Abstract
Men with metastatic prostate cancer (PCa) who are treated with androgen deprivation therapies (ADT) usually relapse within 2–3 years with disease that is termed castration-resistant prostate cancer (CRPC). To identify the mechanism that drives these advanced tumors, paired-end RNA-sequencing (RNA-seq) was performed on a panel of CRPC bone marrow biopsy specimens. From this genome-wide approach, mutations were found in a series of genes with PCa relevance including: AR, NCOR1, KDM3A, KDM4A, CHD1, SETD5, SETD7, INPP4B, RASGRP3, RASA1, TP53BP1 and CDH1, and a novel SND1:BRAF gene fusion. Amongst the most highly-expressed transcripts were ten non-coding RNAs (ncRNAs), including MALAT1 and PABPC1, which are involved in RNA processing. Notably, a high percentage of sequence reads mapped to introns, which were determined to be the result of incomplete splicing at canonical splice junctions. Using quantitative PCR (qPCR) a series of genes (AR, KLK2, KLK3, STEAP2, CPSF6, and CDK19) were confirmed to have a greater proportion of unspliced RNA in CRPC specimens than in normal prostate epithelium, untreated primary PCa, and cultured PCa cells. This inefficient coupling of transcription and mRNA splicing suggests an overall increase in transcription or defect in splicing.
Keywords: RNA, splicing, prostate cancer, RNA-seq, transcription
Introduction
With over 230,000 new patients and nearly 30,000 deaths annually, prostate cancer (PCa) is the second-most common cause of cancer-related deaths in men in the United States (1). Although greater than 75% of patients with early-stage PCa can be cured with surgical and/or radiation treatment, the remainder ultimately recur with metastatic disease. Androgen deprivation therapy (surgical castration or the administration of luteinizing hormone–releasing hormone agonists) is the standard treatment for metastatic PCa (2), but most tumors eventually relapse despite castrate androgen levels (castration-resistant prostate cancer, CRPC). It has now become clear that androgen receptor (AR) is substantially reactivated in a large proportion of these relapsed tumors through increased intratumoral androgen synthesis, in conjunction with other mechanisms that may enhance AR expression and activity, and many of these tumors will respond to agents that further suppress androgen synthesis (CYP17A1 inhibitors such as abiraterone) or new AR antagonists (such as enzalutamide). Unfortunately these men generally relapse within 1–2 years, and rising serum PSA in most cases suggests that AR is again active in these resistant tumors.
We reported previously on an analysis of gene expression in CRPC bone marrow metastases using Affymetrix oligonucleotide microarrays and immunohistochemistry, which showed increased expression of enzymes mediating androgen synthesis and alterations in the expression of additional genes linked to tumor progression (3). We hypothesize that additional mechanisms mediating progression to CRPC will also contribute to tumor progression after treatment with new hormonal agents including abiraterone and enzalutamide. Therefore, in this study we have used paired-end RNA-seq to assess more comprehensively the transcriptome of eight CRPC bone marrow metastases that had been examined previously on Affymetrix U133A microarrays.
Materials and Methods
Tissue samples
All tissue samples in this study were obtained with consent from PCa patients in compliance with the Beth Israel Deaconess Medical Center Institutional Review Board. CRPC biopsies were obtained from the posterior iliac crest and snap frozen as previously described (3–5). Frozen sections stained with H&E were examined histologically and 4–6 6 μm ribbons with >90% tumor and minimal bone marrow elements were treated with TRIzol (Invitrogen, Carlsbad, California) for purification of total RNA.
To obtain non-neoplastic prostate epithelium we examined snap-frozen samples from radical prostatectomies in patients with low volume PCa, and collected sections with 20–80% normal prostate epithelium and no evident tumor on histology. DNase-treated RNA was extracted from 10 6 μm ribbons using the RNeasy Plus Micro Kit (Qiagen). To obtain primary PCa, samples from radical prostatectomies were fixed in PaxGene (Qiagen), processed into paraffin and sectioned at 5 μm onto Arcturus polyethylene naphthalate metal-framed slides (Molecular Machines & Industries, Zurich, Switzerland). Approximately 50,000 cells in Gleason pattern 3 and Gleason pattern 4 glands identified by a board-certified pathologist were captured onto caps using 20-micron infrared pulses and excised from the adjacent tissue using the ultraviolet laser on an ArcturusXT Nikon Eclipse Ti-E microdissection system. DNase-treated RNA was extracted using the PaxGene Tissue RNA Kit (Qiagen).
Library Preparation and Data Analysis
50 ng of RNA from CRPC samples was prepared for Illumina paired-end sequencing using the Ovation RNA-Seq System (NuGEN, San Carlos, CA) and FastQ files were aligned to the human genome (version Hg19). Complete descriptions of library preparation methods and sequencing data analysis are provided as Supplementary online material.
