Abstract
In many cases cell function is intimately linked to cell shape control. We utilized endothelial cell branching morphogenesis as a model to understand the role of myosin-II in shape control of invasive cells migrating in 3D collagen gels. We applied principles of differential geometry and mathematical morphology to 3D image sets to parameterize cell branch structure and local cell surface curvature. We find that Rho/ROCK-stimulated myosin-II contractility minimizes cell-scale branching by recognizing and minimizing local cell surface curvature. Utilizing micro-fabrication to constrain cell shape identifies a positive feedback mechanism in which low curvature stabilizes myosin-II cortical association, where it acts to maintain minimal curvature. The feedback between myosin-II regulation by and control of curvature drives cycles of localized cortical myosin-II assembly and disassembly. These cycles in turn mediate alternating phases of directionally biased branch initiation and retraction to guide 3D cell migration.
Introduction
During migration in tissue in vivo or in culture in a 3D extracellular matrix (ECM), endothelial cells, fibroblasts, and tumor cells exhibit a characteristic complex shape that consists of a spindle-shaped cell body and arboreal, branched protrusions extending into the surrounding microenvironment 1–3. This branched morphology is critical to invasion and path-finding during angiogenesis, tissue repair, and metastasis. Endothelial cell branching morphogenesis is mediated by regulation of the acto-myosin cytoskeleton by both mechanical and biochemical cues 2,4–6. Previous studies have shown that actin polymerization dynamics power plasma membrane protrusion to drive branch formation, while myosin-II contractility inhibits branching 4,7. While much is known about the biophysical mechanism by which actin polymerization drives membrane protrusion to effect shape change 8, the basic principles by which myosin-II contractility locally effects membrane geometry to inhibit cell branching and control global cell shape is unknown. Three central questions remain unresolved regarding the control of 3D cell shape by myosin-II. First, how is the molecular-scale activity of myosin-II motors related to the cell-scale shape? Second, does cell shape feedback to regulate actomyosin? And third, how is actomyosin spatially and temporally controlled to mediate branching dynamics and guide invasive migration?
We utilized 4D imaging, computer vision and differential geometry to quantify cell shape and invasive migration of endothelial cells in 3D collagen ECMs. We found that myosin-II motor activity regulates micro-scale cell surface curvature to control cell-scale branch complexity and orientation. Myosin-II preferentially assembles onto cortical regions of minimal surface curvature while also acting to minimize local curvature. Perturbations of Rho-ROCK signaling or myosin-II ATPase function disrupt curvature minimization and branch regulation, but do not prevent curvature-dependent cortical assembly of myosin-II. Myosin-II contractility also controls branch orientation, possibly through differential association of myosin to outer low-curvature and inner high-curvature surfaces of branches, linking local curvature control to global directional control of migration. Thus, cell surface curvature minimization is a core mechanism that translates the molecular activity of myosin-II at the cortex into dynamic shape control for guiding invasive cell migration in 3D.
Results
Cell surface segmentation for defining quantifiable morphological parameters
To determine how myosin-II controls cell shape and branching morphogenesis in a 3D microenvironment, we utilized primary aortic endothelial cells (AECs) embedded in collagen gels. This recapitulates key morphologic and dynamic features of endothelial tip cell migration during angiogenesis in vivo 4. To visualize the shape of the cell surface, including thin cell protrusions, we used time-lapse 3D spinning disk confocal microscopy to image AECs derived from transgenic mice ubiquitously expressing Td-tomato-CAAX to label the plasma membrane (Figure 1A, B, Supplemental Figure 1A; Supplemental Movie 1). We developed a robust methodology for the complete segmentation and numerical representation of the cell surface. To allow accurate segmentation of both dim, thin protrusions as well as the bright, thick cell body, we combined a 3D Gaussian partial-derivative kernel surface filtering algorithm with a self-adjusting high intensity threshold that allowed the processing of variable image conditions without user intervention (Figure 1C, Supplemental Methods and Supplemental Figure 1B–I). The resulting cell surface representations were used for quantification of two types of features that describe cell morphology during migration in 3D: (1) the “morphological skeleton” (Supplemental Movie 2) to quantify cell-scale aspects of branching topology (Figure 1D); and (2) the local cell surface curvature to quantify morphology nearer to the molecular length scale of actomyosin contractile units 9.
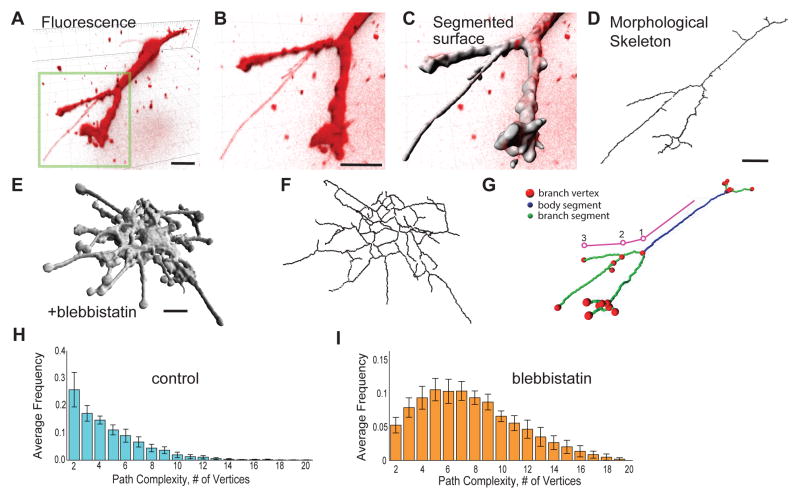
Figure 1. Quantification of cell morphological skeleton shows that myosin-II limits branch complexity in 3D.
(A–B) Living AEC in a 3D collagen gel expressing Td-tomato-CAAX imaged by spinning disk confocal microscopy displayed as a 3D shadow projection. Bars = 10μm. (B) Rotated higher magnification of (A) to show thin branches. (C) Result of computational segmentation, with dim, thin protrusions accurately segmented. (D) Thinning of segmented volume to create a morphological skeleton. Segmented surface (E) and morphological skeleton (F) of AEC treated with 20 μM blebbistatin to inhibit myosin-II. Bars = 10 μm. (G) Parameterization of branch structure into a single main body segment (blue), branch segments (green), and vertices (red dots). Branch path complexity is calculated by following any path from a point on the skeleton at the body center to a terminal branch segment and counting vertices along the path; one example path shown in pink. (H, I) Distribution of branch path complexity in control (H) and blebbistatin-treated (I) AECs. Data pooled from n=18 cells for control, n=16 cells for blebbistatin. Error bars show +/− 1.96 SEM (approximate 95% confidence interval).
