SUMMARY
Decreases in the diversity of enteric bacterial populations are observed in patients with Crohn’s disease (CD) and ulcerative colitis (UC). Less is known about the virome in these diseases. We show that the enteric virome is abnormal in CD and UC patients. In-depth analysis of preparations enriched for free virions in the intestine revealed that CD and UC were associated with a significant expansion of Caudovirales bacteriophages. The viromes of CD and UC patients were disease- and cohort-specific. Importantly, it did not appear that expansion and diversification of the enteric virome was secondary to changes in bacterial populations. These data support a model in which changes in the virome may contribute to intestinal inflammation and bacterial dysbiosis. We conclude that the virome is a candidate for contributing to, or being a biomarker for, human inflammatory bowel disease and speculate that the enteric virome may play a role in other diseases.
INTRODUCTION
Inflammatory bowel disease (IBD) is a complex, remitting and relapsing inflammatory disease with genetic and environmental risk factors. One environmental contributor is thought to be microorganisms that live in the intestine (Gevers et al., 2014; Kostic et al., 2014; Minot et al., 2011; Norman et al., 2014; Virgin, 2014). Of these microorganisms, bacteria have gained the greatest attention and are linked to training mucosal immunity and minimizing mucosal inflammation [reviewed in (Belkaid and Hand, 2014)]. An aberration in either of these immune processes can have detrimental consequences for IBD progression. For example, a reduction of Bacteroidetes and Firmicutes and expansion of normally less abundant bacterial taxa (dysbiosis), as well as changes in bacterial microbiome function, have been associated with both Crohn’s disease (CD) and ulcerative colitis (UC) (Kostic et al., 2014; Stappenbeck et al., 2011). Importantly, household contacts without IBD can also exhibit signs of bacterial dysbiosis (Joossens et al., 2011). These individuals have increased intestinal permeability compared to healthy community controls (Hollander et al., 1986), suggesting that the bacterial microbiome is heavily influenced by the household environment. Investigations have also shown that the home environment is a primary determinant of the individual’s bacterial microbiome, and that humans are the primary vector of bacterial transmission between people living within the same household (Lax et al., 2014). Exchange of viruses between humans within a household has not been thoroughly investigated. Nevertheless, investigations of the bacterial microbiome and the enteric virome in IBD are likely to be optimized by the investigation of household controls rather than matched controls from different households.
Emerging data indicate that the viral component of the microbiome, termed the virome, can profoundly influence host physiology (Handley et al., 2012; Norman et al., 2014; Virgin, 2014). Recent advances in sequencing technology have led to the discovery of a diverse enteric human virome consisting of bacteriophages as well as eukaryotic viruses (Breitbart et al., 2003; Finkbeiner et al., 2008; Minot et al., 2013; Minot et al., 2012; Minot et al., 2011; Reyes et al., 2010). Importantly, evidence that eukaryotic viruses can interact with IBD risk genes to alter intestinal disease comes from studies of mice carrying mutations in IL-10 or Atg16L1, indicating that members of the virome may contribute to IBD (Basic et al., 2014; Cadwell et al., 2010; Irving and Gibson, 2008; Sun et al., 2011). Bacteriophages may also play a direct role in intestinal physiology or change the bacterial microbiome through predator-prey relationships (Barr et al., 2013; Duerkop et al., 2012; Reyes et al., 2013; Willner et al., 2009; Willner et al., 2012).
The virome, much of which is comprised of bacteriophages, contains the most diverse genetic elements on earth, and is only beginning to be characterized at the sequence level (Virgin, 2014). In the absence of disease, enteric bacteriophage populations exhibit significant diversity between individuals and are temporally stable (Minot et al., 2013; Reyes et al., 2010). Bacteriophages in the healthy human intestine are predominantly temperate dsDNA Caudovirales or ssDNA Microviridae that latently infect their bacterial hosts and generate few viral progeny that may infect and kill other bacteria (Minot et al., 2013; Minot et al., 2011; Reyes et al., 2010; Waller et al., 2014). However, environmental stimuli, such as nitric oxide and antibiotics, induce the production of infectious bacteriophages that lyse their bacterial host and infect neighboring cells bearing specific receptors (Lindsay et al., 1998; Maiques et al., 2006; Zhang et al., 2000; Zhang and LeJeune, 2008). This process releases infectious virions into the intestine, which can be purified and analyzed. Alterations in bacteriophage abundance have been suggested in CD (Lepage et al., 2008; Perez-Brocal et al., 2013; Wagner et al., 2013); however, these studies did not characterize the enteric virome in detail and did not control for factors within households that may influence the microbiome.
Here we characterized the normal human and IBD enteric virome by metagenomic sequencing of the DNA of virus-like particle (VLP) preparations from fecal samples obtained from UC and CD patients and controls. Throughout the text we refer to two ecological metrics, richness (the number of taxa counted per sample) and diversity. Diversity measures both richness and the relative abundance (or evenness) of the taxa present; changes in diversity can result from alterations in either richness or evenness. Detailed analysis of purified virions in VLP preparations and bacterial 16S ribosomal RNA sequences from a longitudinal patient cohort compared to household controls revealed the expected decrease in bacterial richness and diversity accompanied by a striking IBD-associated increase in bacteriophage richness. These findings were validated in two independent and geographically distinct patient cohorts that contained matched controls. The taxonomic sub-structure of the enteric virome and bacterial microbiome in CD and UC showed geographic variation in the specific bacteria and viruses detected. Together these data support a model where IBD-associated increases in bacteriophage richness are not merely accounted for by an increase in their bacterial host cells. We observed both positive and negative correlations between specific viral and bacterial taxa. These data demonstrate, for the first time, that unique changes in the bacteriophage component of the enteric virome occur in CD and UC, raising the possibility that these changes may contribute to disease pathogenesis, perhaps through a predator-prey relationship between bacteriophages and their bacterial hosts. These data provide a rationale for considering virome diagnostics for IBD and manipulation of the enteric virome as a novel therapeutic strategy for the management of IBD and emphasize the need for a greater understanding of trans-kingdom interactions within the microbiome for other diseases associated with changes in the bacterial microbiome (Duerkop and Hooper, 2013; Norman et al., 2014; Virgin, 2014).
