-specific CD8+ T cells.
As a control for HIV-specific CD8+ T cells, we also measured the caspase-8 MFI in cytomegalovirus (CMV)-specific CD8+ T cells and total CD8+ T cell populations (Figure 2H). Analysis of 18 HLA-A*0201 CPs revealed significantly higher caspase-8 activities present in CD8+ T cells targeting the HLA-A*0201-restricted HIV SL9 epitope in comparison to those directed against the HLA-A*0201-restricted CMV-specific epitope NV9 (amino acids 495–503; P < 0.01) and the total CD8+ T cell population (P < 0.0001). In addition, caspase-8 activity in both HIV tetramer+ CD8+ T cells and the total CD8+ T cell population of HIV+ patients was significantly higher than in CD8+ T cells from seronegative controls (P < 0.05), further indicating a relationship between antigenic exposure and caspase-8 activity.
Increased caspase-8 activity is associated with impaired HIV-specific CD8+ T cell proliferation
We next assessed the relationship between caspase-8 activity and proliferative capacity in HIV tetramer+ CD8+ T cells (Figure 2I). Within a group of 13 individuals, we found a significant inverse correlation between the percentage of cells that had undergone at least one cell division (Tetramer+ CFSE Lo) and caspase-8 MFI (P = 0.0001). We further characterized the Casp8+ HIV tetramer+ CD8+ T cell pool using a panel of phenotypic markers associated with CD8+ T cell memory/effector status (Figures 2J and 2L–2N). Similar to previous work (Hess et al., 2004), the total HIV tetramer+ CD8+ T cell population exhibited an effector/effector memory phenotype characterized by low expression of CD45RA and CCR7, and high expression of CD27 (Figure 2J). Analysis of Casp8+ HIV tetramer+ CD8+ T cells however, revealed significantly higher expression of CD45RA (P < 0.01) and CCR7 (P < 0.05) relative to Casp8 negative (Casp8−) cells (Figures 2L–2O). The increased percentage of CD45RA+ CCR7− CD27+ within the Casp8+ population was consistent with an effector memory/terminal effector memory phenotype (Temra), further associating elevated caspase-8 activity with persistent activation and diminished CD8+ T cell proliferative potential. The modest, but significant elevation in CCR7 expression suggested that memory CD8+ T cells, which have been shown to express low amounts of the caspase-8-inducing cell surface marker CD95 (Gattinoni et al., 2011), likely also contribute to the Casp8+ HIV tetramer+ CD8+ T cell pool.
Antigenic stimulation modulates expression and spatial localization of caspase-8
Stimulation of the TCR leads to rapid upregulation and translocation of caspase-8 activity to the plasma membrane (Koenig et al., 2008; Misra et al., 2007), which is essential to CD8+ T cell proliferation through suppression of necroptosis. Thus, we examined whether proliferative differences between EC and CP HIV-specific CD8+ T cells could be attributed to their differential ability to upregulate membrane-associated caspase-8 activity following antigenic stimulation. We established our experimental approach using a highly active KK10-specific CD8+ T cell clone from an HLA-B*2705 EC. Incubation of the clone with an anti-CD3 antibody for 30 min resulted in increased caspase-8 activity (Figure S3A). Furthermore, stimulation of the clone with immobilized anti-CD3 and anti-CD28 antibodies resulted in upregulation and translocation of active caspase-8 molecules to the plasma membrane, as determined by confocal microscopy and co-localization with the membrane dye FM4-64 (P < 0.01; Figure S3B,C). Using the same approach, peptide stimulation of EC HIV tetramer+ CD8+ T cells led to a significant increase in caspase-8 activity (P < 0.05), while peptide-stimulation of CP cells resulted in a significant decrease in caspase-8 activity (P < 0.05; Figure 3A,B).
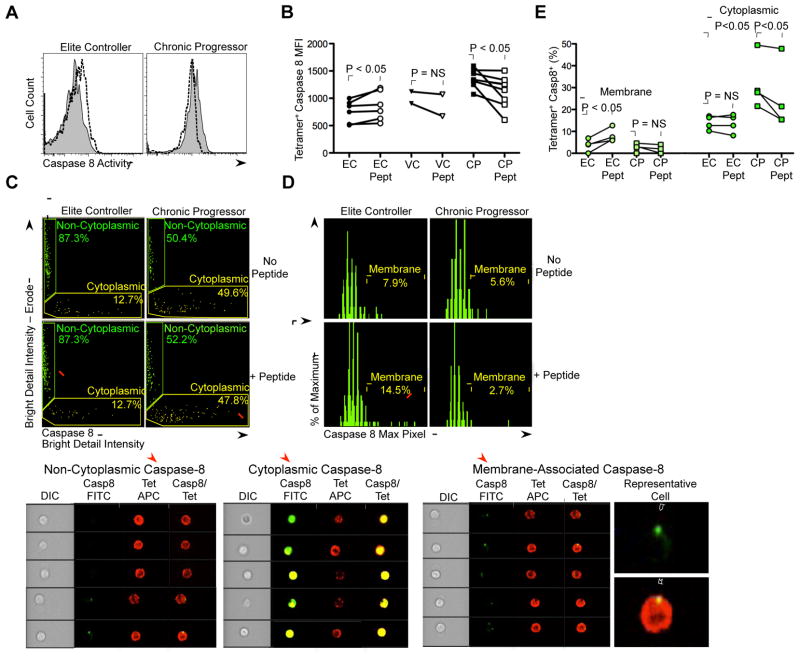
Figure 3. Effect of peptide stimulation on caspase-8 activity and localization in HIV-specific CD8+ T cells.