Myosin-II activity inhibits dendritic branching
To investigate how myosin-II activity regulates global branch organization, we derived from the morphological skeleton the number of branch tips and the branch complexity. Inhibition of myosin-II ATPase activity with blebbistatin induced a complex cell morphology with an extensive network of thin branches (Figure 1E, F), as noted previously 4. As a measure of the branch complexity, we analyzed the number of branch vertices along the path of each branch from the center of the skeleton outwards (Figure 1G). This showed that blebbistatin treatment promoted dendritic branching and increased branch path complexity (Figure 1H–I) indicating that myosin-II reduces cell branch number by minimizing the number of branch vertices. Furthermore, the dendritic nature of blebbistatin-treated cell morphology indicates that myosin-II inhibition allowed branching to occur not only on the cell body, but at any point on the branch tree (Figure 1E), suggesting that the regulation of branched cell shape occurs via myosin-II acting locally at all points on the branched cell surface.
Myosin-II suppresses branch tips and bases by minimizing local cell surface curvature
To determine how myosin-II controls cell shape locally, we mapped cell surface curvature (Figure 2A–C, Supplemental Methods Figure 2A–F) and determined the role of myosin-II in its regulation. At each point on the cell surface, we determined the principal curvatures, κ1 and κ2, being the minimal and maximal values of curvature along two orthogonal axes, respectively, in the plane tangential to the surface. We specified a minimal curvature scale that excluded small surface fluctuations due to image noise and nano-scale processes like endo- and exocytosis (Supplemental Methods and Supplemental Figure 2E, F). We verified the accuracy of our curvature measurements by imaging and analysis of fluorescent microspheres of known size embedded in collagen gels (Supplemental Figure 2G–I), and excluded artifacts due to the microscope point-spread function geometry via simulations (Supplemental Material and Supplemental Figure 2J–M).
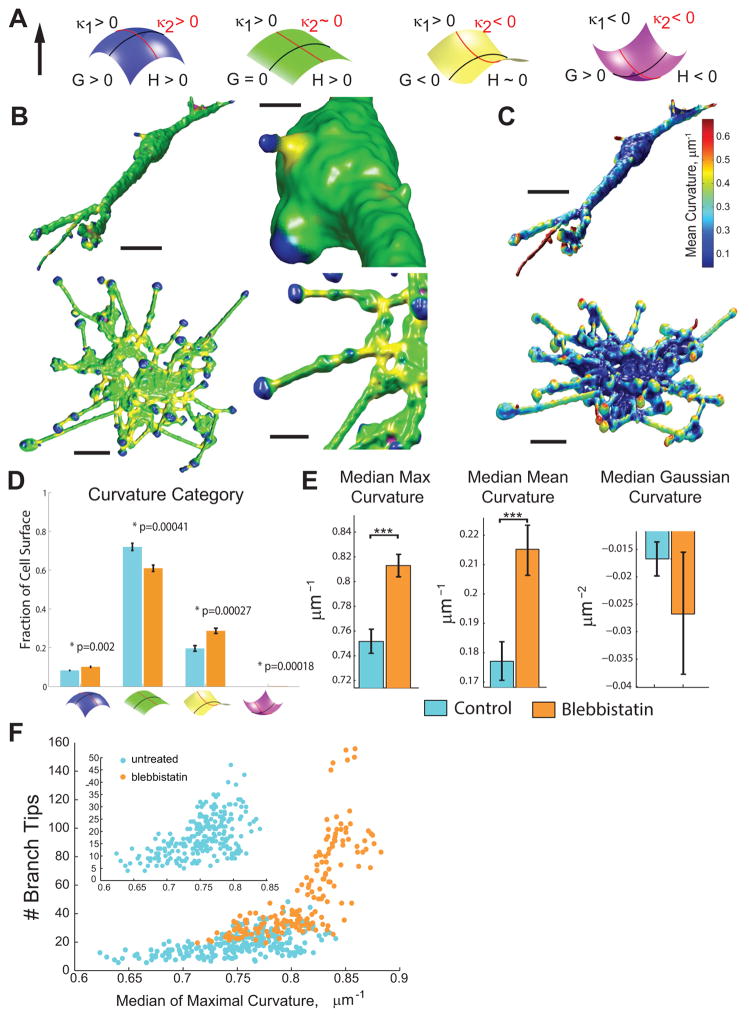
Figure 2. Myosin-II regulates cell surface curvature locally.
(A) Schematic of the classification scheme for cell surface curvatures. Local curvature of a surface point is parameterized by two principal curvatures in orthogonal tangential directions, κ1 and κ2, where κ1=1/r1 (black lines) and κ2=1/r2 (red lines). Blue: A surface with both κ1 and κ2 > 0 resulting in a mean curvature, H=(κ1+ κ2)/2 >0 and a Gaussian curvature G = κ1*κ2 > 0, referred to as a hill. Green: cylindrical surface (κ1 > 0 and κ2 ~ 0), referred to as a tube. Yellow: κ1 and κ2 are of opposite signs, referred to as a saddle. Purple: concave surface with κ1 and κ2 < 0, referred to as an invagination. (B) Examples of different classes of surface curvatures mapped onto cell surface using the color scheme in (A). Top row: control AEC; Bottom row: blebbistatin-treated AEC. Bars = 10 μm on left, 3 μm in middle. (C) Mean curvature values mapped to cell surface of control (top) and blebbistatin treated (bottom) cells from (B). Mean curvature value scale bar shown on right. (D) Comparison of the fraction of the total cell surface in each of the surface curvature categories described in (A) for controls (light blue, n=16 cells) and cells treated with 20 μm blebbistatin (orange, n=16 cells). Error bars show S.E.M., p-values given above bars tested using Wilcox-rank sum test. (E) Comparison of mean values of median maximal local curvature (maximum of |κ1|, |κ2|), absolute Gaussian curvature (“G”), and mean curvature (“H”) for multiple cells in control (light blue, n=16 cells) or blebbistatin-treated (orange, n=16 cells) conditions. Error bars show +/−S.E.M., *** indicates p<0.01 (p=5.37×10−5 for max, p=1.38×10−3 for mean and p=.396 for Gaussian curvature using Welch’s two-tailed t-test). (F) Time lapse imaging and analysis of the number of branches versus median maximum curvature for live AECs. Each point represents values for an individual cell at one time point. Control (blue, n=246 time points, 14 cells) and myosin inhibition (orange, n=570 time points, 27 cells) conditions. Inset shows data for control cells only.