RESULTS
Virome Alterations are Observed in Multiple Cohorts
To initially define the enteric virome associated with IBD, we performed metagenomic sequencing of stool filtrates using the Roche 454 platform on three independent cohorts consisting of IBD and non-IBD household controls (Tables S1 and S2; Cambridge, United Kingdom (UK); Chicago, USA; and Los Angeles, USA). On average, we obtained 32,591 +/− 27,531 sequences (number +/− SD) that were 282 +/− 47 nucleotides in length from 72 fecal samples (Table S3; 12 household controls, 18 CD and 42 UC). Sequences were demultiplexed, quality filtered, and assigned taxonomy (Supplemental Information). The majority of sequences obtained were assigned to the human host or to bacterial taxa (Table S3). Consistent with previous reports, bacteriophages of the Caudovirales order and Microviridae family were the most abundant viral taxa identified in all three cohorts (Figure 1A), (Minot et al., 2011; Reyes et al., 2010). Other viruses were detected in a limited number of samples and represented an average of five percent or fewer of the total viral sequences. An analysis of the relative abundances of sequences from all three cohorts revealed an inverse correlation between the Caudovirales and Microviridae (Figure 1B). This inverse correlation was also present when controls, CD, and UC were analyzed separately (data not shown). Comparing household controls to IBD samples within the UK cohort revealed that this disproportionate representation of bacteriophage abundance was associated with IBD (Figure 1C). Disparate ratios of Caudovirales and Microviridae were also observed in patients from Los Angeles. This suggestive correlation between disease and a change in sequences from the enteric virome was striking given the geographical and environmental diversity of the cohorts.
Figure 1. Virus Taxonomic Assignment and Imbalance in IBD.
A) Relative abundance of sequences assigned to the indicated viral taxa. Error bars represent the mean +/− SD. B) Correlation plot of the Caudovirales and Microviridae relative abundance for all samples. Linear regression +/− 95% confidence interval and Spearman correlation coefficient are shown. C) Microviridae and Caudovirales relative abundance for United Kingdom household controls, UC and CD (top); Los Angeles and Chicago IBD (bottom). The bars indicate the median and interquartile range. Statistical significance was determined by the Mann-Whitney test. See also Tables S1, S2, and S3.
In-Depth Analysis of Free Virions in the Enteric Virome in IBD
These initial observations prompted us to perform an in-depth analysis of the virome by metagenomic sequencing of VLPs purified from the feces of patients and controls from 17 IBD households in the UK (Figure 2A, Tables S1 and S2). VLP purification enriches for free virions (Reyes et al., 2012; Thurber et al., 2009). To further refine the relationship between IBD and the enteric virome and to take into account prior data indicating the bacterial microbiome and virome are similar within households (Lax et al., 2014; Reyes et al., 2010), we compared IBD patients to matched household controls (Figure 2A). This is particularly important for virome analysis given the high inter-personal variation of viruses (Reyes et al., 2010). Samples were collected from both the IBD patient and household control at the time of a clinical flare of disease (Supplemental Information). In total, 21 household control samples and 52 IBD samples (24 active disease and 28 inactive disease samples; 36 total UC and 16 total CD) were used to isolate VLPs for sequencing. We validated observations in two additional cohorts from Chicago and Boston that contained CD and UC patients and matched healthy control subjects (described in greater detail below; Tables S1 and S2).
Figure 2. In-depth, Longitudinal Cohort Graphical Timeline.
A) United Kingdom IBD stool samples and non-IBD household control stools were collected at the onset of symptoms (flare) and collected as symptoms resolved where indicated. The length of each IBD flare is indicated by the red shaded oval. The samples are annotated by household ID and sample number. B) Chicago UC samples were collected first during inactive disease and again when symptoms exacerbated. The samples are annotated by the subject and sample number. See also Tables S1 and S2.
For the UK cohort, we obtained 1,111,569 +/− 493,164 paired-end sequences per sample with an average sequence quality of 36.5 +/− 3.7 (Table S4; Experimental Procedures). Quality control trimming resulted in an average of 2% reduction in the number of sequences to 1,094,360 +/− 503,337 with an average reduction in sequence length from 250 to 241.7 +/− 3.7 bases and an average increase in quality score of 1.0 +/− 0.03. There were no statistically significant differences in the number of total or quality-controlled sequences obtained between control and IBD patient samples (Figure S1A). Sequences were clustered at 95% identity to remove similar sequences and to generate sequences termed unique hereafter. Clustering resulted in an average reduction of 89% to 112,192 +/− 71,820 unique sequences per sample. Interestingly, despite the fact that there were no significant differences in the total sequences obtained, we detected a significant increase in unique reads in CD patients compared to either household controls or UC patients. (Figure S1A).
Increased Caudovirales Taxonomic Richness Associated with IBD
Unique sequences were mapped to a custom virus protein database as described in the Experimental Procedures. We were able to assign viral taxonomy to an average of 17,022 +/− 15,725 sequences (15%) (Figure S1A), the majority being to Caudovirales and Microviridae bacteriophages (Figure S1B). Sequences were also assigned to many less-abundant viral taxa, including bacteriophages whose annotated hosts include bacteria commonly found in human fecal samples and common eukaryotic viruses (Figure S1B). Analysis of our VLP sequences revealed a low level of contamination with human sequences (0 – 4%). Possible contamination with bacterial sequences was confounded by the presence of integrated prophages in full genome sequences of bacteria. We mapped a subset of our total VLP enriched sequences from the UK cohort to the recently discovered crAssphage genome and detected it in 71% of our samples (Dutilh et al., 2014) (Table S4). The percentage of sequences that mapped to this virus varied greatly (range 1% – 89%) and did not correlate with disease status or drug treatment.