Cell Lines
VCaP and LNCaP cells were obtained from the American Type Culture Collection (ATCC) and passaged for fewer than six months after receipt. VCS2 (6) and C4-2 (7) cells were derived from VCaP and LNCaP cells, respectively. Subconfluent cultures of VCaP, LNCaP, VCS2 and C4-2 cells grown in the presence of androgen (5–10% fetal bovine serum) were used as a source of control RNA. Cell lines’ identities were routinely validated by examining cell morphology, verifying AR mRNA expression, and sequencing for expected AR mutations (in LNCaP and LNCaP derived C4-2 cells) and/or TMPRSS2:ERG translocation (in VCaP and VCaP derived VCS2 cells). DNase-treated RNA was extracted using the RNeasy Plus Mini Kit (Qiagen).
Results
RNA-seq gene expression analysis is concordant with previous microarray analysis
We had previously analyzed on Affymetrix U133A microarrays a panel of 33 CRPC bone marrow biopsies in comparison with a series of primary PCa (3). However, the additional information that can be gained by paired-end RNA-seq led us to re-analyze a subset of these CRPC samples, which were selected based on very low contaminating hematopoietic or stromal cell content (>90% tumor by H&E) and availability of adequate RNA. For each of the 8 samples selected, 50 ng of total RNA was amplified into double-stranded cDNA and Illumina paired-end adaptors were ligated onto the library for 76 cycles of paired-end sequencing (samples 49 and 66) or 101 cycles of paired-end sequencing (samples 24, 28, 39, 55, 71 and 74) (see Supplementary Methods).
Although RNA from the previously-analyzed primary PCa was not available, we were still interested in whether gene expression data from the RNA-seq and the previous Affymetrix U133A microarrays were consistent. Therefore, we re-analyzed the Affymetrix raw data to perform a transcript-level normalization and performed a correlation analysis between the intensity values of these arrays with the RPKM from our RNA-seq data (see Supplementary Methods). Considering approximately 13,000 transcripts (Supplementary Table S1), our analysis showed a statistically significant, positive correlation between gene expression values measured from the same CRPC sample on both platforms (Supplementary Fig. S1). Our observation of r values less than 0.7 may be attributed to the 3-prime bias intrinsic in the U133A microarray, whereas our random priming, whole transcriptomic RNA-seq approach resulted in consistent coverage across transcripts (8) and better detection of low abundance transcripts (9). Spearman r values increased when only the last exon RPKM was used for correlation analysis (data not shown). Nonetheless, this result indicated that gene expression values were not platform-dependent, and supported our previous conclusions regarding gene expression differences between the primary PCa and CRPC samples (3).
Mutation analysis reveals potential drivers of tumor development or progression
Across the 8 CRPC samples we found an average of 131 protein-coding, somatic mutations (either frameshift, nonsense, or missense) with at least 20% variant reads at 20× coverage that were screened against the SNP databases as described in the supplementary methods (Table 1 and Supplementary Table S2). Among the mutations that were likely drivers of tumor progression, we found mutations in AR that we had previously reported in these tumors (4). These were an H875Y mutation in CRPC 39 and T878A mutation in CRPC 55 and 71 (Hg19 annotation; equivalent to H874Y and T877A, respectively, in the former Hg18 annotation).
Table 1.
Spectrum of genetic alterations detected in CRPC.
| Sample | Total | Somatic | >10 Coverage | >20 Coverage | Protein Coding | ||
|---|---|---|---|---|---|---|---|
| >10% Allele | >20% Allele | Missense | Nonsense | Frameshift | |||
| 24 | 132923 | 2133 | 1074 | 440 | 122 | 7 | 25 |
| 28 | 120300 | 2608 | 1599 | 740 | 122 | 1 | 26 |
| 39 | 70115 | 2139 | 1248 | 546 | 86 | 0 | 27 |
| 49 | 101200 | 2903 | 781 | 293 | 87 | 0 | 3 |
| 55 | 102631 | 2024 | 1123 | 496 | 67 | 0 | 27 |
| 66 | 142647 | 3584 | 984 | 318 | 103 | 1 | 6 |
| 71 | 136153 | 2460 | 1468 | 671 | 95 | 2 | 42 |
| 74 | 108171 | 2647 | 1762 | 799 | 132 | 7 | 66 |
The total number of variants is indicated, with anticipated (somatic) variants filtered as present in the COSMIC database or not represented in the dbSNP, HapMap, or 1000Genomes databases. Amongst the higher confidence 10% and 20% sequence read fractions are protein-coding mutations of missense, nonsense and frameshift variants.
We observed additional novel mutations in genes that have been previously reported as being mutated in PCa (10–12). These included an R398W mutation in NCoR1 (Nuclear Receptor Corepressor 1) in CRPC 66, which may decrease its corepression of AR (13), a premature stop codon at position 546 in KDM3A (Lysine Specific Demethylase 3A) in CRPC 74, a frameshift mutation in KDM4A (Lysine Specific Demethylase 4A) in CRPC 28, frameshift mutations in the lysine methyltransferase genes SETD5 and SETD7 (in CRPC 71 and 74, respectively), as well as a missense mutation in SETD5 in CRPC 49. We also found a premature stop codon in a RasGEF, RASGRP3, at codon 204 in CRPC 28, and an L319V mutation in a RasGAP, RASA1 in CRPC 39. The RASGRP3 truncation would preserve the Ras binding REM domain and its exchange function CDC25 domain while deleting key regulatory regions in the C-terminus, which may lead to enhanced Ras activity, while the RASA1 mutation in the PH domain could affect its membrane localization and thus ability to inactivate Ras (14,15). We also detected potential loss-of-function mutations in the tumor suppressor proteins encoded by CHD1, TP53BP1, and INPP4B, which have been reported previously as mutated in PCa (10–12). Finally, we observed an R800P mutation in CDH1 (E-cadherin) in CRPC 74, which may interfere with the ability of the cytoplasmic domain to bind and regulate signaling through β-catenin (16).