Next, we categorized local surface curvature into four geometric classes relevant to cell shape features (Figure 2A, Supplemental Figure 3A–D and Supplemental Methods), and examined the role of myosin-II in their regulation. These included hills (κ1 and κ2 are both positive), tubes (κ1 is positive and κ2 is close to zero), saddles (κ1 is positive and κ2 is negative), and invaginations (κ1 and κ2 are both negative). Mapping these classes on cell surface renderings showed that most of the surface of the cell body and branches were tube-like (Figure 2B, D, Supplemental Movie 3, Supplemental Figure 3C, D), hills localized primarily to branch tips, saddles localized to branch bases, and – although rare – invaginations localized to junctions of multiple branches oriented at acute angles to each other (Figure 2B, Supplemental Figure 3C, D). To determine how myosin-II activity contributed to these shape features, we analyzed AECs treated with blebbistatin. This showed that compared to control, inhibition of myosin-II increased the fraction of hills, invaginations, and saddles, demonstrating that myosin-II not only regulates overall branching, but also suppresses salient surface features like branch tips and bases (Figure 2B, bottom row, Supplemental Movie 4, Figure 2D).
We then determined the role of myosin-II in controlling the magnitude cell surface curvature. Curvature at any point on 3D surfaces can be described by either “mean curvature” (H= (κ1+κ2)/2), “Gaussian curvature” (G= κ1*κ2), or “maximal curvature” (maximum of |κ1|, |κ2|). Mapping mean curvature on control cells allowed visualization of the low mean curvature at branch bases, which increased as branches extended away from the cell body and their diameter decreased, and was highest at branch tips (Figure 2C). Mapping mean curvature on blebbistatin-treated cells that displayed extensive thin dendritic branches suggested that inhibition of myosin-II increased curvature over the entire cell surface (Figure 2C). To test this, we quantified the maximal, mean, and Gaussian curvatures at all points on the surface of control and blebbistatin-treated cells and determined the per-cell median values. This showed that blebbistatin treatment significantly increased the median maximal and mean surface curvatures per cell (Figure 2E). Gaussian curvature was also increased by myosin-II inhibition, although not significantly because in both control and blebbistatin treated cells, most surface points belong to the tube class, where κ2 is ~0 and hence the product κ1*κ2 is insensitive to changes in the tube diameter, which are reflected in changes of κ1. Together, this shows that myosin-II activity minimizes local cell surface curvature.
Our observations that myosin-II inhibition affects both branch structure and surface curvature suggest that these global and local morphological parameters are interdependent. Qualitatively, this is not surprising, because the cell boundary describes a closed surface geometry. We tested directly this by correlating the median of the maximal curvature with the branch number on a per-cell basis, irrespective of treatment with blebbistatin (Figure 2F). Since the cell is largely tube-like, we focused on maximal curvature because it best represents the inverse of tube diameters. We found that both within the natural variation of myosin-II activation level in control cells as wells as in myosin-II-inhibited cells, branch number and surface curvature correlated over a wide range of values (Spearman’s ρ= 0.77, p < 1×10−6), confirming that these two independently measured parameters are correlated. We conclude that our multi-scale measurements are indeed interdependent.
Cell surface curvature negatively regulates myosin-II cortical localization
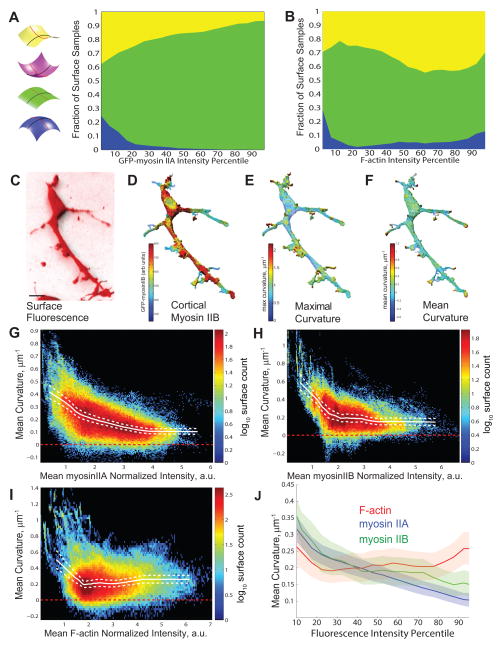
Our finding that myosin-II regulates cell surface curvature suggests that myosin-II localization could be linked to curvature. Therefore, we determined the whether myosin-II cortical localization and local surface curvature in freely migrating cells. AECs from knock-in transgenic mice expressing GFP-myosin-IIA or GFP-myosin-IIB under endogenous promoters 4,10,11 were fixed in collagen gels and stained with Cell Tracker CM-DiI and fluorescent phalloidin to label the plasma membrane and F-actin, respectively (Supplemental Figure 3F, H). We determined the local cortical myosin-IIA and F-actin intensities and corresponding local surface curvature, carefully controlling for sampling artifacts (Supplemental Methods, Supplemental Figure 4A–L), and then categorized these surface curvatures. This showed that the highest myosin-IIA concentration was associated with tube-like and saddle-shaped surfaces, with hills exhibiting very low myosin-IIA (Figure 3A). In contrast, F-actin exhibited a broad range of concentrations in tubes, saddles, and hills that are presumably the tips of actin-based protrusions (Figure 3B, Supplemental Figure 3E–H). Mapping cortical GFP-myosin-IIB intensity and local mean curvature on cell surface renderings (Figure 3C–F) confirmed these results for GFP-myosin-IIA.
Figure 3. Cortical myosin-II associates with minimal surface curvature.
(A, B) Fraction of the population of surface points of a given cortical mean myosin-II-GFP (A) or F-actin (B) intensity, whose curvature is categorized according to the color scheme shown (n=268844 points on 25 cells for A, n=230475 points on 25 cells for B). Note that there were a negligible number of points in the “invagination” category. (C–F) AEC in 3D collagen gel expressing Td-tomato-CAAX and myosin-IIB-GFP. Bar = 10μm. (C) 3D shadow projection of Td-tomato-CAAX fluorescence. (D–F) Color-encoded (highest= red, lowest= blue) normalized values of (D) mean cortical myosin-IIB-GFP intensity; (E) local absolute maximum curvature; and (F) local mean curvature; mapped onto the segmented cell surface. (G–I) Distributions of mean cortical GFP-myosin-IIA, GFP-myosin-IIB, or phalloidin stained F-actin intensity as a function of mean surface curvature for all surface points (n=268,844 points from 15 cells for G; n=167,449 points from 14 cells, for H, and n=230,475 points from 15 cells for I). Color scale indicates log10 of number of surface point samples; solid white line, mean curvature value for each intensity value; dashed line, bootstrapped 95% confidence intervals of the mean. (J) Mean curvature plotted versus mean intensity as a percentile rank for cortical F-actin, myosin-IIA and myosin-IIB, as for solid lines in (G–I). Faded color background areas indicate the 95% confidence limits by subsampled bootstrap for each curve (Supplemental Methods).