(A) Representative data showing modulation in MFI of caspase-8 activity following 1h peptide stimulation (1 ug/mL) in KK10-specific CD8+ T cell responses from an HLA-B*2705 EC and CP. Filled plot (gray) represents caspase-8 MFI in the absence of KK10 peptide and dashed line represents caspase-8 MFI in the presence of KK10 peptide. (B) Summary data of change in caspase-8 activity following peptide stimulation in ECs (n = 6), VC (n = 2) and CPs (n = 8). Filled symbols represent HLA-B*2705 KK10-specific responses and open symbols represent HLA-B*5701 KF11-specific responses. Statistical comparisons were made using the Wilcoxon matched pairs test. (C) Representative plots of Imagestream analysis of KK10 Tetramer+ CD8+ T cells from an HLA-B*2705 EC and CP in the presence and absence of KK10 peptide (1 μg/mL). The Imagestream plots depict total cellular caspase-8 activity on the x-axis (Caspase-8 Bright Detail Intensity; Cytoplasmic, yellow) and caspase-8 activity localized in a circular ring located around the edge of the cell on the y-axis (Caspase-8 Bright Detail Intensity – Erode; Non-cytoplasmic, green). Representative images corresponding to non-cytoplasmic and cytoplasmic caspase-8 cells are shown in the lower panels. (D) Representative histograms of Imagestream analysis of non-cytoplasmic caspase-8 KK10 Tetramer+ CD8+ T cells. The histograms depict the maximum pixels of caspase-8 activity on the x-axis, which successfully differentiates caspase-8+ KK10 Tetramer+ CD8+ T cells with membrane-associated caspase-8 activity from those with negative caspase-8 activity. Representative images of membrane-associated caspase-8 positive cells are shown in the lower panels. (E) Summary of percentage of membrane-associated caspase-8 and cytosolic caspase-8 positive KK10-specific CD8+ T cells within a group of ECs (n = 4) and CPs (n = 4). Statistical analyses were carried out using the paired t test (intra-patient group comparison, −/+ peptide) and the Mann-Whitney test (inter-patient group comparison). See also Supplemental Figure 3.
To assess the localization of caspase-8 activity within HIV tetramer+ CD8+ T cells, we used a technique that integrates quantitative image analysis with flow cytometry into a single platform (Imagestream) (Figures 3C, 3D). We found that CPs had a higher percentage of HIV tetramer+ CD8+ T cells with cytoplasmic caspase-8 activity (P < 0.05), but also a decrease in this cell population following peptide stimulation (P < 0.05; Figure 3E). In contrast, EC HIV tetramer+ CD8+ T cells were found to have a significant increase in membrane-associated caspase-8 activity following peptide stimulation (P < 0.05; Figure 3E). Collectively, these data suggest that HIV-specific CD8+ T cells from ECs and CPs can be differentiated by their ability and inability, respectively, to upregulate and translocate caspase-8 to the plasma membrane following antigenic stimulation.
Antigenic stimulation induces necrosis of HIV-specific CD8+ T cells
Given the decreased ability of CP HIV-specific CD8+ T cells to upregulate membrane-associated caspase-8, we hypothesized that their reduced proliferative capacity could be due to the activation of necroptosis following TCR engagement. Thus, we evaluated the responses of 10 HLA-B*2705 and HLA-B*5701 ECs and 10 HLA-B*2705 and HLA-B*5701 CPs. We found that 6d cognate peptide stimulation of ECs resulted in a 3.8-fold increase in HIV tetramer+ CD8+ T cells (P < 0.01; Figures 4A,C). However, the same conditions led to a 1.4-fold decrease in the frequency of tetramer+ CD8+ T cells in CPs (P = 0.0015). These data indicate that antigenic stimulation of CP HIV-specific CD8+ T cells results in significant cell loss.
Figure 4. Effect of peptide stimulation on HIV-specific CD8+ T cell expansion and induction of cellular necrosis.
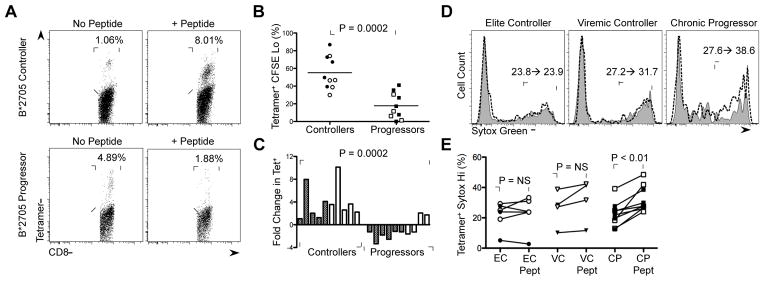
(A) Representative data of change in percentage of KK10-specific CD8+ T cells upon peptide stimulation in a HLA-B*2705 EC and CP. (B) Percentage of CFSE lo Tetramer+ CD8+ T cells in HLA-B*2705 KK10-specific (filled) and HLA-B*5701 KF11-specifc (open) CD8+ T cell responses in ECs and CPs. Statistical comparisons were made by the Mann-Whitney test. (C) Summary data of fold-change in Tetramer+ CD8+ T cells upon peptide stimulation in HLA-B*2705 KK10-specific (filled) and HLA-B*5701 KF11-specific (open) CD8+ T cell responses in ECs and CPs presented in (B). Statistical comparisons were made by the Mann-Whitney test. (D) Representative data of change in percentage of Sytox green Hi KK10-specific CD8+ T cells upon peptide stimulation in an HLA-B*2705 EC, viremic controller and CP. Filled plot (gray) represents Sytox green staining in the absence of KK10 peptide and dashed line represents Sytox green staining in the presence of KK10 peptide. (E) Summary data of change in percentage of Sytox green Hi HIV-specific CD8+ T cells following peptide stimulation in ECs (n = 6), VC (n = 4) and CPs (n = 8). Statistical comparisons were made using the Wilcoxon matched pairs test. See also Supplemental Figure 4.
To evaluate the characteristics of HIV tetramer+ CD8+ T cell loss, we utilized the necrosis-specific dye Sytox green (Vanden Berghe et al., 2010). Analysis of the live culture from 20 HIV+ individuals (Figure S4A), using standard forward scatter and side scatter parameters, revealed no significant difference in the percentage of Sytox green Hi cells between patient groups (Figures S4B, S4C). However, when the entire culture was analyzed (Figures S4A–S4C), significant increases in the percentage of Sytox green Hi HIV tetramer+ CD8+ T cells were appreciated in peptide-stimulated CP samples (P < 0.01), but not in EC or VC samples (Figures 4D,E). We therefore assessed apoptotic cell death by quantifying the MFI of executioner caspase-3 activity in the presence and absence of cognate peptide (Figure S5) and observed a significant increase in caspase-3 activity in peptide-stimulated HIV tetramer+ CD8+ T cells from ECs, but not CPs (P < 0.01; Figures S5A, S5B), which is consistent with its previously defined non-apoptotic role as an early activation marker during CD8+ T cell expansion (Alam et al., 1999; McComb et al., 2010).