Paired-end sequencing of metastatic CRPC reveals expression of novel fusion genes
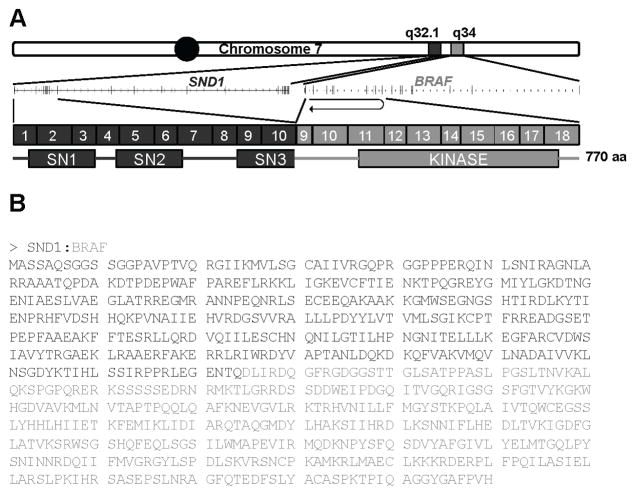
We performed post-processing for the discovery of fusion genes using both an annotation-dependent algorithm (ChimeraScan) and an annotation-independent algorithm (deFuse) (see Supplementary Methods). We found only three high-confidence fusions detected by both algorithms, each of which was novel (Table 2). The first of these predicted fusions, SND1:BRAF (Fig. 1), is a potential driver of tumorigenesis in CRPC 28, having fused the kinase domain of B-Raf (contained within exons 9–18) to the three Staphylococcal Nuclease homolog domains of Snd1. Lacking the regulatory Ras-binding domain (exons 3–7) and inhibitory serine phosphorylation site (exon 8) in wild-type B-Raf, this fusion kinase has been detected once previously in the gastric cancer cell line GTL16, and was noted to promote cancer cell growth via uncontrolled and increased activation of downstream MAP kinases (17). BRAF rearrangements to other genes have been observed previously in PCa (18), and this particular fusion puts the B-Raf kinase domain under control of the SND1 promoter, which is active in a majority of PCa (19).
Table 2.
Fusion and splice site location for 3 novel fusion transcripts detected by deFuse and ChimeraScan.
| Sample | 5′ Gene | 3′ Gene | Fragments | Type | 5′ Splice | 3′ Splice | Frame (5′/3′) |
|---|---|---|---|---|---|---|---|
| 28 | SND1 | BRAF | 27 | Intrachromosomal | chr7:127361454 | chr7:140487384 | Coding/Coding |
| 49 | EPB41L5 | PCDP1 | 9 | Intrachromosomal | chr2:120844816 | chr2:120317265 | Coding/Coding |
| 66 | PHF20L1 | LRRC6 | 12 | Intrachromosomal | chr8:133790157 | chr8:133584728 | Coding/Coding |
For each fusion shown, information is provided indicating the CRPC identifier, number of fusion/splice spanning fragments sequenced, as well as the chromosomal coordinates for the novel splice junction.
Figure 1. SND1:BRAF fusion transcript detected in CRPC 28.
(A) Schematic representation of the fusion between SND1 and BRAF on chromosome 7. SND1 exons, SND1 SN domains, BRAF exons, and the BRAF kinase domain are indicated. (B) Predicted amino acid sequence for the SND1:BRAF fusion protein. Amino acids originating from SND1 are represented in red, while amino acids contributed by BRAF are blue.
We also detected with high confidence two additional putative fusions genes, EPB41L5:PCDP1 (Supplementary Fig. S2) and PHF20L1:LRRC6 (Supplementary Fig. S3). However, it is unknown whether the fusion of their respective functional domains would confer oncogenic activity, and these genes have not been previously documented as upregulated or fused in cancer (20–23). Fusion between TMPRSS2 and ERG or ETV1, which occur in approximately half of all PCa, were notably absent from the list of predicted fusions (24). Consistent with this result, clustering of these 8 CRPC and other CRPC sets in the Affymetrix microarray dataset (GEO Accession ID GSE32269) revealed that the 8 CRPC samples we sequenced are fusion-negative (Supplementary Fig. S4). Interestingly, ChimeraScan (but not deFuse) detected with high probability a fusion between TMPRSS2 and ETV4 in CRPC 74 (Supplementary Table S3), which occurs with far less frequency than the TMPRSS2:ERG or TMPRSS2:ETV1 fusions (24).