To further analyze the distribution of cortical myosin-IIA, IIB, and F-actin relative to surface curvature, we generated 2D histograms of mean curvature versus cortical intensity of GFP-myosin-IIA (Figure 3G), GFP-myosin-IIB (Figure 3H) and F-actin (Figure 3I) over all surface points. This revealed a strong negative correlation between myosin-IIA and mean curvature and a slightly weaker negative correlation between myosin-IIB and mean curvature (myosin-IIA, Spearman’s ρ= −0.50, p < 1×10−6; myosin-IIB, Spearman’s ρ= −0.32 p < 1×10−6, Figure 3G, H, Supplemental Figure 4A–M and Supplemental Methods). In contrast, F-actin was associated with a broad range of mean surface curvatures, with a weak positive correlation (Spearman’s ρ= 0.17 p < 1×10−6, Figure 3I, J, Supplemental Figure 3E–H). We conclude that myosin-II localization in cells in 3D is not dominated by the total F-actin content at the cortex, but is specifically associated with cell surface areas of lower curvature.
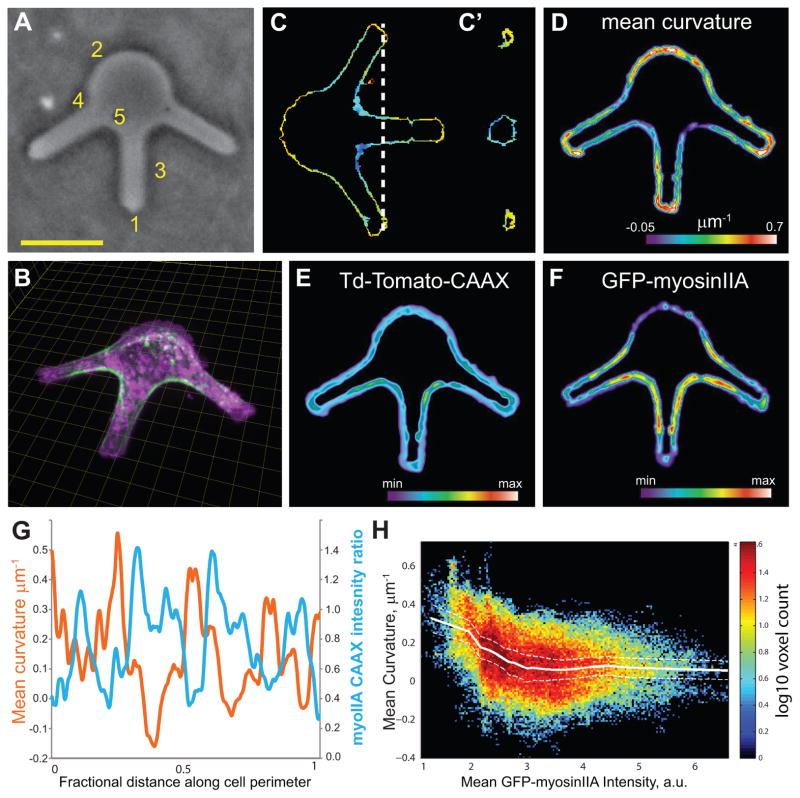
Our findings indicate that cortical myosin-II concentration correlates with cell surface curvature, suggesting that curvature could regulate myosin-II localization. To test this directly, we externally enforced 3D cell shape and analyzed local cortical myosin-II concentration with respect to surface curvature. Endothelial cells expressing GFP-myosin-IIA and tdTomato-CAAX were cultured in micro-fabricated agarose chambers assembled on the surface of collagen-coated coverslips (Figure 4A, B). The walls of the chamber were non-adherent, yet constrained cells to a simple branched shape and imposed varied surface geometries (Figure 4A). Cross-sections of branches showed that the lateral constraint imposed cylindrical geometries as the cell filled the chamber (Figure 4C, C′). Registering multiple constrained cells in 3D allowed determination of the relationship between average imposed surface curvature (Figure 4D) and average cortical GFP myosin-IIA recruitment (Figure 4F). We discounted the dorsal and ventral cell surfaces and limited our analysis to surfaces constrained by the non-adherent agarose walls. This showed that the CAAX plasma membrane marker localized relatively uniformly to the cell surface independent of local curvature (Figure 4E). In contrast, GFP-myosin-IIA was absent from high curvature branch tips, low along the unbranched rear of the cell body, but was enriched in areas of low mean curvature at the bases of branches and along their shafts (Figure 4F). Plotting the ratio of normalized intensities of GFP-myosin-IIA or membrane label and the local mean curvature as a function of position along the confined cell edge confirmed this, and indicated a strong negative correlation between myosin-IIA localization and curvature (Spearman’s ρ = −0.71, p < 1×10−5; Figure 4G and Supplemental Figure 5A). Quantification of GFP-myosin-IIA intensity versus mean curvature for all points on the confined surfaces of multiple cells showed a similar inverse correlation (Spearman’s ρ = −0.23, p < 1×10−6, Figure 4H) as was observed for cells migrating freely in 3D collagen ECMs, suggesting that low mean curvature is sufficient to recruit myosin-IIA. In contrast, the membrane label showed no correlation between intensity and curvature (Spearman’s ρ = −0.093, p=8.8 e-164, Supplemental Figure 5B). In summary, myosin-II is recruited to minimally curved cortical surfaces in freely migrating cells, and by imposing cell shape externally, we show that local cell surface curvature negatively regulates myosin-II localization to the cortex. Together with the finding that disruption of myosin-II activity increased cell surface curvature, these results suggest a positive feedback loop in which myosin-II minimizes local curvature to promote its recruitment to the cortex, thus maintaining low cell surface curvature.
Figure 4. Externally imposed local curvature modulates myosin-IIA localization to the cortex.
Human umbilical vein endothelial cells expressing Td-Tomato-CAAX and myosin-IIA-GFP were plated into microfabricated agarose chambers to constrain their 3D shape. (A) Phase-contrast image of 30 μm deep agarose chambers, the vertical walls of which enforce five regimes of decreasing mean curvature: High positive mean curvature at tips (1) to lowest mean curvature at saddle-like structures (5). Mag bar= 20μm, applies to (A, C,D, E). (B) Cells expressing GFP-myosin-IIA (green) and Td-Tomato-CAAX (purple) were subjected to 3D spinning disk confocal microscopy, and maximal intensity projection of a 3D stack (with rotation) is shown; grid spacing is 5μm. (C) X–Y cross-section of an example cell with segmented local mean surface curvature calculated from cell surface renderings of Td-Tomato-CAAX and displayed according to the heat map shown in D. (C′) shows YZ cross-section at position indicated by dashed line in (C). (D–F) Averaged measurements at constrained surfaces of multiple cells (n=6). (D) Mean Curvature (color scale in μm−1; length of color bar is 40 μm); (E) Td-Tomato-CAAX intensity; (F) GFP-myosin-IIA intensity. In (E) and (F); color scales for both are in arbitrary units. (G) Averaged GFP-myosin-IIA:iTd-Tomato-CAAX intensity ratio (blue), and mean curvature (red) as a function of position along the perimeter of an agarose chamber (n= 6 cells). (H) Mean cortical GFP-myosin-IIA intensity as a function of mean surface curvature for all confined surface points of multiple cells in individual chambers. Color scale indicates log10 of number of surface point samples, n= 95126 surface points sampled from 6 cells. Solid white line is mean curvature for each intensity bin; dotted white lines show bootstrapped 95% confidence interval of mean.