Blockade of necroptosis reverses defective HIV-specific CD8+ T cell proliferation
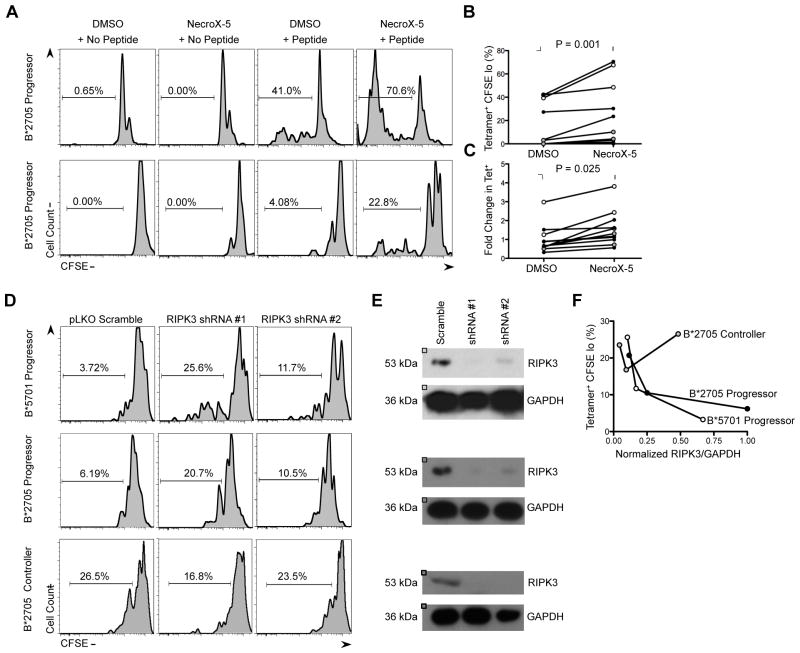
We next examined the role of necroptosis in defective HIV-specific CD8 T cell proliferation through utilization of necrosis inhibitor NecroX-5 (Kim et al., 2010), which has previously been shown to be effective in reversing RIPK1-RIPK3 induced necroptosis (Roca et al., 2013). CFSE-labeled PBMCs were treated with NecroX-5 or DMSO for 1h prior to 6d stimulation with cognate peptide, an example of which is shown for 2 HLA-B*2705 chronic progressors (Figure 5A). In these cases, stimulation with KK10 peptide resulted in a 1.7-fold and 5.5-fold increase in the percentage of HIV tetramer+ CFSE Lo cells following NecroX-5 pre-incubation. Similar assays were performed on a total of 12 chronic progressors, and a significant increase in the percentage of HIV tetramer+ CFSE Lo cells (P < 0.001) and the expansion of HIV tetramer+ CD8+ T cells (P = 0.025) was observed in the presence of peptide and NecroX-5, in comparison to the proliferation and expansion of HIV tetramer+ cells following stimulation with peptide and DMSO (Figures 5B,C).
Figure 5. Blockade of necroptosis restores HIV-specific CD8+ T cell proliferation.
(A) Representative data of increased proliferation of CFSE-loaded KK10 tetramer+ CD8+ T cells from two HLA-B*2705 CPs in the presence of NecroX-5 (1uM). (B) Summary data of change in proliferation of HIV tetramer+ CD8+ T cells in the presence of NecroX-5 in CPs (n = 12). Filled symbols represent HLA-B*2705 KK10 tetramer+ esponses and open symbols represent HLA-B*5701 KF11 tetramer+ responses. Statistical analyses were made using the Wilcoxon matched pairs test. (C) Summary data of expansion of HIV tetramer+ CD8 T cells in the presence of NecroX-5 in CPs (n = 12). Statistical analyses were made using the Wilcoxon matched pairs test. (D) Representative data of increased proliferation of CFSE-loaded KK10 tettramer+ CD8+ T cells from an HLA-B*5701 CP, an HLA-B*2705 CP and an HLA-B*2705 EC were transduced with lentiviral vectors encoding control shRNA (pLKO Scramble) or two sequence-independent shRNA constructs specific for necroptosis activating gene RIPK3 (RIPK3 shRNA #1 and #2). (E) Primary CD8+ T cells from the three patients in (D) were assessed for RIPK3 protein expression by western blotting following lentiviral transduction and 7d puromycin selection. GAPDH was used as a loading control. (F) Correlation between level of RIPK3 silencing with percentage of HIV tetramer+ CFSE lo CD8+ T cells in patients presented in (D). Expression of RIPK3 was normalized to GAPDH control.
To further verify the role of necroptosis in dysfunctional HIV-specific CD8+ T cell proliferation, we depleted the necroptosis mediator RIPK3 in primary CD8+ T cells using RNA interference (RNAi). Lentiviruses encoding two distinct short hairpin RNAs (shRNAs) against RIPK3 and a control shRNA (pLKO Scramble) were generated and used to transduce primary CD8+ T cells from a B*2705 CP, a B*5701 CP and a B*2705 EC. Lentiviral transduced CD8+ T cells exhibited >92% viability following puromycin selection. Silencing of RIPK3 expression was assessed by immunoblotting (Figure 5E). For assessment of HIV-specific CD8+ T cell proliferation, transduced CD8+ T cells were reconstituted with autologous non-CD8+ cells, labeled with CFSE and stimulated for 6d with cognate peptide. RIPK3 silencing resulted in a dose-dependent increase in the percentage of CFSE Lo HIV tetramer+ CD8+ T cells in both CP patients, however there was no effect in the EC patient (Figure 5F). These data collectively indicate that blockade of necroptosis, by either chemical or genetic means, results in an improvement in HIV-specific CD8+ T cell proliferation in patients with progressive disease.
DISCUSSION
While HIV-specific CD8+ T cell proliferation has been shown to be one of the strongest correlates of immune control in HIV infection (Ndhlovu et al., 2013), the cellular pathways that account for these impaired proliferative responses remain poorly understood. In this study, we utilized high-resolution gene expression profiling and functional immune assays on EC and CP HIV-specific CD8+ T cells to identify and characterize the roles of caspase-8 and necroptosis in dysfunctional CD8+ T cell proliferation. To control for differences between patient groups, such as the differential enrichment of protective and risk alleles, we focused our transcriptional profiling studies exclusively on CD8+ T cells specific for the well-characterized HLA-B*2705 Gag p24 KK10 epitope. Analysis of the transcriptional profiles of antigen-stimulated KK10 tetramer+ CD8+ T cells from ECs and CPs revealed the differential expression of caspase-8 mRNA, while GSEAs with LCMV-specific CD8+ T cells from acute and chronic infection validated the statistical reliability of these gene expression datasets. Application of the protein-protein network analysis platform DAPPLE, which has previously identified key players in complex disease states such as Rheumatoid Arthritis and Crohn’s disease (Rossin et al., 2011), further implicated the potential association between caspase-8 and defective HIV-specific CD8+ T cell proliferation.