Non-coding RNAs expressed in CRPC
RNA-seq permitted us to examine the expression of genes for which probes were not present on the microarrays performed previously. A complete list of genes and their computed RPKM values is provided in supplementary online data (Supplementary Table S4). Interestingly, amongst the top-expressing 100 transcripts by mean RPKM across all 8 CRPC samples (Supplementary Table S5) were 10 previously annotated noncoding RNAs (ncRNAs), all of which were also present in the list of the top 100 genes determined by median RPKM (Supplementary Table S6). The most highly-expressed transcript, the ncRNA MALAT1 (CR595720), is a long noncoding RNA (lncRNA) that has been implicated in regulating mRNA splicing (25) and its expression was recently found to be associated with prostate cancer progression, including CRPC (26). Also on this list is the lncRNA PABPC1, which interacts with poly-A-mRNA binding proteins and is important for RNA decay in response to poly-A shortening. Its upregulation in PCa has been suggested to be in response to an increased number of improperly-spliced or improperly-processed transcripts (27).
We observed that our list of highly expressed ncRNAs did not contain any of the non-coding PCa associated transcripts (PCAT’s) recently reported such as SChLAP1 (28) and PCAT-1 (29) although they were expressed in a subset of samples at lower levels (see Supplementary Table S4). To identify any additional highly expressed lncRNA, we next performed novel lncRNA discovery using CuffLinks, accepting any novel unannotated transcript greater than 200 nucleotides with at least two exons. A complete list of novel lncRNA’s and their mean FPKM values is provided in supplementary online data (Supplementary Table S7).
Pathways Upregulated in CRPC
In order to determine whether the coding or non-coding RNAs abundantly expressed in CRPC may play a significant physiological role in promoting cancer progression, we performed differential expression analysis of these samples against RNA-seq performed on 240 primary prostate cancers sequenced as part of The Cancer Genome Atlas (TCGA). In a combined dataset of both the TCGA and CRPC samples, unsupervised hierarchical clustering of 1,465 transcripts with the widest range of expression across all samples separated the TCGA and CRPC samples into two distinct groups (Supplementary Fig. S5A). The average RPKM difference between CRPC and TCGA samples for these 1,465 transcripts are listed in Supplementary Table S8.
To determine whether these other differentially regulated transcripts indicated any disease-driving pathways, we used Gene Set Enrichment Analysis to identify pathways enriched in CRPC vs. primary cancer (TCGA). Pathways identified as enriched in CRPC included cell adhesion molecules and MAP kinase signaling (Supplementary Fig. S5B), although the small number of input genes precluded reaching a statistically-significant P value for these pathways.
Transcripts in metastatic CRPC contain high frequency of intronic reads
We anticipated that this RNA-seq analysis would also add to the previous Affymetrix U133A analysis by revealing alternatively spliced isoforms for many genes. However, while we expected the RNA-seq analysis of RNA that was not poly-A selected to yield many intronic reads, we found an unexpectedly high level of intronic coverage (Supplementary Table S9) that made discovery of novel splice variants difficult. Examination of the mapping statistics showed that the high percentage of intronic reads was not correlated with the percentage of intergenic reads (which were much lower when corrected for total intergenic DNA), indicating that the intronic reads were not genomic DNA contamination (Supplementary Table S9). Amongst the top 10 genes as determined by intronic read depth in two samples examined in detail (CRPC 49 and CRPC 66) (Table 3), we found known markers of PCa including AR, KLK3, KLK2, and STEAP2, all of which are regulated by AR (30,31). However, these genes also had high levels of exonic reads, indicating they were highly expressed. Moreover, global assessment of intronic sequence coverage in CRPC 49 and CRPC 66 (Supplementary Tables S10 and S11, respectively) showed high levels of intronic sequence for a broad spectrum of genes, and this was correlated with their exonic read depth (see below, Supplementary Fig. 7).
Table 3.
Ten top-ranking genes with retained introns in CRPC 49 and 66.
| mCRPC 49 | mCRPC 66 |
|---|---|
| OR51E2 | KLK2 |
| KLK2 | KLK3 |
| TMEFF2 | AR |
| AR | HFM1 |
| STEAP2 | AMACR |
| KLK3 | TPT1 |
| TPT1 | SNORA31 |
| SNORA31 | SHROOM1 |
| SAT1 | HNRNPC |
| SAT | HNRPC |
Genes are ranked in descending order based on their intronic RPB measurement.
