Abstract
Inflammation has been associated with fatigue during and after various types of breast cancer treatments. We examined whether prior chemotherapy was associated with DNA methylation patterns that could explain persisting inflammation and/or fatigue in women treated for breast cancer. Prior to breast radiation therapy, DNA was extracted from peripheral blood mononuclear cells (PBMCs) of 61 Stage 0-IIIA breast cancer patients who had received partial mastectomy with or without chemotherapy. DNA methylation was assessed at >485,000 CpG sites across the genome along with fatigue and plasma inflammatory markers previously associated with fatigue. Compared to non-chemotherapy-treated, women who had received chemotherapy exhibited significantly decreased methylation at eight CpG sites (p < 1.03 × 10−7) including four in exon 11 of transmembrane protein 49 (TMEM49), which demonstrated the largest decreases in methylation. Lower methylation at each identified CpG site was associated with increased plasma soluble tumor necrosis factor receptor 2 (sTNFR2) and interleukin (IL)-6 and mediated the relationship between chemotherapy and increases in these inflammatory biomarkers adjusting for multiple clinical and treatment characteristics. sTNFR2, but not CpG methylation status, was correlated with fatigue. Six months after breast radiation therapy, DNA methylation, inflammatory biomarkers and fatigue assessments were repeated in a subset of subjects (N = 39). Reduced methylation in 4 of the 8 identified CpG sites was still observed in chemotherapy versus non-chemotherapy-treated patients, albeit with some decay indicating the dynamic and potentially reversible nature of the changes. Reduced methylation in these 4 CpG sites also continued to correlate with either increased sTNFR2 or IL-6, but not fatigue. In conclusion, prior chemotherapy treatment was associated with decreased methylation of CpG sites in DNA from PBMCs of breast cancer patients, which correlated with increased inflammatory markers prior to and 6 months after radiation therapy. Persisting epigenetic changes secondary to chemotherapy may be one factor that contributes to inflammation and its consequences including cancer-related fatigue in vulnerable breast cancer patients.
Keywords: Breast cancer treatment, Epigenetics, Inflammation, Fatigue
1. Introduction
Cancer-related fatigue is one of the most common long-term debilitating toxicities experienced by approximately 30% of women previously treated for breast cancer (Bower et al., 2009; Geinitz et al., 2001; Giese-Davis et al., 2011; Kesler et al., 2012; Shapiro and Recht, 2001; Tevaarwerk et al., 2013; Wratten et al., 2004). Increasing data suggest that cancer-related fatigue is associated with a heightened inflammatory response (Bower et al., 2011; Wang et al., 2012). Breast cancer survivors with fatigue exhibit increased inflammatory markers, and similar findings have been reported during breast cancer treatment (Liu et al., 2012; Torres et al., 2013). Inflammatory cytokines have been shown to access the brain and interact with neurocircuits and neurotransmitters that regulate motor activity and motivation leading to fatigue (Raison et al., 2009).
Breast cancer patients often undergo multimodality treatment which is based upon tumor stage and patient tolerance for potentially toxic therapies. Women with breast cancer generally require surgical removal of the tumor, and patients who choose to undergo lumpectomy, also known as breast conserving surgery, are treated with post-lumpectomy breast radiation to significantly decrease the likelihood of disease recurrence. Patients with aggressive subtypes of breast cancer or advanced stage disease are also recommended chemotherapy which may be given before (neoadjuvant) or after (adjuvant) surgery, but almost always before radiation. We and others have shown that chemotherapy-treated breast cancer patients experience significantly more cancer-related fatigue than non-chemotherapy-treated subjects (Torres et al., 2013). For example, in our previous study of women assessed immediately before, during and 6 weeks after whole breast radiation, prior chemotherapy was associated with increased markers of inflammation including plasma concentrations of soluble tumor necrosis factor receptor 2 (sTNFR2; a marker of TNF activity) and interleukin (IL)-6, both of which correlated with fatigue and have been associated with fatigue in breast cancer patients during and after treatment in previous studies (Bower et al., 2011; Liu et al., 2012; Torres et al., 2013). Interestingly, we found that increased fatigue severity in chemotherapy-treated patients prior to radiation therapy was similar regardless of whether patients received chemotherapy before (neoadjuvant) or after (adjuvant) surgery and thus was independent of time since the last cycle of treatment (up to 22 weeks) (Torres et al., 2013). Similar results were found with inflammatory markers (Torres et al., 2013). These findings indicated that inflammation and fatigue in chemotherapy-treated patients lingered for several months after chemotherapy treatment, raising the possibility that persistent alterations in the inflammatory response may be involved.
Although a number of studies, including our own, have shown that breast cancer and its treatment are associated with activation of inflammatory pathways (Bower et al., 2011; Liu et al., 2012; Torres et al., 2013), a central question is how does inflammation persist into survivorship. One possibility is that cancer or its treatment leads to epigenetic changes that predispose to chronic inflammation. Epigenetic alterations as a consequence of multiple chemotherapy regimens have been reported in numerous genes in DNA samples extracted from both breast cancer and peripheral blood cells (Ari et al., 2011; Avraham et al., 2012; Sharma et al., 2012; Swisher et al., 2009). However, the impact of chemotherapy-induced epigenetic changes has not been related to systemic inflammation and/or behavior.