Myosin-II assembly at the cortex in response to low curvature is independent of contractility
We then sought to determine how cell surface curvature negatively regulates myosin-II localization. We hypothesized three possible mechanisms by which curvature could control local assembly of cortical myosin-II: 1) curvature-dependent upstream signaling; 2) tension-dependent regulation of myosin-II assembly; or 3) curvature-dependent stabilization of cortical binding. To test whether myosin-II was regulated by signaling to control its local assembly/activation, we first determined if myosin-II phosphorylation was spatially regulated. We immuno-localized myosin-II regulatory light chain phosphorylated on S19 (pS19-MRLC) in cells expressing GFP-myosin-IIA 12,13. Co-localization analysis in 3D revealed no substantial difference in localization of the two markers, suggesting that differences in myosin-IIA localization or its association with curvature could not be accounted for by local regulation of phosphorylation (Supplemental Figure 6C). We perturbed signaling upstream of pS19-MRLC and determined the effects on curvature-dependent localization of myosin-II. We inhibited either ROCK, which directly phosphorylates S19-MRLC 14, with Y-27623 (Figure 5A), or its upstream activator, Rho, with exoenzyme C3 (Figure 5B) in cells expressing GFP-myosin-IIA. Like blebbistatin, these agents increased cell branching and branch path complexity (Supplemental Figure 6A), suggesting that activation of myosin-II via Rho-ROCK signaling regulates branching. In spite of their effects on cell shape changes, these inhibitors had no effect on the inverse correlation between cortical myosin-IIA localization and local mean surface curvature, although, as expected, the curvature distribution was shifted to higher values (Figure 5D, E) compared to control. Similar results were found for analysis of cells expressing GFP-myosin-IIB, where treatment with Y-27623 or ML-7 (that blocks myosin light chain kinase) both increased cell branching and curvature, but preserved the negative correlation between cortical GFP-myosin-IIB intensity and mean curvature (Supplemental Figure 6B). None of the treatments altered the relationship between actin localization and curvature (Figure 5H). Thus, myosin-II assembles at the cortex in a curvature-dependent manner, independent of signals that control regulatory light chain phosphorylation. This suggests that local signaling and activation of myosin-II, although required for cell shape regulation, is not responsible for localized myosin-II assembly at the cortex.
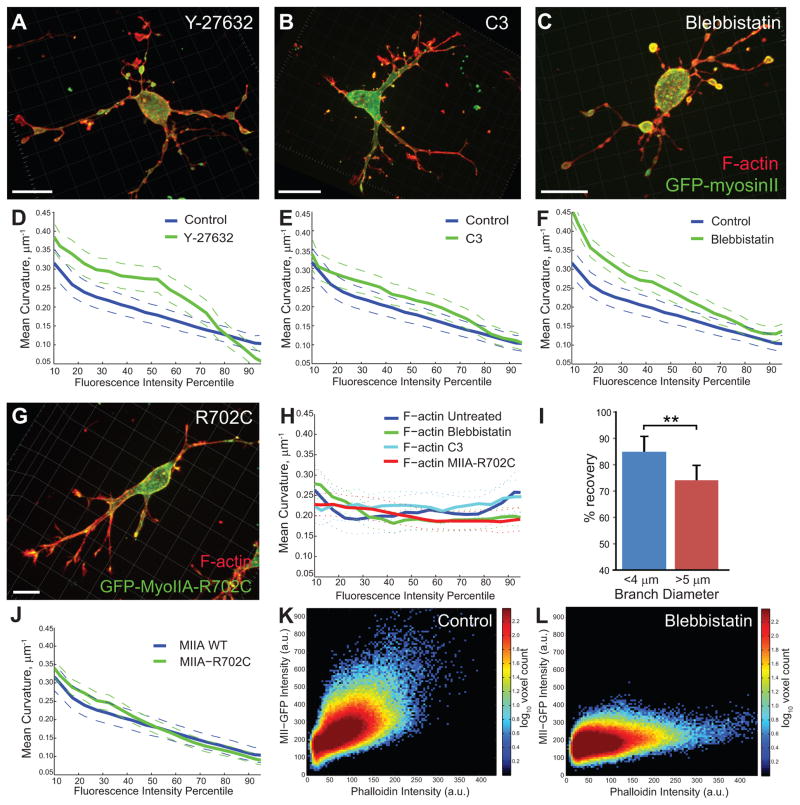
Figure 5. Curvature-dependent myosin-II association with the cell cortex is independent of myosin-II motor activity.
(A–C) AECs expressing GFP-myosin-IIA (green) in a 3D collagen gel exposed to 2 mM y-27632 (A), 100 μg/ml C3-exoenzyme (B), or 20 μM blebbistatin (C) were fixed and stained with fluorescent phalloidin (red). Maximal intensity projections of 3D reconstruction are shown. (D–F) Mean curvature plotted versus mean intensity as a percentile rank for cortical myosin-IIA in the presence (green) or absence (blue) of inhibitors indicated. Control data is re-plotted on each graph for comparison. Solid lines show mean curvature value for each intensity value; dashed lines show bootstrapped 95% confidence about the mean. n=7 cells, 268,844 points for control; n=7 cells, 393282 points for Y-27632; n=7 cells, 379475 points for C3; and n=15 cells, 968398 points for blebbistatin. (G) An AEC expressing GFP-myosin-IIA-R702C (green) in a 3D collagen gel was fixed and stained with fluorescent phalloidin (red). Maximal intensity projection of 3D reconstruction is shown. (H) Mean curvature plotted versus cortical F-actin mean intensity as a percentile rank for all conditions in A–G,J. (I) AECs in a 3D collagen gel expressing myosin-IIA-GFP were subjected to localized fluorescence photobleaching and recovery and imaged by 3D spinning disk confocal microscopy. Percent fluorescence recovery at the cortex in small (<4μm) and large (>5μm) diameter branches (n =18 for small branches, n=18 for large branches, error bars show standard deviation, p<0.001 using Students two-tailed t-test) is shown. (J) Mean curvature plotted versus mean intensity as a percentile rank for wild-type GFP-myosin-IIA (blue) or GFP-myosin-IIA-R702C (green). n=19 cells, 797766 points for R702C. Note wild type data is reproduced from (C) control data for comparison. Bars in micrographs equal 10 μm. (K, L) Direct comparison of local cortical F-actin and myosin-IIB in the presence (L) and absence (M) of blebbistatin shown in 2D per-voxel distributions. While the ratio (slope) of myosin-II to F-actin is lowered by blebbistatin, much of the myosin-II remains associated with F-actin at the cortex.