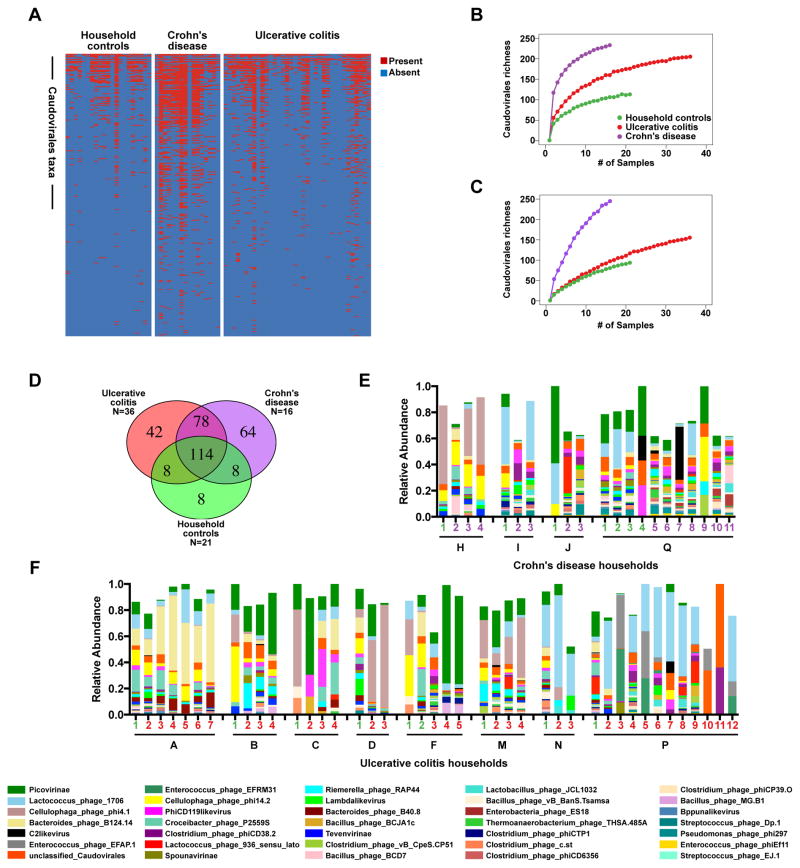
Interestingly, we observed an increase in the richness of bacteriophages, specifically, members of the Caudovirales in IBD (Figures 3A and 3B). It is unlikely that these differences can be attributed to an uneven number of samples or unique sequences in control and disease groups since the rate of acquisition of new bacteriophage taxa in disease samples rapidly outpaces new taxa acquisition in control samples (Figure 3B). No increases in Microviridae richness or diversity were observed (Figure S2A), indicating that bacteriophage expansion was restricted to certain taxa, and that our methods do not systematically increase bacteriophage richness in IBD samples compared to controls. Some IBD samples had many fewer Microviridae than controls, further supporting that the viromes were different between the two disease states (Figure S2A).
Figure 3. Bacteriophage Expansion is Associated with CD and UC.
A) Presence-absence heat map of the sequences assigned to Caudovirales taxa in VLP preparations from UK household control, CD and UC stool samples. B and C) Rarefaction curves of Caudovirales richness versus an increasing number of sub-samplings with replacement. B) Caudovirales richness based on individual sequences. C) Richness based on assembled Caudovirales contigs. The curves represent the average of 500 iterations at each depth of samples. D) Venn diagram of the Caudovirales taxa in household control, CD and UC samples. N = the number of samples within each sub-group. E and F) Plots of the relative abundance of the 35 most abundant Caudovirales taxa in the UK UC and CD households. Bars are annotated by the household ID and sample number. Green numbers = household controls; purple numbers = CD; red numbers = UC. See also Figure S1, Tables S4, S5, and S6.
Taxonomic assignment of de novo assembled contigs longer than 1,000 nucleotides also indicated that Caudovriales were enriched in IBD (Figure 3C). These data indicated that the VLP preparations contained partial or complete virus genomes and that the overwhelming majority of those assignable genomes were from Caudovirales bacteriophages. Taken together, Caudovirales sequences and assembled contigs were differentially expanded in CD and UC compared to household controls and were therefore capable of having disparate effects on the microbiome and immune responses.
Disease-Specific Changes in the Enteric Virome
In addition to differences in bacteriophage richness between IBD and household control samples, we also observed striking differences in richness and the types of bacteriophage taxa observed between CD and UC samples (Figure 3D). A substantial number of taxa were observed amongst all samples; however, each disease type harbored unique bacteriophages. These differences in Caudovirales taxa could be the result of multiple factors including: health or disease, household, duration of cohabitation, disease activity, age, sex, and/or drug treatment. We therefore performed a multivariate analysis of Caudovirales abundance using MaAsLin (Multivariate Association with Linear Models) on VLP sequences compared to their household controls (Morgan et al., 2012). MaAsLin identified 35 different Caudovirales that were significantly associated with different households (43 total associations) (Tables S5 and S6). This relationship was also revealed by plotting Caudovirales relative abundances, which indicate conservation within households (Figure 3E). This finding is consistent with a previous report (Reyes et al., 2010). Five of the Caudovirales identified by MaAsLin were significantly associated with disease, including sequences most closely related to Lactococcus, Lactobacillus, Clostridium, Enterococcus, and Streptococcus bacteriophages (Table S6). The increased Caudovirales richness in CD samples relative to household controls corresponded with increased bacteriophage diversity (Figure S2B). This was not observed in UC, further highlighting differences in the virome between UC and CD.