We therefore validated the upregulation of caspase-8 mRNA in CP HIV tetramer+ CD8+ T cells by qRT-PCR and confirmed its functional significance through assessment of caspase-8 activity by flow cytometry. The localization of active caspase-8 within the cytoplasm of CP cells provided an explanation for the observed increase in caspase-8 transcripts, as caspase-8 gene expression is self-induced by cytoplasmic death effector domains (Yao et al., 2007). The significant correlations between the caspase-8 MFI in HIV tetramer+ CD8+ T cells and markers of disease progression (viral load, CD4+ T cell count) and T cell exhaustion (e.g. PD-1) suggested a strong relationship between the level of active caspase-8 in HIV-specific CD8+ T cells and the degree of antigenic exposure. This was further supported by the significant decrease in caspase-8 activity following the initiation of HAART. These data were consistent with observations from other chronic infections such as hepatitis B virus (HBV) and hepatitis C virus (HCV), which also have elevated levels of caspase-8 activity within their antigen-specific and bulk CD8+ T cell populations (Radziewicz et al., 2008; Arends et al., 2011; Peppa et al., 2013).
The negative correlation between the caspase-8 MFI in HIV tetramer+ CD8+ T cells and the percentage of CFSE Lo cells following 6d peptide stimulation was consistent with our CD8+ T cell phenotyping studies, which revealed an increase in the percentage of Temra cells (CD45RA+, CCR7−, CD27+) within the Casp8+ pool. In addition, the significant upregulation of membrane-associated caspase-8 activity following peptide stimulation in highly proliferative HIV tetramer+ CD8+ T cells from ECs was consistent with the essential role of caspase-8 in mediating CD8+ T cell proliferation (Salmena et al., 2003; Leverrier et al., 2011). This was in contrast to the elevated cytoplasmic caspase-8 activity in CP HIV tetramer+ CD8+ T cells and the inability of these cells to significantly upregulate membrane-associated caspase-8 activity following peptide stimulation. Given that membrane-associated caspase-8 has been shown to form a heterodimer with c-FLIP(L) to promote proliferation through nuclear factor-κ B (NF-κB) activation (Misra et al., 2007), this suggested that the proliferative defect of CP HIV tetramer+ CD8+ T cells could simply be due to decreased NF-κB translocation. However, the decrease in the percentage of cytoplasmic caspase-8+ HIV tetramer+ CD8+ T cells in CPs following peptide stimulation also suggested that TCR engagement likely triggers increased cell turnover, which we observed in an additional set of poorly proliferative CPs.
We initially presumed that the increased cell loss was due to excessive AICD (Green et al., 2003), in light of previous work demonstrating that CP HIV-specific CD8+ T cells have increased apoptotic effector caspase-3 activity and surface annexin V expression, and decreased amounts of anti-apoptotic molecule Bcl-2 (Petrovas et al., 2004; Yan et al., 2013). These notions however were not consistent with the lack of association between the amount of Fas-induced apoptosis in HIV-specific CD8+ T cells and either viral load or disease progression (Regamey et al., 1999; Mueller et al., 2001). In addition, we observed no significant difference in ex vivo caspase-3 activity between HIV tetramer+ CD8+ T cells and actually found that EC cells significantly increased caspase-3 activity following peptide stimulation. This result highlighted recent observations implicating caspase-3 and annexin V as early markers of CD8+ T cell expansion (Fischer et al., 2006; Alam et al., 1999; McComb et al., 2010) and suggested that the increased caspase-3 activity in CP cells could potentially be due to chronic antigenic stimulation. Furthermore, Bcl-2 expression has been shown to be a marker of memory CD8+ T cells (Dunkle et al., 2013), which may explain its increased expression in ECs (Yan et al., 2013), given their known enrichment for HIV-specific CD8+ T cells that display a strong central memory phenotype (Ndhlovu et al., 2013).
Thus, given the dual role of pro-apoptotic molecules in CD8+ T cell activation, we considered the potential contribution of the programmed cell death pathway necroptosis to HIV tetramer+ CD8+ T cell loss, which has been shown to mediate CD8+ T cell homeostasis when apoptosis is not completely effective (Osborn et al., 2010; Han et al., 2011). Necroptosis plays a critical role in regulating the baseline CD8+ T lymphocyte pool, as gene-knockout mice that lack necroptosis (Casp8−/− Ripk3−/−) are characterized by a phenotype of severe lymphadenopathy, splenomegaly and increased accumulation of aberrant B220+ CD3+ T lymphocytes (Oberst et al., 2011). Our results utilizing the necrosis-specific cell dye Sytox green demonstrated that CPs are enriched for necrotic HIV-specific CD8+ T cells following antigenic stimulation. The involvement of necroptosis was further confirmed by the significant improvement in HIV tetramer+ CD8+ T cell proliferation following pre-treatment with either necrosis inhibitor NecroX-5 or silencing of necroptosis-mediating enzyme RIPK3. These data revealed that necroptosis significantly contributes to the proliferative defect of CP HIV-specific CD8+ T cells, and suggests that chronic HIV infection may potentially drive the normal homeostatic function of necroptosis to a pathogenic state due to persistent antigenic stimulation. The result is a rate of cell turnover that cannot be compensated for by the generation of new virus-specific CD8+ T cells, and consequently a ‘net deficit’ in cell proliferation. Of note, experiments with necroptosis inhibitor Necrostatin-1 did not reproduce the results of NecroX-5, potentially due to its off-target inhibitory effects on the Erk and Jnk pathways that mediate T cell activation (Cho et al., 2011) and its effects on the indoleamone-2,3-dioxygenase (IDO)-kynurenine pathway, a known regulator of CD8+ T cell function (Liu et al., 2009). This effect was confirmed by the reduction in HIV-specific CD8+ T cell proliferation of an elite controller pre-treated with Necrostatin-1.