Because tension in a surface also decreases curvature, we tested whether acto-myosin-mediated cortical tension regulated the curvature-dependence of myosin-II localization. We inhibited motor activity either pharmacologically or with a mutation in myosin-IIA heavy chain, R702C, which reduces ATPase activity and causes hematologic and kidney defects in humans and mice 15,16. Analysis of the curvature-dependence of GFP-myosin-IIA or GFP-myosin-IIB cortical localization in AECs treated with blebbistatin (Figure 5C) showed that even in the presence of blebbistatin, local myosin-II intensity inversely correlated with mean curvature (Figure 5F, Supplemental Figure 6B). We performed similar analysis on AECs derived from mice heterozygous for GFP-R207C-myosin-IIA. Examination of AECs in 3D suggested this mutant induced a slight elevation in cell branching (Figure 5G). However, analysis of localization relative to mean curvature demonstrated that GFP-myosin-IIA-R702C was still localized to the cortex with curvature dependence (Figure 5J). Again, these perturbations did not affect the curvature-independence of cortical F-actin (Figure 5H). Thus, our data show that motor activity is not required for myosin-II’s curvature-dependent localization, suggesting that the association of myosin-II with low-curvature cortical surfaces is independent of actomyosin-mediated cortical tension.
We then sought to determine whether curvature regulated the stability of cortical binding by myosin-II. We analyzed the relationship between curvature and the stability of GFP-myosin-IIA association with the cortex by fluorescence recovery after photobleaching. We measured the fraction of fluorescence recovery in a bleached region on the cortex and compared this value in branches of large (low maximal curvature, d>5 μm) and small (higher maximal curvature, d<4 μm) diameter. This showed that a larger fraction of GFP-myosin-IIA was stably associated with the cortex (less recovery of fluorescence) in low- compared to high-curvature branches (Figure 5I, p<0.001). Hence, myosin-IIA association with the cortex is stabilized by lower curvature
The perturbation experiments above provide insight into the mechanism of cortex binding by myosin-II. Blebbistatin blocks myosin-II ATPase activity by locking myosin-II in a weakly bound state 15. Although blebbistatin did not affect the inverse correlation between myosin-II localization and curvature (Figure 5F), analysis of F-actin and myosin-IIA co-localization showed that myosin-IIA was only partially displaced from cortical F-actin by blebbistatin (Figure 5K, L). This suggests that actin binding is critical to cortical association of myosin-II. Together, these data show that curvature regulates myosin-II’s localization at the cortex not by local control of signaling or contractility, but by stabilizing myosin-II’s association with low-curvature regions of the cortex, possibly mediated by F-actin binding.
3D cell migration is mediated by branching/debranching cycles that are driven by cycles of myosin-II cortical dissociation/association
Our results show that myosin-II regulates and responds to cell surface curvature. To approach how this mediates dynamic control of cell shape during migration in 3D, we analyzed the temporal relationships between parameters of cell movement, branching, cell surface curvature, and myosin-II recruitment to the cortex. We determined the morphological skeleton and local surface curvature over time in AECs expressing Td-tomato-CAAX during migration in 3D collagen (Figure 6A, Supplemental Movie 5). Tracking of the cell center (Supplemental Figure 6D–F and Supplemental Movie 6)) showed that cells moved in a pulsatile manner, speeding up and slowing down at varying intervals (Figure 6B). Autocorrelation analysis demonstrated that cell speed fluctuations were not due to positional noise, but represented stochastic pulses in cell instantaneous velocity (Supplemental Figure 6G). Analysis of cell center movement relative to the evolution of the morphological skeleton in 3D over time showed that branch number and cell center velocity appeared out of synch (Figure 6B). Indeed, temporal cross-correlation between total branch radius (branch number weighted by branch radius) and cell center velocity indicated significant negative correlations at all negative time lags, but a strong positive correlation peak at a positive time lag of ~1200 sec. This indicates that branch number increases during periods of low cell velocity, and branch pruning precedes rapid cell body advance by ~20min (Figure 6C). Similarly, total branch complexity (sum of all branch vertices per time point) also fluctuated out of synch with cell center velocity (Figure 6D). However, in this case, the temporal cross-correlation exhibited a peak of negative correlation at zero time-shift, indicating that dendritic branching impedes cell movement (Figure 6E). In support of this, for individual cells, the mean total branch complexity over time was inversely correlated with mean speed over time (Figure 6F). Thus, AECs migrate in a 3D microenvironment by alternating between phases of dendritic branch expansion, which inhibits cell movement, followed by debranching to allow cell body advance.
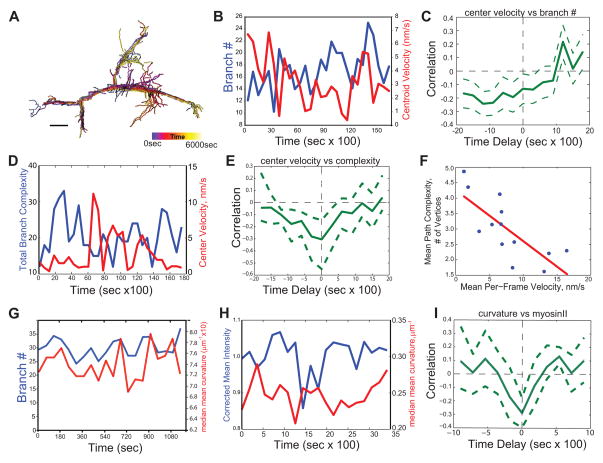
Figure 6. Myosin-II dynamically associates with the cortex to control cycles of AEC branching and movement by local minimization of surface curvature to guide cell migration.