Inverse Correlation Between IBD-Associated Changes in the Virome and Bacterial Microbiome
We next assessed whether the increase in bacteriophage richness observed in CD and UC was associated with a parallel change in bacterial populations. To answer this question we performed bacterial 16S rRNA gene sequencing (Tables S1 and S2). On average, we obtained 64,107 +/− 38,570 sequences that were clustered at 97% identity into 56,096 +/− 32,774 operational taxonomic units (OTUs) per sample (Table S7). As expected, CD and UC were associated with a significant reduction in bacterial diversity and bacterial richness compared to household controls (Figures 4A and 4B). However, we also observed significant similarity in the bacterial microbiome within IBD households as determined by permutation multivariate analysis of variance of the weighted UniFrac distances (Figures 4B and 4C; ADONIS p = 0.001, 999 permutations) (McArdle and Anderson, 2001). Interestingly, in patients that were sampled longitudinally the bacterial diversity did not recover during periods of disease inactivity in either CD or UC (Figures 4B and 4C).
Figure 4. Alterations in the Bacterial Community Composition in IBD.
A) Alpha diversity (left) and bacterial species richness (right) based on 16S rRNA gene sequences in the stool of household controls, CD and UC patients. Statistical significance was determined by the Kruskal-Wallis test with Dunn’s correction comparing all samples to all samples. ** = p > 0.01. B and C) Plots of Alpha diversity normalized to the diversity in household controls (top) and relative bacterial family abundance (bottom) of UK B) CD households and C) UC households. Green numbers = household controls; purple numbers = CD; red numbers = UC. See also Tables S5, S7, and S8.
Like the shifts observed for bacteriophages, the changes in bacterial diversity could be multifactorial. We therefore performed a multivariate analysis of bacterial abundance using MaAsLin (Morgan et al., 2012). We identified 18 different bacterial taxa that were significantly associated with disease or disease activity (Tables S5 and S8). The vast majority of the significant OTUs were of the Bacteriodetes and Firmicutes phyla, including significant differences in members of the bacterial families Ruminococcaceae, Lachnospiraceae, Bacteriodaceae and Prevotellaceae. Therefore, and as expected, disease-specific alterations in bacterial taxa were observed in our cohort; however, the variation was also largely linked to specific households, supporting previous studies of the microbiome that have indicated similarities among IBD patients and their household contacts (Joossens et al., 2011).
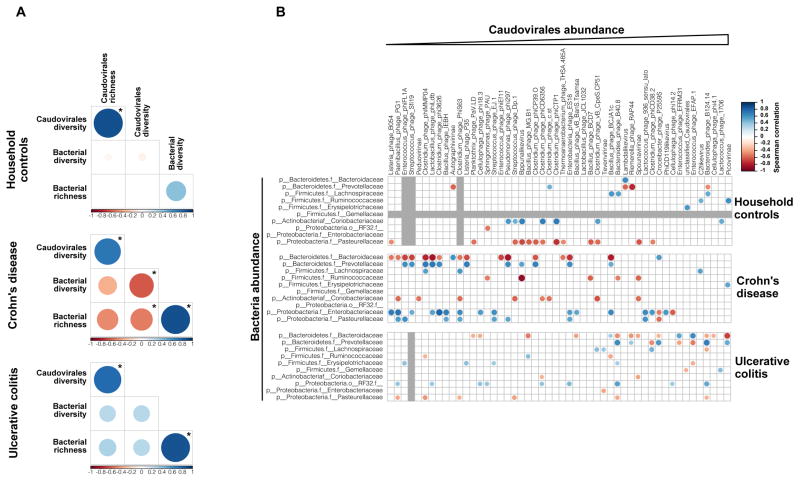
Importantly, the observed reduction in bacterial diversity was inversely related to the expansion of Caudovirales bacteriophages in IBD. Comparing the bacterial and Caudovirales bacteriophage communities in household controls, CD and UC samples indicated clear differences between the disease states (Figure 5A). Among the UK cohort, significant positive correlations were observed in 5 out of 6 possible comparisons between bacterial diversity or richness and Caudovirales richness or diversity. However, a significant inverse correlation was observed in CD samples between Caudovirales diversity and both bacterial richness and diversity, which suggests that bacteriophage expansion was not simply the result of increases in their bacterial hosts (Figure 5A).
Figure 5. Disease-Specific Bacteria-Caudovirales Patterns in IBD.
A) Spearman correlation plot of Caudovirales richness, Caudovirales Shannon diversity, bacterial alpha diversity, and bacterial species richness for UK household control, CD, and UC samples. Statistical significance was determined for all pair-wise comparisons; those with p values < 0.05 are indicated. Positive values (blue circles) indicate positive correlations and negative values (red circles) indicate inverse correlations. The size and shading of the circle indicates the magnitude of the correlation where darker shades are more correlated than lighter shades. B) Spearman correlation plots of the relative abundances of the 50 most abundant Caudovirales taxa and bacterial families identified to be significantly associated with disease. UK household control (top), CD (middle), and UC (bottom) samples. The gray bars indicate any taxa that were not detected in the cohort sub-group. Statistical significance was determined for all pair-wise comparisons; only significant (p value < 0.05) are displayed. See also Figure S2.
To further characterize the relationships between Caudovirales and bacterial taxa, we calculated the Spearman correlation between the Caudovirales taxa and the bacterial families found to be significantly altered in disease by MaAsLin analysis. Similar to the overall diversity and richness correlations, inverse correlations between Caudovirales and the significantly altered bacterial taxa are prevalent in UK CD patients (Figure 5B). In particular, the Bacteroidaceae bacterial families were inversely correlated with several Caudovirales taxa in CD. This corresponded with a reduction in the relative abundance of these bacterial taxa in CD patients compared to household controls. In contrast, the Caudovirales were positively correlated with the Enterobacteriaceae and Pasteurellaceae bacterial families in CD; these bacterial families were increased in abundance in CD patients (Figures 5B). Positive correlations were also observed between the Caudovirales and Prevotellaceae in CD; however, there were no changes in the relative abundance of Prevotellaceae in CD, further indicating that we were not merely sequencing prophages. Fewer positive or negative correlations were observed in UC patients despite the significant expansion of Caudovirales bacteriophages and decreased bacterial diversity in these patients (Figures 3B and 4A), further suggesting the existence of disease-specific elements of the virome-bacterial microbiome relationship in UC versus CD.