Collectively, our data suggest that chronic stimulation of HIV-specific CD8+ T cells leads to an increase in the basal level of caspase-8 activity within the cellular cytoplasm. However, given that processed cytoplasmic caspase-8 homodimers are structurally incapable of forming heterodimers with c-FLIP(L) (Boatright et al., 2004), the accumulation of caspase-8 in the cytoplasm may actually result in the depletion of available caspase-8 molecules capable of re-engaging c-FLIP(L) and driving proliferation. As shown in previous work, the pool of available full-length caspase-8 required for CD8+ T cell activation is in fact reduced in cells that have elevated levels of cytoplasmic caspase-8 due to increased caspase-8 proteolysis (Leverrier et al., 2011). Thus, we propose that CP HIV-specific CD8+ T cells actually exhibit a pseudo-caspase-8 deficiency phenotype, which is consistent with the striking similarity between CP cells and CD8+ T cells from patients with homozygous inactivating caspase-8 mutations (i.e. robust proliferation following stimulation with PMA and ionomycin, but poor activation following TCR stimulation) (Chun et al., 2002; Migueles et al., 2008).
In summary, this report is the first to identify caspase-8 and necroptosis as a novel correlate and key mediator, respectively, of dysfunctional HIV-specific CD8+ T cell proliferation in patients with progressive disease. Caspase-8 and necroptosis have already been implicated in the immunopathology of Crohn’s disease (Gunther et al., 2011), systemic inflammatory response syndrome (Duprez et al., 2011) and chronic lymphocytic leukemia (Liu et al., 2012). Thus, chronic HIV infection may represent another case where caspase-8 and necroptosis disrupt the finely tuned immune response, resulting in excessive HIV-specific CD8+ T cell loss and ineffective immune control. These findings have potential implications for additional states of persistent antigenemia, such as HBV and HCV infection, and malignancy. Ultimately, necroptosis may represent a novel therapeutic target to enhance T cell immunity by improving the proliferative potential of antigen-specific CD8+ T cell responses.
EXPERIMENTAL PROCEDURES
Study Subjects
Peripheral blood was obtained from HIV-infected people and HIV-negative subjects in Boston, Massachusetts, after institutional review board approval and written informed consent was obtained. Only cryopreserved PBMCs were used. All study subjects were HLA typed to four-digit resolution by molecular methods (Kiepiela et al., 2004). Untreated subjects and people with controlled viremia on antiviral therapy were enrolled. ‘Elite controllers’ were defined as people who spontaneously controlled viremia to below 50 RNA copies per mL plasma in the absence of therapy, and ‘viremic controllers’ were defined as subjects with a viral load of more than 50 and less than 2000 RNA copies per mL plasma in the absence of treatment. Plasma viral loads were measured by the HIV-1 Amplicor Monitor Ultrasensitive method (Roche).
Microarray Data Acquisition and Analysis
Sorted tetramer+ CD8+ T cells were pelleted, resuspended in Cell Lysis Buffer (Ambion) and immediately snap frozen in liquid nitrogen prior to storage at −80°C. RNA was extracted using the RNAqueous Micro Kit (AM1931; Ambion) according to the manufacturer’s instructions. Concentrations of total RNA were determined with a NanoDrop spectrophotometer. RNA purity was determined by Bioanalyzer 2100 traces (Agilent Technologies). Total RNA was amplified by the WT-Ovation Pico RNA Amplification system (NuGEN) according to the manufacturer’s directions. Whole-genome transcriptional profiling was performed using WG-DASL microarrays (Illumina) by the Core Facility at the Washington University School of Medicine (St Louis, MO). Data retrieved from the Illumina software were background corrected (Illumina beadstudio software) and quantile normalized using the Arraystar normalization function. Analysis was restricted to genes with significant expression (P < 0.01). Differentially expressed genes were detected using the empirical Bayes (eBayes) adjusted t test. Multiple testing was corrected with the Benjamini-Hochberg false discovery rate (FDR). Differentially expressed genes were defined as being at least 1.5-fold different after log2 transformation and having an FDR q-value of <0.05. To correct for batch effect differences between peptide-stimulated controller and progressor HIV tetramer+ CD8+ T cells, the ratio G method was utilized with bulk CD8+ T cells used as an internal control as described previously (Luo et al., 2010).
CFSE Proliferation Assay
PBMCs were first suspended at 106/ml in PBS and incubated at 37°C for 7 min with 0.5 μM carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes). After the addition of serum and washes with PBS, cells were suspended at 106/ml in medium (RPMI 1640 supplemented with glutamine, 10% human FCS, penicillin, and streptomycin). No exogenous cytokines were added to the medium. Individual HIV peptides were added at a final concentration of 10 ng/mL per peptide. On day 6, cells were harvested, washed with PBS, and stained with APC-labeled dextramers (Immunodex), mAbs (anti-CD3 Alexa 700, anti-CD8 APC Cy7; BD Biosciences), and viability dye (Violet; Molecular Probes). Cells were then washed and fixed in 1% paraformaldehyde and subjected to flow cytometric analysis. For reversal experiments, CFSE-labeled PBMCs were incubated with either NecroX-5 (1 uM; Enzo Life Sciences) or DMSO for 1h prior to stimulation with HIV peptide.
Statistical Analysis
The generation of dot plots, nonparametric statistical analysis, analyses of variance (Kruskal-Wallis, Repeated Measures) and correlations (Spearman) were performed using the statistical programs in Graphpad Prism V.5. Differences between groups were evaluated by using a Mann-Whitney t test or ANOVA as indicated. Paired analyses were performed using the Wilcoxon matched pairs rank test.
Supplementary Material
Figure S1, related to Figure 1 and Experimental Procedures: Flow cytometry gating strategy and sorting strategy of HIV-specific CD8+ T cells. (A) Whole PBMCs were stained with HIV tetramer, a viability dye and anti-CD8 antibody. Frequency of tetramer+ CD8+ T cells was determined by gating on viable CD8+ T cells. (B) PBMCs were incubated in the presence or absence of KK10 peptide (10 ng/mL) for 6 days. Cells were stained with B*2705 KK10-tetramer and anti-CD8 antibody and three cell subsets (Tetramer+ Peptide-Stimulated, Tetramer+ Non-Peptide Stimulated, and Bulk CD8 T cells) were isolated for the four B*2705 controller and four B*2705 progressors presented in Figure 1A. Total RNA was harvested for each cell subset (24 total) and analyzed by transcriptional profiling.