Living AECs migrating in a 3D collagen gel expressing Td-Tomato-CAAX (and myosin-IIA-GFP where indicated) were imaged by time-lapse 3D spinning disk confocal microscopy. (A) Evolution of 3D morphological skeleton, color encoded according to elapsed time, (bar = 10μm). (B) Time courses of branch number (number of branch tips, blue) and instantaneous cell centroid velocity (red) for a representative AEC. (C) Temporal cross-correlation analysis of branch number versus cell center velocity (data pooled from n=8 cells, > 180min each). Positive delay indicates that change in branch number precedes change in velocity. (D) Time courses of cell branching complexity (total number of branch vertices, blue) and centroid velocity (red) for a representative migrating AEC. (E) Temporal cross-correlation analysis of cell branching complexity versus centroid velocity (data pooled from n=8 cells, > 180 min each). Positive delay indicates that change in velocity precedes change in cell branching complexity. (F) Mean cell branching complexity versus mean velocity for individual AECs (blue circles, means determined from 100 min time-series per cell). Red line, best linear fit; slope = −.21 vertices/nm/s, slope 95% confidence interval −.32 to −.11. (G) Time courses of branch number (number of branch tips, blue) and median mean curvature (red) for a representative AEC. (H) Plot of photobleach-corrected mean intensity of myosinIIA-GFP at the cortex (blue) versus the median mean curvature (red) of a region of a branching AEC over time. (I) Temporal cross-correlation analysis of local mean cortical myosin-IIA-GFP intensity versus local mean curvature (data pooled from n=11 regions from 10 cells, 20 time points each). Positive delay indicates that curvature change follows myosin-II change. In C, E, and I: Solid line, correlation; dashed lines, 95% confidence interval determined by 1000 bootstrap estimations of correlation functions (see Supplemental Methods).
Because cell branching and cell surface curvature are linked (Figure 2F), the above result predicts that curvature would fluctuate similarly to branching during migration. In individual cells, as branch number varied over time, the median mean surface curvature varied in synch (Figure 6G). Since myosin-II localization and curvature are inversely correlated (Figures 3 and 4), this further predicts that myosin-II association with the cortex would fluctuate out-of-phase with curvature during migration. Time-lapse 3D imaging (60 sec intervals) of GFP-myosin-IIA and tdTomato-CAAX in AECs migrating in 3D collagen gels and analysis of mean surface curvature and cortical GFP-myosin-IIA intensity showed that mean cortical GFP-myosin-IIA intensity and median mean curvature both fluctuated over time (Figure 6H, Supplemental Movie 7). Temporal cross-correlation between myosin-IIA intensity and median mean curvature within dynamically active regions of the cell cortex (Supplemental Movie 8) displayed a significant negative correlation at zero time lag, indicating that cortical myosin-IIA concentration increased and curvature decreased in synchrony (Figure 6I). Hence, during cell migration, cycles of exploratory branching followed by debranching and subsequent cell body advance are associated with cycles of myosin-II dissociation from the cortex that allow plasma membrane curvature, followed by myosin-II cortical recruitment that promotes curvature minimization.
Myosin-II controls branch orientation to mediate directional cell movement in 3D
In addition to regulating cell branching, myosin-II activity also promotes directional persistence of migration in 3D 4,7. Therefore we analyzed the role of myosin-II in controlling branch directionality in AECs expressing TdTomato-CAAX during migration in 3D collagen. From the morphological skeleton, we determined branch angle relative to the direction of cell center movement. Plotting mean branch angle over time showed that, similar to branch number (Figure 6B) and branch complexity (Figure 6D), mean branch angle also fluctuated over time (Figure 7A). Rose plots of mean branch angle relative to the direction of cell movement showed that branches with near normal angles relative to the cell axis were rare, indicating that branch angle was biased along the axis of cell movement (Figure 7B). Treatment of cells with blebbistatin removed this bias, and fully randomized branch angle (Figure 7B). Together, our results indicate that cells migrate by cyclically varying branch angle relative to the direction of cell movement, and maintain directionality by myosin-II-mediated minimization of branch angle.
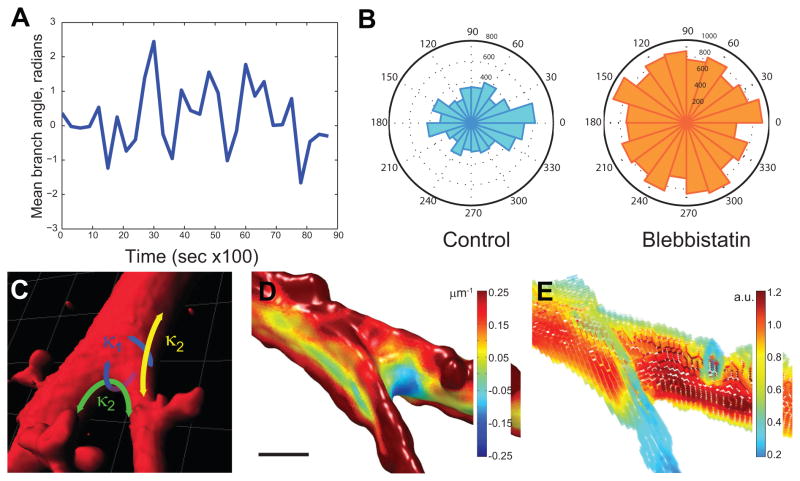
Figure 7. Myosin-II biases branch orientation in the direction of migration and shows differential association with inner high curvature and outer low curvature saddles at a branch base.
(A) Quantification of branch angle over time. An example cell was imaged every 300 seconds over time, and for each time point, a morphological skeleton was extracted as described in the Supplemental Methods. All branch vectors for a given time point were summed to derive a mean branch angle relative to the migration direction over all time points. (B) Radial histograms of branch angle relative to the direction of cell migration in the presence or absence of 20 μM blebbistatin; total branches indicated as radii shown for each condition over, n= 13 cells, 286 time points for control, n=11 cells, 129 time points for blebbistatin. (C) A typical branch geometry shown in a surface rendering of a branch region in AECs, with two sets of principle curvatures, one parallel to a plane containing the branch angle (κ2, green and yellow) and one perpendicular (κ1, blue). While the out-of-plane curvatures of the two saddles are comparable to each other and are dominated by the radii of the branches, the in-plane curvatures differ significantly with the saddle at the inner surface forming a higher negative curvature than the saddle at the outer surface. (D) Example of acute branch junction, where long region of low maximal curvature occurs at the outside of branch in the plane of the branch angle; a much smaller region of low maximal curvature occurs inside the branch junction. Color scale indicates local maximal curvature. (E) Heat map of GFP-myosin-IIA at cortex in the same branch junction. Color scale map indicates arbitrary units of GFP-myosin-IIA.