Independent IBD Cohorts for Validation of Virome Findings
We considered the possibility that our observations in UK IBD patients were either geographically determined or not reproducible. Therefore, we performed bacterial 16S and VLP sequencing of stool samples from two additional cohorts of CD and UC patients (Figure 2, Tables S1 and S2). In these cases, household controls were not available, and so matched healthy controls were used. The first validation cohort was from Chicago and included 23 healthy control samples and 25 IBD samples (18 UC and 7 CD), including several samples that were part of the initial 454 analyses (Figure 1). For five of the Chicago UC patients we were able to acquire samples both during inactive and active disease (Figure 2B). Two household controls that matched UC patients from Chicago were included in these analyses as healthy subjects. The second validation cohort was from Boston and included 10 healthy control samples and 25 IBD samples (11 UC and 14 CD).
Relationship Between Caudovirales Richness and IBD Across Cohorts
As expected, significant reductions in bacterial diversity and richness were observed in CD and UC patients compared to healthy controls from both the Chicago and Boston validation cohorts (Figures 6A – 6C). MaAsLin analysis of bacterial taxa revealed significant associations with IBD and disease activity in both Chicago and Boston cohorts, although many fewer bacterial taxa were significantly associated with IBD than observed in the UK cohort, which included household controls (Tables S5 and S8).
Figure 6. Validation of Bacterial Dysbiosis in Two Additional Cohorts.
A and B) Faith’s phylogenetic alpha diversity (left) and bacterial species richness (right) based on 16S rRNA gene sequences in the stool of A) Chicago healthy controls, CD, and UC patients and B) Boston healthy controls, CD, and UC patients. Statistical significance was determined by the Kruskal-Wallis test with Dunn’s correction comparing all samples to all samples. * = p > 0.05, ** = p > 0.01. C) Plot of alpha diversity normalized to the average diversity in healthy subjects (top) and relative bacterial family abundance (bottom) for the Chicago and Boston cohorts. Healthy control, UC, and CD samples are indicated. Longitudinal samples from the same subject are grouped together. See also Tables S5, S7, and S8.
As observed in the UK cohort (Figure 3), a significant expansion of Caudovirales bacteriophages was observed in IBD patients from both validation cohorts (Figure 7). However, the specific relationships between bacteriophage richness and disease varied between cohorts. In Chicago, CD patients were Caudovirales rich compared to healthy controls (Figures 7A and 7B). This was evident when richness was assessed for both individual sequences and Caudovirales contigs. In the Boston cohort, both CD and UC patients had increased Caudovirales, with UC contigs being more enriched than CD (Figures 7A and 7B). We also detected unique Caudovirales taxa in CD and UC samples from both the Chicago and Boston cohorts (Figure 7C), which was anticipated given our observations in the UK samples and the high inter-individual virome diversity reported previously (Reyes et al., 2010). Multivariate analysis of the relative abundance of Caudovirales in the Chicago and Boston samples revealed several disease-specific associations (Tables S5 and S6). We were unable to complete a correlation analysis to associate bacteriophage to bacteria abundances (as in Figure 5) due to the inadequate number of bacterial taxa associated with disease diagnosis as determined by MaAsLin analysis (Table S8). Therefore we were unable to validate specific relationships between bacteriophage and bacterial taxa across our cohorts. Together these data indicate that Caudovirales bacteriophages were expanded in both CD and UC patients compared to household or healthy controls from three independent cohorts.
Figure 7. Validation of Caudovirales Expansion in Two Additional Clinical Cohorts.

A and B) Rarefaction curves of Caudovirales richness versus an increasing number of sub-samplings with replacement for the Chicago (top) and Boston (bottom) cohorts. A) Caudovirales richness based on individual sequences. B) Richness based on Caudovirales contigs. C) Venn diagram of the Caudovirales taxa in healthy control, CD and UC samples from Chicago (top) and Boston (bottom). N = the number of samples within each sub-group. See also Tables S4, S5, and S6.
Eukaryotic Viruses in IBD Cohorts
We also took advantage of the available data from the three IBD cohorts to analyze sequences from eukaryotic viruses. Anellovirus sequences were more prevalent in IBD samples compared to healthy controls (Table S4; anellovirus positive: Household controls = 0%, Healthy controls = 4.7%, CD = 27%, UC = 29%). However, anellovirus sequences from fecal samples were not detected in all patients and did not correlate with disease activity or drug treatment.
DISCUSSION
In this paper we demonstrate that disease-specific changes in the enteric virome occur in both major forms of IBD, Crohn’s disease and ulcerative colitis. This was observed in a cohort of patients in comparison to household controls with increased power to detect disease-associated changes in the metagenome, and validated in two independent and geographically distinct cohorts containing matched controls. The primary change in the virome associated with IBD was a significant expansion of the taxonomic richness of Caudovirales bacteriophages. Importantly, while this change was observed in both CD and UC, the viruses responsible for the change appeared to differ between the two diseases, suggesting that the virome is specific for CD versus UC. Comparison across cohorts revealed that enteric viromes were unique between individuals and between cohorts, which is consistent with previous reports on the incredible diversity of the human gut virome (Minot et al., 2011; Reyes et al., 2010). While the variability of gut bacteria and viruses in non-household controls and IBD patients makes it more difficult to observe specific relationships between individual bacteriophages and individual bacterial taxa, an increase in bacteriophage richness was consistently associated with IBD despite a decrease in bacterial richness and diversity.
Our data are consistent with reports that detected more Caudovirales bacteriophage sequences in intestinal washes and biopsy tissues of pediatric CD patients compared to non-inflammatory controls (Wagner et al., 2013) and enumerated more Caudovirales virions in CD biopsy washes by microscopy (Lepage et al., 2008). We believe that our observations reflect an expansion of infectious bacteriophages in IBD for two reasons. First, we sequenced VLP preparations enriched for virions. Second, an expansion of temperate bacteriophages integrated in bacterial genomes would be predicted to positively correlate with their bacterial hosts while we observe an inverse correlation. However, our data do not rule out the possibility that bacterial species harboring specific prophages are also expanded. Analysis of this will require full sequencing of the bacterial microbiome to complement analysis of bacterial taxa via analysis of 16S ribosomal RNA sequences.