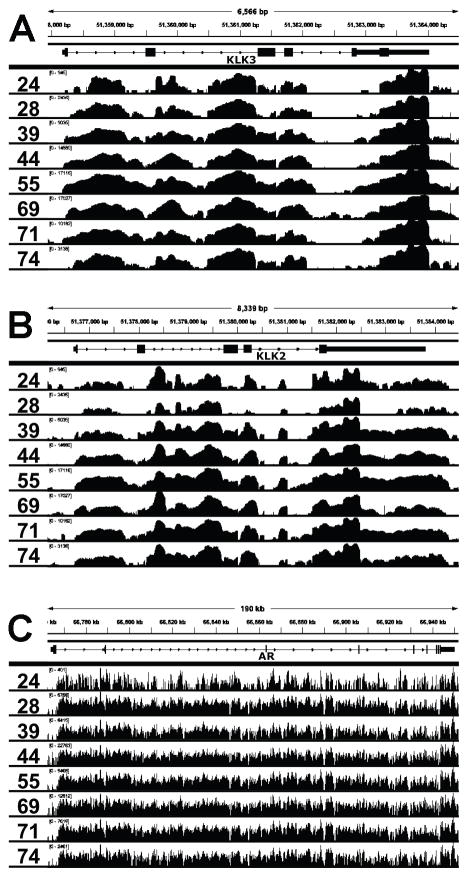
Inspection of the Bowtie-mapped reads in the Integrative Genome Viewer (IGV) for all eight CRPC samples similarly revealed substantial intronic coverage for KLK3, KLK2, and AR (Fig. 2A–C) and for STEAP2 (Supplementary Fig. S6A). As anticipated from the mapping statistics, we observed much lower levels of intergenic reads between and outside of KLK2 and KLK3 (Supplementary Fig. S6B), further indicating only a low level of gDNA contamination. We also observed high intronic read depth in many other genes that are not AR-regulated, such as CDK19 and CPSF6 (Supplementary Fig. S6C–D), further showing that this phenomenon was not limited to AR regulated genes. To globally assess whether intronic read depth was related to overall gene expression, we plotted the log10 transformed values for the exonic RPKM versus the log10 transformed values for the intronic RPKM for all genes across all eight CRPC samples (Supplementary Fig. S7). The observed strong positive correlation indicated that the level of intron reads for most genes was proportional to the overall expression of the gene.
Figure 2. Extensive intronic coverage in a subset of genes.
Quality-filtered read coverage for (A) KLK3, (B) KLK2, and (C) AR for all 8 CRPC mRNA samples sequenced.
Metastatic CRPC cells undergo inefficient splicing
We next addressed whether the high frequency of intronic reads reflected unspliced introns versus introns that were spliced but not degraded. Therefore, for each splice site in every gene we computationally counted the total number of fragments spanning the site that was spliced (exon-to-exon reads) versus fragments that were not spliced (exon-to-intron reads). We then calculated the percentage of reads corresponding to an unspliced junction out of the total number of reads for that junction (spliced plus unspliced) (Supplementary Table S12). Based on these calculations across all samples, we determined that approximately 28% of the splice junctions were not spliced. It should be noted that the absolute number of reads that mapped completely within an exon or within an intron were approximately equal (see Supplementary Table S9). However, this is not inconsistent with the above estimate of 28% unspliced mRNA as the greater length of introns relative to exons increases the likelihood that an RNA-seq read from an unspliced transcript will map to an intron versus an exon.
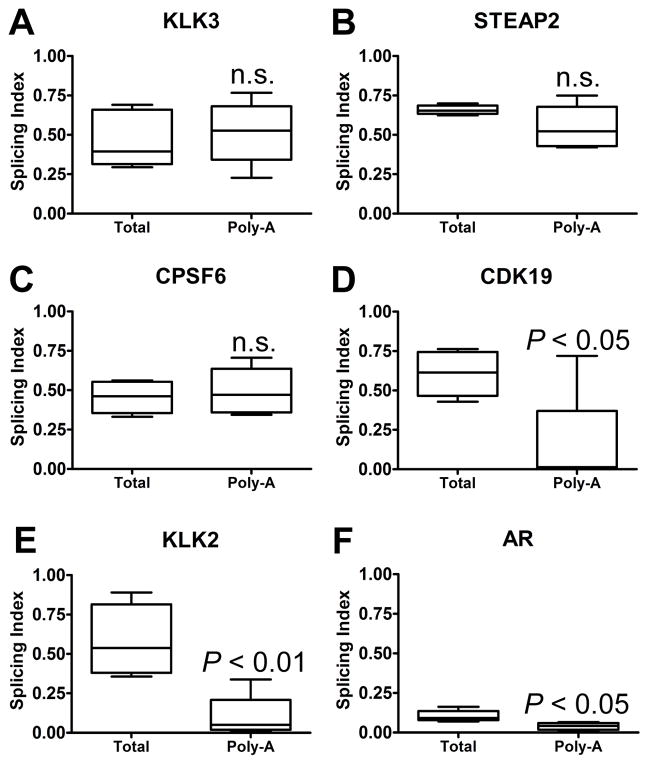
We next wanted to determine the extent to which the unspliced introns reflected nascent mRNA that was not yet polyadenylated. To address this question, we isolated the polyadenylated fraction of mRNA from the total RNA pool in four samples and performed whole transcriptome amplification using the same method employed for whole cellular RNA. We then used a series of PCR primer pairs in a qRT-PCR scheme (Supplementary Fig. S8) to amplify either spliced or unspliced junctions in a group of highly expressed genes that had high frequencies of intronic reads (AR, KLK2, KLK3, STEAP2, CPSF6, and CDK19) (see Supplementary Table S13 for primer sequences). Similar to our computational approach above, we calculated a relative splicing index for each junction based on amplification with exon-intron primers versus amplification with exon-exon plus exon-intron primers. This relative splicing index, which reflects the ratio of unspliced to total junctions (unspliced plus spliced), was then averaged for each gene and was further normalized across the samples based on amplification with primers within exons. Finally, we compared the results for the poly-A versus unfractionated total cellular RNA. For KLK3, STEAP2, and CPSF6 (Fig. 3A–C) there were no significant differences between the poly-A and total cellular RNA fractions, indicating that a substantial fraction of the poly-A mRNA for these genes is unspliced. In contrast, the splicing index values for CDK19, KLK2, and AR were lower in the poly-A fraction, indicating that a proportion of the unspliced junctions for these genes were contained in nonpolyadenylated nuclear RNA (Fig. 3D–F).