Figure S2, related to Figure 1: Differentially expressed genes and gene ontology analysis following peptide stimulation in HIV-specific CD8+ T cells from elite controllers and chronic progressors (A) The top 200 differentially expressed genes in peptide-stimulated and non-peptide stimulated KK10-specific CD8+ T cells in B*2705 controllers. Each column represents an individual sample and each row an individual gene, colored to indicate normalized expression (blue = increased expression, yellow = decreased expression). (B) Controller and progressor networks reveal a functional distinction between patient groups. We used GOrilla (http://cbl-gorilla.cs.technion.ac.il/) to calculate the overrepresented gene ontology terms (http://www.geneontology.org) within the controller and progressor differentially expressed and induced gene networks. A selected list of terms with p-value < 1 × 10−3 was visualized. The asterisk (*) denotes those GO terms within the progressor network in which caspase-8 was involved.
Figure S3, related to Figure 3: Active Caspase-8 upregulation and translocation upon TCR engagement in a KK10-specific CTL clone. (A) Histogram illustrating total MFI of Caspase-8 activity in KK10-specific CTL clones at 30 min stimulated with Isotype antibody (gray), anti-CD3 antibody (red) or anti-FAS antibody (blue). (B) KK10-specific CTL clones were imaged on Poly-L-Lysine coated coverslips at 30 min, and were stimulated with Isotype control antibody (upper panels) or anti-CD3 and anti-CD28 antibodies (lower panels). Active Caspase-8 (Green) and FM4-64 plasma membrane dye (Red) were acquired by confocal microscopy. Arrows illustrate active caspase-8 activity at the plasma membrane, indicating its translocation. (C) Quantitative measurement of MFI of active caspase-8 by confocal microscopy. The MFI of active caspase-8 per cell in the presence of anti-CD3 and anti-CD28 was increased as compared to isotype control stimulated CTLs. P-value was determined using unpaired t-test (two-tailed). (D) Schematic of Boolean approach to assess for cytoplasmic and membrane-associated caspase-8 activity utilized in Figure 3C–E.
Figure S4, related to Figure 4: Assessment of necrotic death by Sytox Green staining. (A) Representative plot of purified CD8+ T cells with gating of live and dead populations based on forward and side scatter properties. (B) CD8+ T cells were stimulated in the presence or absence of HIV peptide and stained with Sytox Green dye, in addition to tetramer and anti-CD8 antibody. As shown, gating on live cells results in relatively low levels of necrotic Sytox Hi cells. However, gating on the entire culture reveals a significant increase in the number of Sytox Hi cells. (C) Summary data of percentage of Sytox Hi cells in the live and entire cultures within each patient group.
Figure S5, related to Figure 4: Assessment of caspase-3 activity within peptide-stimulated HIV-specific CD8+ T cells from controllers and progressors. (A) Representative data showing modulation in MFI of caspase-3 activity following 3d peptide stimulation (1 ug/mL) in KK10-specific CD8+ T cell responses from an 2 HLA-B*2705 elite controllers and 2 HLA-B*2705 chronic progressors. Filled plot (gray) represents caspase-3 MFI in the absence of KK10 peptide and dashed line represents caspase-3 MFI in the presence of KK10 peptide. (B) Summary data of change in caspase-3 activity following peptide stimulation in controllers (n = 5) and chronic progressors (n = 5). Statistical comparisons were made using the Wilcoxon matched pairs test.
Table S1, related to Figure 1: Patient characteristics of HLA-B*2705+ elite controllers and chronic progressors selected for cell sorting and transcriptional profiling. Clinical and molecular features of the patients utilized for our transcriptional profiling studies.
Table S3, related to Figure 1: List of Direct Interacting Partners within the Controller and Progressor Networks as identified by DAPPLE Analysis. This table delineates the genes involved in the direct EC and CP networks which are involved in driving overall network connectivity.
Acknowledgments
We are grateful to Judy Lieberman and Jennifer Sims for comments on the manuscript and to Deanna Nguyen for assistance with experiments. This study was supported by funds from the Howard Hughes Medical Institute (B.D.W), the Ragon Institute (B.D.W) and NIH grants AI105343, AI112521, AI082630, AI095608, HHSN266200500030C (E.J.W).
Footnotes
Accession Numbers
Microarray data sets are available through the Gene Expression Omnibus (GSE56775, GSE56971).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam A, Cohen LY, Aouad S, Sekaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. The Journal of experimental medicine. 1999;190:1879–1890. doi: 10.1084/jem.190.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends JE, Hoepelman AI, Nanlohy NM, Hoppener FJ, Hirsch KR, Park JG, van Baarle D. Low doses of the novel caspase-inhibitor GS-9450 leads to lower caspase-3 and -8 expression on peripheral CD4+ and CD8+ T-cells. Apoptosis : an international journal on programmed cell death. 2011;16:959–966. doi: 10.1007/s10495-011-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS. Activation of caspases-8 and -10 by FLIP(L) The Biochemical journal. 2004;382:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. The Journal of experimental medicine. 2011;208:633–641. doi: 10.1084/jem.20110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, McQuade T, Zhang H, Zhang J, Chan FK. RIP1-dependent and independent effects of necrostatin-1 in necrosis and T cell activation. PloS one. 2011;6:e23209. doi: 10.1371/journal.pone.0023209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- Day CL, Kiepiela P, Leslie AJ, van der Stok M, Nair K, Ismail N, Honeyborne I, Crawford H, Coovadia HM, Goulder PJ, et al. Proliferative capacity of epitope-specific CD8 T-cell responses is inversely related to viral load in chronic human immunodeficiency virus type 1 infection. Journal of virology. 2007;81:434–438. doi: 10.1128/JVI.01754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle A, Dzhagalov I, Gordy C, He YW. Transfer of CD8+ T cell memory using Bcl-2 as a marker. Journal of immunology. 2013;190:940–947. doi: 10.4049/jimmunol.1103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T, Declercq W, Libert C, Cauwels A, Vandenabeele P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Ekert PG, Silke J, Vaux DL. Caspase inhibitors. Cell death and differentiation. 1999;6:1081–1086. doi: 10.1038/sj.cdd.4400594. [DOI] [PubMed] [Google Scholar]