Potential Role of Bacteriophages in IBD
In the human gut and in many ecosystems, the predominant bacteriophages are tailed, dsDNA Caudovirales and non-tailed, ssDNA Microviridae (Breitbart et al., 2003; Reyes et al., 2010). The biology of bacteriophages has been extensively reviewed (Brüssow et al., 2004; Clokie et al., 2011; Fortier and Sekulovic, 2013). The expansion in Caudovirales bacteriophage richness observed here could arise from the induction of prophage from commensal microbes or reflect the introduction of new viruses acquired from the environment, for example from food or contact with other people including household contacts. These changes might have significant consequences for the bacterial microbiome. For example, bacteriophages are primary drivers of bacterial diversity and fitness in different ecosystems (Brüssow et al., 2004). In the gut, bacteriophages are responsible for the horizontal transfer of genetic material among bacterial communities including those for pathogenesis (e.g. Cholera toxin, Pertussis toxin, Shiga toxin) and antibiotic resistance (Brüssow et al., 2004; Maiques et al., 2006; Zhang and LeJeune, 2008). Widespread bacteriophage induction, mutation, or introduction from external sources could effectively shuffle the deck of bacterial fitness and resistance genes. Secondly, the activation of latent prophages leads to the lysis of their bacterial hosts and can have profound ecological consequences (Weitz and Wilhelm, 2012). The intestinal microbiome has been shown to be sensitive to bacteriophage invasion, leading to changes in the abundance of specific gut bacterial species (Reyes et al., 2013). Lysis of bacteria would also be expected to release proteins, lipids, and nucleic acids that serve as pathogen-associated molecular patterns (PAMPs) and antigens that trigger inflammatory signaling cascades to induce cytokines, cellular infiltration and tissue damage. The development of animal models to test these predator-prey relationship(s) in IBD pathogenesis and intestinal inflammation will certainly be an important area for future investigation.
Another potential consequence of a change in enteric bacteriophages might result from direct interactions between these viruses and the mammalian host. For example, bacteriophages are able to translocate from the GI lumen to systemic sites in animals (Górski et al., 2006), CD patients and healthy controls (Parent and Wilson, 1971). Bacteriophages are also capable of inducing humoral immune responses (Uhr et al., 1962). Further, in vitro stimulation of macrophages with bacteriophage particles induces MyD88-dependent pro-inflammatory cytokine production (Eriksson et al., 2009). Chronic intestinal inflammation is the most basic element of IBD pathology, leading to the destruction of intestinal tissue and increased epithelial permeability. This leads to increased systemic exposure to the flow of microbial immunogens, potentially including those from bacteriophages and lysed bacterial cells, to maintain and further exacerbate inflammation. For these reasons, bacteriophages may serve as antigens or innate immune ligands that stimulate host immunity and inflammation.
The “Dark Matter” of the Virome
The metagenomic sequencing of samples enriched for intact virions has led to an appreciation of the incredible richness of enteric viruses in humans. Importantly, the methods we used here would not be expected to detect either enveloped viruses or RNA viruses, and so there may be much to learn about these aspects of the IBD virome. Prior attempts to characterize the virome using these methods were only able to assign 60% to 87% of VLP sequences or contigs to anything within sequence databases (Minot et al., 2013; Minot et al., 2011; Reyes et al., 2010). Across our cohorts, we were only able to assign on average 14% of VLP sequences to a viral database. The apparent discrepancy between our study and previous ones may be explained by our use of higher throughput Illumina-based sequencing, sequence databases or taxonomic assignment criteria. The percent identity of those VLP sequences that were assigned in this study varied greatly in a subset of sequences that were analyzed (40% to 100% identity; data not shown). This is a major issue for future studies as we were only able to report on the bacteriophages that are most closely related to taxa in the database. It is likely that additional viruses are present in our sequencing datasets but are not detected due to this limitation. An implication of this is that current databases lack sufficient depth for us to be able to link specific bacteriophages to individual bacteria or disease. This limitation will only be overcome by significant expansion of the bacteriophage sequences and annotations in available databases. This will not be a simple task and will require a global, coordinated effort to improve virus databases; it is clear that the Human Microbiome Project (http://www.hmpdacc.org/), an effort of similar scale, has improved the ability to classify and study bacteria. As databases improve, the resolution of our picture of bacterial microbiome-virome associations will improve. Many of the sequences that we did not assign using our viral database map to bacterial genomes (data not shown), an assignment complicated by the fact that many bacterial genome sequences contain prophages that have not been independently annotated. Addressing the misannotation of integrated prophages as ‘bacterial’ sequences will require development of new tools and significant improvement of viral databases containing a much greater diversity of fully annotated complete viral genomes.
Virome Implications for Disease Pathogenesis, Treatment and Monitoring
The primary therapeutic goal in IBD is to limit inflammation and halt or even reverse tissue destruction. In many cases, pharmacologic or bio-molecule therapies fail, and surgical resection of inflamed/damaged tissue is required. Thus, novel approaches are required to optimize IBD management. One potential approach is the manipulation of enteric bacteria through probiotics and prebiotics, which have had limited success in humans thus far (Butterworth et al., 2008; Naidoo et al., 2011; Shanahan and Quigley, 2014). Fecal transplantation from healthy donors or with defined bacterial cultures is an approach that is gaining traction due to its success in the treatment of recurring Clostridium difficile infections (van Nood et al., 2013). However, early attempts at curing bacterial dysbiosis in UC through fecal transplantation have not proven successful (Angelberger et al., 2013; Kump et al., 2013), perhaps due to the instability of the donor microbiome in IBD patients. It is intriguing to speculate that a disease-associated, taxonomically-rich virome in the recipient may interact with donor microbes to limit probiotic or fecal transplantation efficacy. Defining the virome before and during probiotic or fecal transplantation will be required to assess this possibility. It is notable that in animal models, eukaryotic viruses change intestinal biology and inflammation by acting in concert with host bacteria in a manner dependent on host genetics (Basic et al., 2014; Cadwell et al., 2010). Thus both eukaryotic viruses and bacteriophages may have a role in IBD through interactions with the bacterial microbiome. It will be important to more completely understand the interactions between bacteria and viruses and viruses and the host to be able to develop and personalize these approaches to managing IBD.