Figure 3. Poly-adenylated RNA contains unspliced introns.
The splicing index was calculated for (A) KLK3, (B) STEAP2, (C) CPSF6, (D) CDK19, (E) KLK2, and (F) AR in CPRC samples before and after OligoTex purification for poly-adenylated (Poly-A) species. Measurement was performed in triplicate, and the average values for each CRPC are depicted on box plots.
Splicing efficiency in CRPC is decreased relative to primary PCa
It did not appear that the apparently substantial unspliced mRNA was due to biases in the whole transcriptome amplification methods we employed, as we observed high levels of intronic reads and of exon-intron junctions. Moreover, examination of transcripts in the bone marrow biopsy samples that were derived from hematopoietic or stromal cells, such as HBB (hemoglobin beta) (Supplementary Fig. S9A) and SPP1 (osteopontin) (Supplementary Fig. S9B), respectively, showed very few intronic reads or exon-intron junctions, indicating that inefficient splicing was a property of the tumor cells. Nonetheless, we next addressed possible biases by comparing cDNA generated from amplified versus unamplified RNA. For this analysis we used RNA from CRPC 66, for which we had an adequate amount of extracted RNA. Portions of the RNA were used to generate single-stranded or double-stranded amplified libraries (NuGEN) or to generate cDNA directly without amplification using conventional reverse transcriptase with a pool of oligo-dT and random oligonucleotide primers. We then assessed the AR splicing index by amplification with primers corresponding to exons 4–5, exons 5–6, exon 4 to intron 4, and exon 5 to intron 5. Significantly, we observed a higher AR splicing index, indicative of more unspliced mRNA, in conventionally synthesized cDNA compared to the whole transcriptome amplified libraries (Supplementary Fig. S9C), further supporting the conclusion that a substantial proportion of transcripts in the CRPC samples were not spliced.
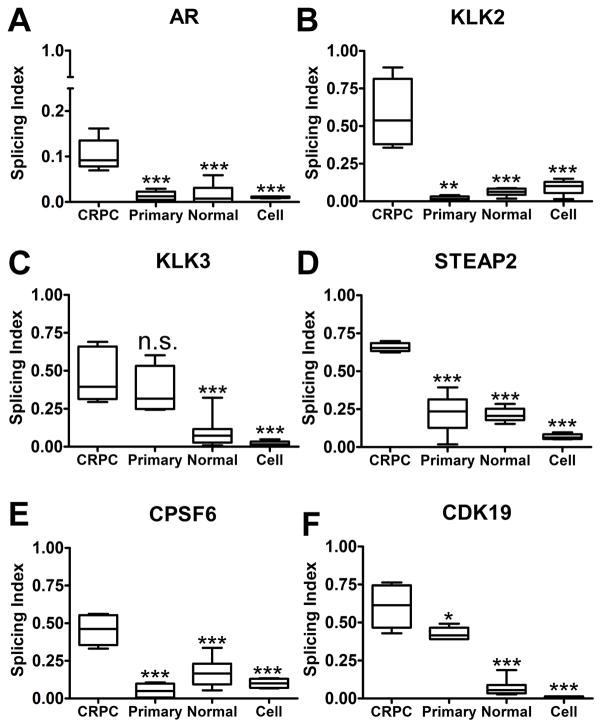
Finally, we addressed whether the inefficient splicing we observed was a general feature of PCa. For this analysis we isolated whole cellular RNA from 6 cases of laser-capture microdissected untreated primary PCa (Gleason score 7), 10 cases of normal prostate epithelium, and 4 PCa cell lines (LNCaP, C4-2, VCaP, and VCS2). The RNA was then subjected to whole transcriptome amplification as for the metastatic CRPC samples, and we determined the splicing index for the six gene panel. Significantly, the median splicing index was higher for all six genes in the CRPC samples when compared to primary PCa, normal epithelium, or cell lines, indicating that splicing is less efficient in metastatic CRPC versus normal prostate or primary PCa (Fig. 4).
Figure 4. CRPC samples express more unspliced mRNA than primary prostate cancers, normal prostatic epithelium, or cultured prostate cancer cell lines.