- Fischer K, Voelkl S, Berger J, Andreesen R, Pomorski T, Mackensen A. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood. 2006;108:4094–4101. doi: 10.1182/blood-2006-03-011742. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. A human memory T cell subset with stem cell-like properties. Nature medicine. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nature medicine. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunological reviews. 2003;193:70–81. doi: 10.1034/j.1600-065x.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- Gunther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H, Waldner MJ, Hedrick SM, Tenzer S, Neurath MF, Becker C. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nature immunology. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- Hess C, Altfeld M, Thomas SY, Addo MM, Rosenberg ES, Allen TM, Draenert R, Eldrige RL, van Lunzen J, Stellbrink HJ, et al. HIV-1 specific CD8+ T cells with an effector phenotype and control of viral replication. Lancet. 2004;363:863–866. doi: 10.1016/S0140-6736(04)15735-8. [DOI] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. The Journal of experimental medicine. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Koo SY, Ahn BH, Park O, Park DH, Seo DO, Won JH, Yim HJ, Kwak HS, Park HS, et al. NecroX as a novel class of mitochondrial reactive oxygen species and ONOO(−) scavenger. Archives of pharmacal research. 2010;33:1813–1823. doi: 10.1007/s12272-010-1114-4. [DOI] [PubMed] [Google Scholar]
- Koenig A, Russell JQ, Rodgers WA, Budd RC. Spatial differences in active caspase-8 defines its role in T-cell activation versus cell death. Cell death and differentiation. 2008;15:1701–1711. doi: 10.1038/cdd.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig A, Buskiewicz IA, Fortner KA, Russell JQ, Asaoka T, He YW, Hakem R, Eriksson JE, Budd RC. The c-FLIPL cleavage product p43FLIP promotes activation of extracellular signal-regulated kinase (ERK), nuclear factor kappaB (NF-kappaB), and caspase-8 and T cell survival. The Journal of biological chemistry. 2014;289:1183–1191. doi: 10.1074/jbc.M113.506428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature biotechnology. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Leverrier S, Salvesen GS, Walsh CM. Enzymatically active single chain caspase-8 maintains T-cell survival during clonal expansion. Cell death and differentiation. 2011;18:90–98. doi: 10.1038/cdd.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. The Journal of experimental medicine. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu L, Liu K, Bizargity P, Hancock WW, Visner GA. Reduced cytotoxic function of effector CD8+ T cells is responsible for indoleamine 2,3-dioxygenase-dependent immune suppression. Journal of immunology. 2009;183:1022–1031. doi: 10.4049/jimmunol.0900408. [DOI] [PubMed] [Google Scholar]
- Liu P, Xu B, Shen W, Zhu H, Wu W, Fu Y, Chen H, Dong H, Zhu Y, Miao K, et al. Dysregulation of TNFalpha-induced necroptotic signaling in chronic lymphocytic leukemia: suppression of CYLD gene by LEF1. Leukemia. 2012;26:1293–1300. doi: 10.1038/leu.2011.357. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McComb S, Mulligan R, Sad S. Caspase-3 is transiently activated without cell death during early antigen driven expansion of CD8+ T cells in vivo. PloS one. 2010;5:e15328. doi: 10.1371/journal.pone.0015328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon LR, Kaul R, Kimani J, Nagelkerke NJ, Wachihi C, Fowke KR, Ball TB, Plummer FA. HIV-specific CD8+ T-cell proliferation is prospectively associated with delayed disease progression. Immunology and cell biology. 2012;90:346–351. doi: 10.1038/icb.2011.44. [DOI] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature immunology. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RS, Russell JQ, Koenig A, Hinshaw-Makepeace JA, Wen R, Wang D, Huo H, Littman DR, Ferch U, Ruland J, et al. Caspase-8 and c-FLIPL associate in lipid rafts with NF-kappaB adaptors during T cell activation. The Journal of biological chemistry. 2007;282:19365–19374. doi: 10.1074/jbc.M610610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller YM, De Rosa SC, Hutton JA, Witek J, Roederer M, Altman JD, Katsikis PD. Increased CD95/Fas-induced apoptosis of HIV-specific CD8(+) T cells. Immunity. 2001;15:871–882. doi: 10.1016/s1074-7613(01)00246-1. [DOI] [PubMed] [Google Scholar]
- Ndhlovu ZM, Chibnik LB, Proudfoot J, Vine S, McMullen A, Cesa K, Porichis F, Jones RB, Alvino DM, Hart MG, et al. High-dimensional immunomonitoring models of HIV-1-specific CD8 T-cell responses accurately identify subjects achieving spontaneous viral control. Blood. 2013;121:801–811. doi: 10.1182/blood-2012-06-436295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn SL, Diehl G, Han SJ, Xue L, Kurd N, Hsieh K, Cado D, Robey EA, Winoto A. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor-mediated necroptosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13034–13039. doi: 10.1073/pnas.1005997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco Y, McLean AP, Rohrbach J, Porichis F, Kaufmann DE, Kavanagh DG. Simultaneous TCR and CD244 signals induce dynamic downmodulation of CD244 on human antiviral T cells. Journal of immunology. 2013;191:2072–2081. doi: 10.4049/jimmunol.1300435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppa D, Gill US, Reynolds G, Easom NJ, Pallett LJ, Schurich A, Micco L, Nebbia G, Singh HD, Adams DH, et al. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. The Journal of experimental medicine. 2013;210:99–114. doi: 10.1084/jem.20121172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Mueller YM, Dimitriou ID, Bojczuk PM, Mounzer KC, Witek J, Altman JD, Katsikis PD. HIV-specific CD8+ T cells exhibit markedly reduced levels of Bcl-2 and Bcl-xL. Journal of immunology. 2004;172:4444–4453. doi: 10.4049/jimmunol.172.7.4444. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva NS. Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytometry. BMC research notes. 2011;4:517. doi: 10.1186/1756-0500-4-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nature medicine. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziewicz H, Ibegbu CC, Hon H, Osborn MK, Obideen K, Wehbi M, Freeman GJ, Lennox JL, Workowski KA, Hanson HL, Grakoui A. Impaired hepatitis C virus (HCV)-specific effector CD8+ T cells undergo massive apoptosis in the peripheral blood during acute HCV infection and in the liver during the chronic phase of infection. Journal of virology. 2008;82:9808–9822. doi: 10.1128/JVI.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regamey N, Harr T, Battegay M, Erb P. Downregulation of Bcl-2, but not of Bax or Bcl-x, is associated with T lymphocyte apoptosis in HIV infection and restored by antiretroviral therapy or by interleukin 2. AIDS research and human retroviruses. 1999;15:803–810. doi: 10.1089/088922299310700. [DOI] [PubMed] [Google Scholar]
- Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 2013;153:521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ International Inflammatory Bowel Disease Genetics C. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes & development. 2003;17:883–895. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H, Jolin JS, Qi Y, Yassine-Diab B, Johnson RC, Kwon DS, Addo MM, Brumme C, Routy JP, Little S, et al. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. Journal of virology. 2009;83:7641–7648. doi: 10.1128/JVI.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nature medicine. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Vanlangenakker N, Parthoens E, Deckers W, Devos M, Festjens N, Guerin CJ, Brunk UT, Declercq W, Vandenabeele P. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell death and differentiation. 2010;17:922–930. doi: 10.1038/cdd.2009.184. [DOI] [PubMed] [Google Scholar]
- Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nature reviews. Immunology. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- Weil R, Levraud JP, Dodon MD, Bessia C, Hazan U, Kourilsky P, Israel A. Altered expression of tyrosine kinases of the Src and Syk families in human T-cell leukemia virus type 1-infected T-cell lines. Journal of virology. 1999;73:3709–3717. doi: 10.1128/jvi.73.5.3709-3717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nature immunology. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nature immunology. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Price DA, Casazza JP, Ferrari G, Nason M, Chattopadhyay PK, Roederer M, Gostick E, Katsikis PD, Douek DC, et al. Surface expression patterns of negative regulatory molecules identify determinants of virus-specific CD8+ T-cell exhaustion in HIV infection. Blood. 2011;117:4805–4815. doi: 10.1182/blood-2010-11-317297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Sabbaj S, Bansal A, Amatya N, Shacka JJ, Goepfert PA, Heath SL. HIV-specific CD8+ T cells from elite controllers are primed for survival. Journal of virology. 2013;87:5170–5181. doi: 10.1128/JVI.02379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Duan S, Hou D, Heese K, Wu M. Death effector domain DEDa, a self-cleaved product of caspase-8/Mch5, translocates to the nucleus by binding to ERK1/2 and upregulates procaspase-8 expression via a p53-dependent mechanism. The EMBO journal. 2007;26:1068–1080. doi: 10.1038/sj.emboj.7601571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1 and Experimental Procedures: Flow cytometry gating strategy and sorting strategy of HIV-specific CD8+ T cells. (A) Whole PBMCs were stained with HIV tetramer, a viability dye and anti-CD8 antibody. Frequency of tetramer+ CD8+ T cells was determined by gating on viable CD8+ T cells. (B) PBMCs were incubated in the presence or absence of KK10 peptide (10 ng/mL) for 6 days. Cells were stained with B*2705 KK10-tetramer and anti-CD8 antibody and three cell subsets (Tetramer+ Peptide-Stimulated, Tetramer+ Non-Peptide Stimulated, and Bulk CD8 T cells) were isolated for the four B*2705 controller and four B*2705 progressors presented in Figure 1A. Total RNA was harvested for each cell subset (24 total) and analyzed by transcriptional profiling.
Figure S2, related to Figure 1: Differentially expressed genes and gene ontology analysis following peptide stimulation in HIV-specific CD8+ T cells from elite controllers and chronic progressors (A) The top 200 differentially expressed genes in peptide-stimulated and non-peptide stimulated KK10-specific CD8+ T cells in B*2705 controllers. Each column represents an individual sample and each row an individual gene, colored to indicate normalized expression (blue = increased expression, yellow = decreased expression). (B) Controller and progressor networks reveal a functional distinction between patient groups. We used GOrilla (http://cbl-gorilla.cs.technion.ac.il/) to calculate the overrepresented gene ontology terms (http://www.geneontology.org) within the controller and progressor differentially expressed and induced gene networks. A selected list of terms with p-value < 1 × 10−3 was visualized. The asterisk (*) denotes those GO terms within the progressor network in which caspase-8 was involved.
Figure S3, related to Figure 3: Active Caspase-8 upregulation and translocation upon TCR engagement in a KK10-specific CTL clone. (A) Histogram illustrating total MFI of Caspase-8 activity in KK10-specific CTL clones at 30 min stimulated with Isotype antibody (gray), anti-CD3 antibody (red) or anti-FAS antibody (blue). (B) KK10-specific CTL clones were imaged on Poly-L-Lysine coated coverslips at 30 min, and were stimulated with Isotype control antibody (upper panels) or anti-CD3 and anti-CD28 antibodies (lower panels). Active Caspase-8 (Green) and FM4-64 plasma membrane dye (Red) were acquired by confocal microscopy. Arrows illustrate active caspase-8 activity at the plasma membrane, indicating its translocation. (C) Quantitative measurement of MFI of active caspase-8 by confocal microscopy. The MFI of active caspase-8 per cell in the presence of anti-CD3 and anti-CD28 was increased as compared to isotype control stimulated CTLs. P-value was determined using unpaired t-test (two-tailed). (D) Schematic of Boolean approach to assess for cytoplasmic and membrane-associated caspase-8 activity utilized in Figure 3C–E.
Figure S4, related to Figure 4: Assessment of necrotic death by Sytox Green staining. (A) Representative plot of purified CD8+ T cells with gating of live and dead populations based on forward and side scatter properties. (B) CD8+ T cells were stimulated in the presence or absence of HIV peptide and stained with Sytox Green dye, in addition to tetramer and anti-CD8 antibody. As shown, gating on live cells results in relatively low levels of necrotic Sytox Hi cells. However, gating on the entire culture reveals a significant increase in the number of Sytox Hi cells. (C) Summary data of percentage of Sytox Hi cells in the live and entire cultures within each patient group.
Figure S5, related to Figure 4: Assessment of caspase-3 activity within peptide-stimulated HIV-specific CD8+ T cells from controllers and progressors. (A) Representative data showing modulation in MFI of caspase-3 activity following 3d peptide stimulation (1 ug/mL) in KK10-specific CD8+ T cell responses from an 2 HLA-B*2705 elite controllers and 2 HLA-B*2705 chronic progressors. Filled plot (gray) represents caspase-3 MFI in the absence of KK10 peptide and dashed line represents caspase-3 MFI in the presence of KK10 peptide. (B) Summary data of change in caspase-3 activity following peptide stimulation in controllers (n = 5) and chronic progressors (n = 5). Statistical comparisons were made using the Wilcoxon matched pairs test.
Table S1, related to Figure 1: Patient characteristics of HLA-B*2705+ elite controllers and chronic progressors selected for cell sorting and transcriptional profiling. Clinical and molecular features of the patients utilized for our transcriptional profiling studies.
Table S3, related to Figure 1: List of Direct Interacting Partners within the Controller and Progressor Networks as identified by DAPPLE Analysis. This table delineates the genes involved in the direct EC and CP networks which are involved in driving overall network connectivity.