Our data also suggest that the specific expansion of bacteriophages in CD is associated with decreased bacterial diversity. The development of methods that block the infection of their bacterial hosts by these bacteriophages is worth investigation. Furthermore, the identification of disease-specific Caudovirales could prove useful in differentiating CD and UC in the ~10% of cases of IBD where the clinical phenotype is indeterminate between the two. Here we did not observe any significant changes in the virome in IBD patients over time as disease activity changed. However, larger cohorts with more frequent repeated sampling of both IBD patients and household controls are required to more fully assess enteric virome stability in IBD.
While IBD is associated with bacterial dysbiosis, additional, and much more common, diseases are also associated with changes in the bacterial microbiome. These include diabetes, obesity, metabolic diseases and cancer (Larsen et al., 2010; Ley et al., 2005; Nicholson et al., 2012; Qin et al., 2012; Shen et al., 2010; Turnbaugh et al., 2009; Turnbaugh et al., 2006; Zackular et al., 2013). Our data indicate that understanding the bacterial microbiome in these diseases likely requires concurrent analysis of the virome. More speculatively, we question whether bacterial microbiome changes in many disease are secondary to changes in emergence of temperate bacteriophages, or introduction of bacteriophages from food, the environment, or through human or animal contact. Thus, data presented here identifying inverse relationships between the bacterial microbiome and the enteric virome open up a new area of research in IBD, and perhaps other diseases that have been shown to be associated with changes in the bacterial microbiome.
EXPERIMENTAL PROCEDURES
Cohort Description
Stool samples were collected at four gastroenterology clinics: 1) Addenbrooke’s Hospital, University of Cambridge, UK; 2) Cedars-Sinai Hospital, Los Angeles, USA; 3) Rush University Medical Center, Chicago, USA; 4) Massachusetts General Hospital, Boston, USA. A detailed list of the subjects and a full description of each cohort is included in the Supplemental Information.
Virus-Like Particle Enrichment and Sequencing
Virus-like particle preparation
VLPs were enriched from pulverized human stool using a protocol based on previously described methods (Reyes et al., 2010; Thurber et al., 2009). Stool sample filtrates were treated with lysozyme and chloroform to degrade any unfiltered bacterial and host cell membranes. Non-virus protected DNA was degraded by treating with a DNase cocktail followed by heat inactivation of DNases. VLPs were lysed and nucleic acid was extracted.
VLP DNA was amplified and DNA was randomly fragmented by ultrasonication before Illumina library construction. An equimolar pool of 12 samples was sequenced on an Illumina MiSeq instrument.
Sequence processing and analysis
Adapters and low-quality bases were trimmed and reads were clustered at 95% identity. Unique sequences were queried against a customized viral database using BLASTx. Reads were assigned taxonomy using the lowest-common ancestor algorithm as implemented in MEGAN (v5.1.5) (Huson et al., 2011). Absolute read counts for selected viral taxa were exported from MEGAN and imported into R, data were normalized, and richness and diversity were calculated.
De novo contig assembly
Contigs were assembled using the IDBA_UD assembler (v 1.1.0) using minimum and maximum kmer lengths of 20 and 120, respectively (Peng et al., 2012). All contigs larger than 1,000 nucleotides were compared to a viral genome reference sequence database consisting of 5,500 viral genomes available in NCBI as of July 7, 2014 using dc_megablast.
Bacterial 16S rRNA Analysis
16S rRNA gene analysis
Stool total nucleic acid was extracted from aliquots of pulverized human stool, as previously described (Reyes et al., 2013) with minor modifications for lower throughput processing of human stool (Supplemental Information). Primer selection and polymerase chain reaction was performed following previously described methods (Caporaso et al., 2011). The final pooled samples were sequenced on the Illumina MiSeq platform. 16S analysis was done in QIIME (Quantitative Insights Into Microbial Ecology, version 1.8.0) (Caporaso et al., 2010) and OTU relative abundance, diversity and richness plots were generated in GraphPad Prism (version 6.0d).
Supplementary Material
Acknowledgments
HWV, SAH, and JMN were supported by R01 AI084887, R01 OD011170, grants 3132 and 274415 from the Crohn’s and Colitis Foundation, and grant IBD-0357 from the Broad Medical Foundation. TSS was supported by grant 3132 from the Crohn’s and Colitis Foundation. JMN was supported by NIH grant 5T32AI007163-35. MTB was supported by NIH grant 5T32CA009547. BCK was supported by NIH grant T32 HL007317-36. CLM was supported by NIH grant 5T32AI007172-34. AK and EAM were supported by R01 AT007143, DK071838 and AT001628. DPBM was supported by DK062413, DK046763-19, AI067068 and U54DE023789-01, grant HS021747 from the Agency for Healthcare Research and Quality, grant 305479 from the European Union, and The Leona M. and Harry B. Helmsley Charitable Trust. MP was supported by the NIHR Biomedical Research Centre awards to Addenbrooke’s Hospital and University of Cambridge School of Clinical Medicine. DW was supported by the NIH Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research grant U54 AI057160. DG and RJX were supported by U54 DE023798 and R01 DK092405. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR000448 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. We would like to thank the patients and relatives who contributed samples to this collection, and the team of research nurses and coordinators who supported this activity including Claire Dawson, Elizabeth Andersen and Martina Lofthouse (Cambridge, UK) and Gildardo Barron (Los Angeles, USA). We also acknowledge the NIHR Biomedical Research Centre award to support research infrastructure at Addenbrooke’s Hospital/University of Cambridge School of Clinical Medicine. We would also like to thank Jessica Hoisington-Lopez from the Center for Genome Sciences and Systems Biology at Washington University School of Medicine for her sequencing expertise.