Splicing indices were calculated and compared between CRPC, normal prostatic epithelial tissue, laser-capture microdissected primary prostate cancer cells, and established prostate cancer cell lines. Boxplots representing the data within each set are shown for (A) AR, (B) KLK2, (C) KLK3, (D) STEAP2, (E) CPSF6, and (F) CDK19. Boxplots represent the set of average values from three replicate experiments for each biological sample. Statistical significance between samples was measured by the Student’s unpaired t-test (95% confidence interval), and probability of statistical difference is indicated by: * P < 0.05; ** P < 0.01; *** P < 0.005; ns: not statistically significant.
Discussion
This study used RNA-seq to further characterize gene expression in a series of metastatic CRPC samples, and in particular to assess for mutations, gene fusions, ncRNA, and alternative splicing. We detected novel mutations in a series of genes that have been implicated previously in PCa development or progression to metastatic CRPC. These included mutations in genes encoding proteins that regulate transcription (NCOR1, KDM3A, KDM4A, CHD1, SEDT5, and SETD7), PI-3 kinase pathway (INPP4B), and Ras pathway signaling (RASGRP3 and RASA1). Although the functional significance of these mutations has not been determined, NCoR1 can function as a corepressor for AR and its loss could enhance AR activity in CRPC. Alterations in KDM3A, KDM4A, and CHD1 could also affect AR activity, but would likely have broad effects on gene expression. Our observation of novel mutations to SETD5 and SETD7 may result in altered chromatin accessibility during co-transcriptional RNA processing and thus may also contribute to intron retention, a phenomenon recently reported in an RNA-seq analysis of clear cell renal cell carcinoma (32). Mutations we found in RASA1 and RASGRP3, and a novel SND1:BRAF gene fusion, may contribute to the enhanced RAS/RAF/MAPK signaling observed with progression to CRPC (33). Interestingly, although gene fusions are common in PCa, they were infrequent in these samples when we used a high stringency threshold. While we may have failed to detect some abundant fusion gene transcripts, it is also likely that many gene fusions are not drivers of tumor progression, and that their expression may thereby not confer a selective advantage in these advanced tumors.
Amongst the most highly-expressed genes were ten noncoding RNAs, including MALAT1 and PABPC1, and in a subset of our cases we also observed expression of one or more of the recently reported noncoding PCa associated transcripts (PCAT’s) (28,29). In particular, the outlier PCAT predictive of lethal disease (PCAT-114, also referred to as SChLAP1) was expressed in a subset of cases. Interestingly, many of the lncRNA’s that were expressed at high levels, in addition to PCAT-114, are known to be involved in regulating transcription and may contribute to tumor progression (25,27,28).
An unexpected result was the large number of sequence reads that mapped to introns. This appeared to reflect incomplete splicing based on the fraction of reads that spanned exon-intron junctions compared to those that spanned exon-exon junctions. For some genes this may reflect the use of whole cell RNA rather than poly-A RNA, but for others we found that the ratio of exon-intron versus exon-exon junctions was not significantly decreased when we examined poly-A RNA. In either case, this inefficient splicing was greater in the metastatic CRPC samples compared to normal prostate and primary PCa, indicating that it is a feature of metastatic CRPC. It is not clear why this inefficient splicing was not observed in the PCa cell lines as these were derived from metastatic CRPC, but possibilites include a role for the tumor microenvironment or a selective advantage in vitro for subclones that splice more efficiently.
Significantly, genes with the greatest levels of intron retention did not group into any specific biological pathways, but rather were those with the greatest overall expression (see Supplementary Tables S10–S11). Therefore, we suggest that these findings reflect global increases in gene transcription in advanced CRPC and a saturation of the cellular splicing machinery, with subsequent uncoupling of transcription and splicing (34). This hypothesis is consistent with the high level and increased expression of multiple ncRNA involved in transcription and RNA processing with PCa progression (25,27,28). It is also supported by a recent report showing that increased transcription of already-upregulated genes, which correspond to changes in the methylation status of the genome, occurs during progression to CRPC (35). Finally, it is of interest that H3K27me3 levels are decreased with PCa progression, which may contribute to global derepression of gene transcription (36,37).
Alternative splicing can clearly contribute to tumor progression (34,38,39), and the inefficient removal of introns may provide increased substrate for alternative splicing to generate isoforms of some proteins that contribute to tumor progression. Moreover, high levels of intronic RNA also would presumably sequester many micro-RNA species, resulting in dysregulation of multiple miRNA regulated protein expression networks. However, further studies are needed to determine whether inefficient splicing provides a selective advantage driving tumor progression in vivo, and whether these tumors may be vulnerable to agents that suppress rate limiting steps in splicing.
Supplementary Material
Implications.
Inefficient splicing in advanced prostate cancer provides a selective advantage through effects on micro-RNA networks, but may render tumors vulnerable to agents that suppress rate-limiting steps in splicing.