We then sought to understand how myosin-II might control branch angle via local curvature sensing and regulation. In general, branches oriented normal to the cell body have similar curvature all around their bases, while branches oriented at non-normal angles relative to the cell body consist of two opposing saddle structures: one associated with the acute angle at the inner surface and one associated with the obtuse angle at the outer surface, creating potential for curvature asymmetry (Figure 7C). Accordingly, asymmetric accumulation of myosin-II at these sites could enhance overall minimization of curvature in the branch junction, favoring acute angles (Figure 7D, E). On the other hand, myosin-II accumulation at the basis of branches normal to the cell body would be equal in all directions, allowing retraction of the branch to minimize curvature. Inspection of time-lapse movies of GFP-myosin-IIA dynamics confirmed an even recruitment of myosin-IIA to the saddles all around the base of normal angles (Supplemental Movie 9). We observed in time lapse movies that rare normal branches were rapidly retracted, whereas slanted branches with asymmetric myosin accumulation tended to be more stable. While this mechanism will require further investigation to characterize quantitatively, our observed examples suggest that myosin-II could bias branch direction by asymmetric association with and curvature minimization of acute branches and/or retraction of normal branches to minimize overall branch angle relative to the cell body, thus favoring branch formation along the direction of movement.
Discussion
Our development of an image analysis framework for quantifying 3D cell morphology across length scales has revealed how myosin-II acts at the molecular-scale to minimize local cell surface curvature, which in turn regulates the interlinked parameter of cell-scale branching morphogenesis. Combining this analysis with microfabrication to constrain cell shape identifies a positive feedback mechanism in which cell shape regulates myosin-II, such that the motor preferentially binds to and is stabilized by lower curvature cortical regions, where it acts to maintain minimal curvature. This feedback loop of local myosin-II regulation by and activity on curvature mediates cycles of spatially controlled cortical myosin-II assembly and disassembly that correlate with cycles of branch initiation and retraction to guide 3D cell migration.
How might myosin-II motors regulate and respond to surface curvature? We suggest that myosin-II minimizes cell surface curvature through generation of motor-driven tension in an isotropic cortical actin network attached to the plasma membrane, similar to how surface tension drives membrane flattening to minimize curvature in soap bubbles or membrane-bounded organelles 17. The question of how myosin-II responds to curvature, however, is more complex. Our analysis suggests that curvature-dependent recruitment of myosin-II to the cortex is independent of Rho/Rock signaling or contractile activity, but that curvature stabilizes its binding to cortical actin. We speculate that since non-muscle myosin-II mini-filaments exist as linear 300 nm rods with bouquets of actin-binding heads at each end 18, a maximal number of heads would engage an actin network that extends in the plane of the rod (Supplemental Figure 7). Extrapolating this concept suggests that high tube-like curvature would favor maximal head engagement for rods aligned along the zero-curvature axis, thus orienting contractile forces along the branch axis, providing a mechanism for retracting thin branches or generating traction on ECM-adhered branch tips. Alternatively, since curvature and tension in a closed surface at equilibrium are inextricably linked by the Young-Laplace relation, myosin-II recruitment to the cortex could be regulated by tension 19. However, our manipulations of motor activity demonstrate that curvature-sensitivity of myosin-II recruitment is independent of acto-myosin tension. One possible reconciliation of our proposed geometric versus tension-dependent models is that tension-dependent changes in actin organization could alter the optimal myosin-II head engagement to enhance myosin-II recruitment without requiring myosin contractility directly.
Our observations support a model in which curvature-sensitive recruitment and turnover of myosin-II defines the central switch in a directionally-biased branching-debranching cycle that drives directed cell migration in complex 3D environments. The cycle is initiated by local fluctuations in cortical myosin-II concentration. Local concentration depressions allow F-actin polymerization and/or hydrostatic pressure to overcome cortical tension and push out protrusions, which are stabilized by adhesion to the ECM 20. Thin branches with high curvature initially survive because of low myosin-II recruitment and thus low contractility, while more mature branches with lower curvature must maintain stronger polymerization and/or adhesion forces to overcome increasing contraction driven by recruitment of myosin-II to their low curvature cortex. Thick branches that are not stabilized by adhesion are retracted if they are normal to the cell surface by even accumulation of myosin-II around their bases and aligned along their length (Supplemental Movie 9). The migration cycle ends when large scale axial contraction of the low-curvature cell body cortex overcomes the anchoring forces of the adhesion-stabilized branches that are aligned with guidance cues. Thus, curvature-sensitive myosin-II recruitment may provide a robust core mechanism for cell shape control and branch morphogenesis in cell migration across a range of length scales from the local initiation of a branch to global traction generation at the cell scale.
Supplementary Material
Acknowledgments
This work was initiated as a collaborative effort between R.S.F. and G.D. (NIH R21 CA124990). H.L.E. and G.D. are supported by NIH R01 GM090317. R.S.F, R.A., K.A.M. and C.M.W. are supported by the NHLBI Division of Intramural Research.
Footnotes
Author Contributions
H.E., R.S.F.C.M.W. and G.D. designed experiments and wrote the manuscript. R.S.F. performed all imaging and FRAP experiments. H.E. designed, implemented and applied analysis software. R.A. provided transgenic mice. K.M., R.D., L.G. and C.C. performed microfabricated coffin experiments.
References
- 1.Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 3.Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 9.Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- 10.Bao J, Ma X, Liu C, Adelstein RS. Replacement of nonmuscle myosin II-B with II-A rescues brain but not cardiac defects in mice. J Biol Chem. 2007;282:22102–22111. doi: 10.1074/jbc.M702731200. [DOI] [PubMed] [Google Scholar]
- 11.Wang A, et al. Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc Natl Acad Sci U S A. 2010;107:14645–14650. doi: 10.1073/pnas.1004023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolega J. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol Biol Cell. 2003;14:4745–4757. doi: 10.1091/mbc.E03-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terry SJ, et al. Stimulation of cortical myosin phosphorylation by p114RhoGEF drives cell migration and tumor cell invasion. PLoS One. 2012;7:e50188. doi: 10.1371/journal.pone.0050188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amano M, et al. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 15.Hu A, Wang F, Sellers JR. Mutations in human nonmuscle myosin IIA found in patients with May-Hegglin anomaly and Fechtner syndrome result in impaired enzymatic function. J Biol Chem. 2002;277:46512–46517. doi: 10.1074/jbc.M208506200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Mouse models of MYH9-related disease: mutations in nonmuscle myosin II-A. Blood. 2012;119:238–250. doi: 10.1182/blood-2011-06-358853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terasaki M, et al. Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell. 2013;154:285–296. doi: 10.1016/j.cell.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surcel A, Kee YS, Luo T, Robinson DN. Cytokinesis through biochemical-mechanical feedback loops. Semin Cell Dev Biol. 2010;21:866–873. doi: 10.1016/j.semcdb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubow KE, Conrad SK, Horwitz AR. Matrix Microarchitecture and Myosin II Determine Adhesion in 3D Matrices. Curr Biol. 2013 doi: 10.1016/j.cub.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.