Footnotes
Author contributions:
Study conception and design - JMN, SAH, HWV, MP, AK, EAM, DPBM, TSS
Recruitment of study subjects and acquisition of samples - MP, DPBM, PF, AK, EAM, LD, DW, JS, DG, RJX.
Acquisition of data - JMN, SAH, MTB, BCK, LD, CLM, CYL
Analysis and interpretation of data - JMN, SAH, GZ, HWV
Drafting of manuscript - JMN, SAH, A Kambal, HWV
Critical revision - JMN, SAH, HWV, MP, DPBM, AK, EAM, TSS, DW, AK, DG, RJX
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. The American Journal of Gastroenterology. 2013;108:1620–1630. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schroder B, Smoczek A, Jorns A, Wedekind D, Zschemisch NH, et al. Norovirus Triggered Microbiota-driven Mucosal Inflammation in Interleukin 10-deficient Mice. Inflammatory bowel diseases. 2014;20:431–443. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. Journal of Bacteriology. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiology and Molecular Biology Reviews. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth AD, Thomas AG, Akobeng AK. Probiotics for induction of remission in Crohn’s disease. The Cochrane Database of Systematic Reviews. 2008:CD006634. doi: 10.1002/14651858.CD006634.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nature immunology. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GGZ, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nature communications. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson F, Tsagozis P, Lundberg K, Parsa R, Mangsbo SM, Persson MAA, Harris RA, Pisa P. Tumor-specific bacteriophages induce tumor destruction through activation of tumor-associated macrophages. The Journal of Immunology. 2009;182:3105–3111. doi: 10.4049/jimmunol.0800224. [DOI] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier L-C, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013:4. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Wazna E, Dabrowska BW, Dabrowska K, Switała-Jeleń K, Miedzybrodzki R. Bacteriophage translocation. FEMS immunology and medical microbiology. 2006;46:313–319. doi: 10.1111/j.1574-695X.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Annals of Internal Medicine. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome research. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving PM, Gibson PR. Infections and IBD. Nature Clinical Practice Gastroenterology & Hepatology. 2008;5:18–27. doi: 10.1038/ncpgasthep1004. [DOI] [PubMed] [Google Scholar]
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, Gevers D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump PK, Gröchenig HP, Lackner S, Trajanoski S, Reicht G, Hoffmann KM, Deutschmann A, Wenzl HH, Petritsch W, Krejs GJ, et al. Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflammatory bowel diseases. 2013;19:2155–2165. doi: 10.1097/MIB.0b013e31829ea325. [DOI] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science (New York, NY) 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Colombet J, Marteau P, Sime-Ngando T, Dore J, Leclerc M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57:424–425. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Molecular Microbiology. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbé J, Penadés JR. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. Journal of Bacteriology. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome research. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo K, Gordon M, Fagbemi AO, Thomas AG, Akobeng AK. Probiotics for maintenance of remission in ulcerative colitis. The Cochrane Database of Systematic Reviews. 2011:CD007443. doi: 10.1002/14651858.CD007443.pub2. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Virgin HW. Kingdom-agnostic Metagenomics: The Importance of Complete Characterization of Enteric Microbial Communities. Gastroenterology. 2014;146:1459–1469. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent K, Wilson ID. Mycobacteriophage in Crohn’s disease. Gut. 1971;12:1019–1020. doi: 10.1136/gut.12.12.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics (Oxford, England) 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Perez-Brocal V, Garcia-Lopez R, Vazquez-Castellanos JF, Nos P, Beltran B, Latorre A, Moya A. Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin Transl Gastroenterol. 2013;4:e36. doi: 10.1038/ctg.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci U S A. 2013;110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F, Quigley EMM. Manipulation of the Microbiota for Treatment of IBS and IBD-Challenges and Controversies. Gastroenterology. 2014;146:1554–1563. doi: 10.1053/j.gastro.2014.01.050. [DOI] [PubMed] [Google Scholar]
- Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Rioux JD, Mizoguchi A, Saitoh T, Huett A, Darfeuille-Michaud A, Wileman T, Mizushima N, Carding S, Akira S, et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy. 2011;7:355–374. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Nava GM, Stappenbeck TS. Host genetic susceptibility, dysbiosis, and viral triggers in inflammatory bowel disease. Curr Opin Gastroenterol. 2011;27:321–327. doi: 10.1097/MOG.0b013e32834661b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Uhr JW, Dancis J, Franklin EC, Finkelstein MS, Lewis EW. The antibody response to bacteriophage phi-X 174 in newborn premature infants. The Journal of clinical investigation. 1962;41:1509–1513. doi: 10.1172/JCI104606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England Journal of Medicine. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Virgin HW. The Virome in Mammalian Physiology and Disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Maksimovic J, Farries G, Sim WH, Bishop RF, Cameron DJ, Catto-Smith AG, Kirkwood CD. Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflammatory bowel diseases. 2013;19:1598–1608. doi: 10.1097/MIB.0b013e318292477c. [DOI] [PubMed] [Google Scholar]
- Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. The ISME journal. 2014 doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz JS, Wilhelm SW. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 biology reports. 2012;4:17. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PloS one. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Haynes MR, Furlan M, Hanson N, Kirby B, Lim YW, Rainey PB, Schmieder R, Youle M, Conrad D, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. American journal of respiratory cell and molecular biology. 2012;46:127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. mBio. 2013:4. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. The Journal of infectious diseases. 2000;181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeJeune JT. Transduction of bla(CMY-2), tet(A), and tet(B) from Salmonella enterica subspecies enterica serovar Heidelberg to S. Typhimurium. Veterinary Microbiology. 2008;129:418–425. doi: 10.1016/j.vetmic.2007.11.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.