Acknowledgments
Grant Support
Supported by grants from the NIH (T32CA081156 to AGS, R00CA135592 to SC, P01CA163227-01A1 to SPB, DF/HCC-Prostate Cancer SPORE P50CA090381 to SPB, R01HG007538 to WL), Department of Defense Prostate Cancer Research Program (Postdoctoral Training Award W81XWH-13-1-0267 to AGS, Idea Development Awards W81XWH-11-1-0295, W81XWH-08-1-0414, and W81XWH07-1-0443 to SPB, and W81XWH-10-1-0501 to WL), CPRIT (RP110471 to WL), and a Prostate Cancer Foundation Challenge Award (SPB).
Footnotes
Disclosure of Potential Conflicts of Interest
There are no potential conflicts.
Authors’ Contributions
Conception and design: A.G. Sowalsky, S.P. Balk, W. Li
Development of methodology: A.G. Sowalsky, Z. Xia, L. Wang, H. Zhao, W. Li
Acquisition of data: A.G. Sowalsky, S. Chen, G.J. Bubley
Analysis and interpretation of data: A.G. Sowalsky, Z. Xia, L. Wang, H. Zhao, S.P. Balk, W. Li
Writing, review, and/revision of manuscript: A.G. Sowalsky, S.P. Balk, W. Li
Administrative, technical, or material support: A.G. Sowalsky, Z. Xia, L. Wang, H. Zhao, W. Li
Study supervision: S.P. Balk, W. Li
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66(5):2815–25. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 4.Taplin ME, Bubley GJ, Ko YJ, Small EJ, Upton M, Rajeshkumar B, et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 1999;59(11):2511–5. [PubMed] [Google Scholar]
- 5.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332(21):1393–8. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 6.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20(4):457–71. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer. 1994;57(3):406–12. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- 8.Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10(7):623–9. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated Evolution of Prostate Cancer Genomes. Cell. 2013;153(3):666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindberg J, Mills IG, Klevebring D, Liu W, Neiman M, Xu J, et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol. 2013;63(4):702–8. doi: 10.1016/j.eururo.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 13.Linja MJ, Porkka KP, Kang Z, Savinainen KJ, Janne OA, Tammela TL, et al. Expression of androgen receptor coregulators in prostate cancer. Clin Cancer Res. 2004;10(3):1032–40. doi: 10.1158/1078-0432.ccr-0990-3. [DOI] [PubMed] [Google Scholar]
- 14.Aiba Y, Oh-hora M, Kiyonaka S, Kimura Y, Hijikata A, Mori Y, et al. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc Natl Acad Sci U S A. 2004;101(47):16612–7. doi: 10.1073/pnas.0407468101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pamonsinlapatham P, Hadj-Slimane R, Lepelletier Y, Allain B, Toccafondi M, Garbay C, et al. p120-Ras GTPase activating protein (RasGAP): a multi-interacting protein in downstream signaling. Biochimie. 2009;91(3):320–8. doi: 10.1016/j.biochi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee NV, Lira ME, Pavlicek A, Ye J, Buckman D, Bagrodia S, et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells through [corrected] MAPK activation. PLoS One. 2012;7(6):e39653. doi: 10.1371/journal.pone.0039653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuruma H, Kamata Y, Takahashi H, Igarashi K, Kimura T, Miki K, et al. Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am J Pathol. 2009;174(6):2044–50. doi: 10.2353/ajpath.2009.080776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML, et al. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res. 2007;313(19):3959–70. doi: 10.1016/j.yexcr.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 21.DiPetrillo CG, Smith EF. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J Cell Biol. 2010;189(3):601–12. doi: 10.1083/jcb.200912009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimojo H, Sano N, Moriwaki Y, Okuda M, Horikoshi M, Nishimura Y. Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain. J Mol Biol. 2008;378(5):987–1001. doi: 10.1016/j.jmb.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Kott E, Duquesnoy P, Copin B, Legendre M, Dastot-Le Moal F, Montantin G, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am J Hum Genet. 2012;91(5):958–64. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 25.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190(6):2278–87. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Ströbel P, Marx A, Hofmann I. Plakophilin-associated RNA-binding proteins in prostate cancer and their implications in tumor progression and metastasis. Virchows Arch. 2013;463(3):379–90. doi: 10.1007/s00428-013-1452-y. [DOI] [PubMed] [Google Scholar]
- 28.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porkka KP, Helenius MA, Visakorpi T. Cloning and characterization of a novel six-transmembrane protein STEAP2, expressed in normal and malignant prostate. Lab Invest. 2002;82(11):1573–82. doi: 10.1097/01.lab.0000038554.26102.c6. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24(2):241–50. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedlander TW, Roy R, Tomlins SA, Ngo VT, Kobayashi Y, Azameera A, et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012;72(3):616–25. doi: 10.1158/0008-5472.CAN-11-2079. [DOI] [PubMed] [Google Scholar]
- 36.Pellakuru LG, Iwata T, Gurel B, Schultz D, Hicks J, Bethel C, et al. Global levels of H3K27me3 track with differentiation in vivo and are deregulated by MYC in prostate cancer. Am J Pathol. 2012;181(2):560–9. doi: 10.1016/j.ajpath.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6(8):454–60. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 39.Sette C. Alternative splicing programs in prostate cancer. Int J Cell Biol. 2013;2013:458727. doi: 10.1155/2013/458727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.