To further explore the relationship among potential chemotherapy-induced epigenetic changes, inflammation, and fatigue, DNA methylation patterns from peripheral blood mononuclear cells (PBMCs) were assessed along with plasma sTNFR2 and IL-6 and fatigue in an overlapping cohort of the breast cancer patients noted above. These women were enrolled in a prospective study of fatigue after partial mastectomy with or without previous chemotherapy treatment and underwent assessments before and 6 months after completing radiation. The goal of the study was to identify epigenetic differences related to chemotherapy that were associated with increased inflammation and/or cancer-related fatigue.
2. Methods
2.1. Participants and data collection
After obtaining Emory Institutional Review Board approval, consecutive breast cancer patients presenting to the Winship Cancer Institute between March 2010 and November 2011 were approached for participation. Eligible subjects were women with Stage 0-IIIA breast cancer between ages 18–75. All subjects were treated with standard breast conserving surgery and lymph node evaluation and enrolled prior to radiation after providing written informed consent (see Fig. 1 for Flow Diagram). Due to advanced stage or aggressive subtype (e.g. triple negative breast cancer), twenty-two (36%) participating subjects also received neoadjuvant (N = 15) or adjuvant (N = 7) chemotherapy, which was completed before or after surgery, respectively, and prior to radiation treatment and enrollment.
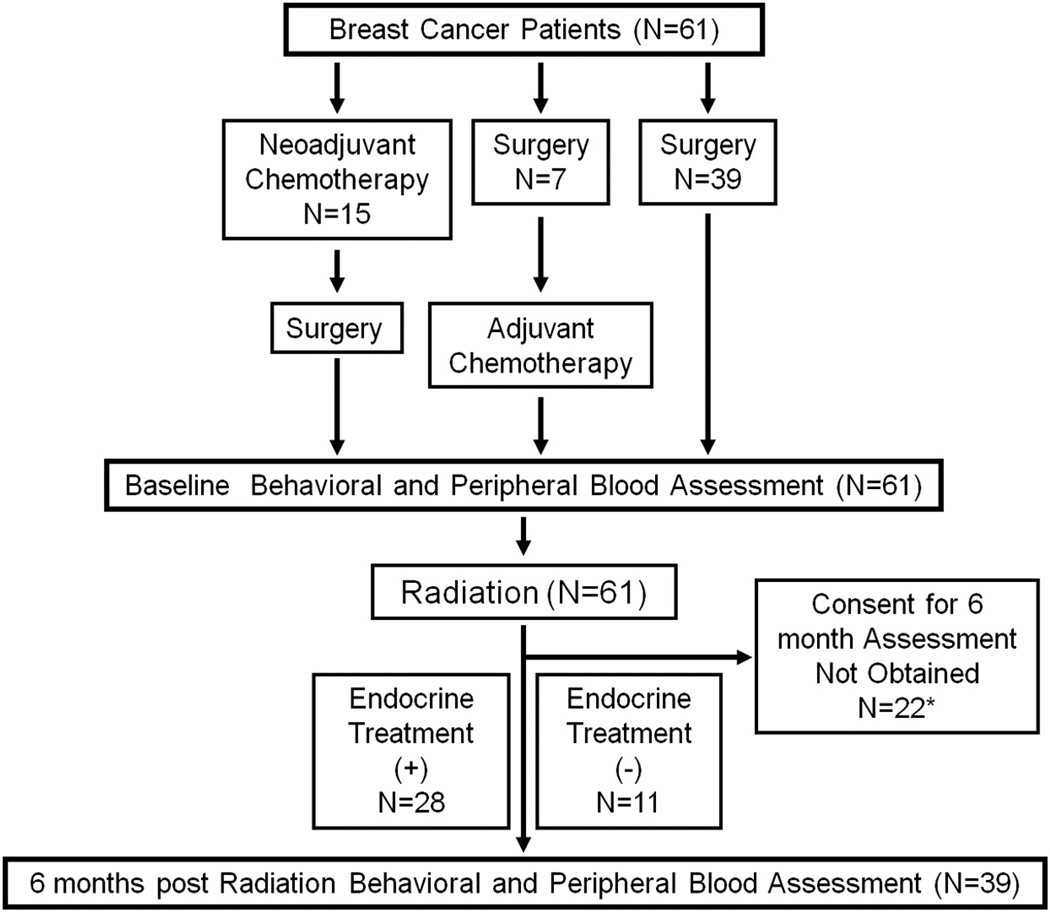
Fig. 1.
Flow diagram of study participants. *Although 61 patients were enrolled at baseline, in order to confirm the observed epigenetic changes, we decided to consent patients for follow-up assessments at 6 months. Once Emory IRB approval for the follow-up was obtained, 39 of the 61 patients were eligible for assessments at this timepoint.
Exclusion criteria included medical conditions that might influence the relationship between fatigue and inflammation including autoimmune or inflammatory disorders, chronic infectious diseases, and uncontrolled cardiovascular, metabolic, pulmonary or renal disease. Subjects with a history of schizophrenia, bipolar disorder or a diagnosis of substance abuse/dependence within the past year were also excluded. Drugs known to affect the immune system (e.g. glucocorticoids, methotrexate), excluding over-the-counter anti-inflammatory medications were not permitted. Enrollment was limited to Caucasians and African Americans, who represent the majority of women treated at Emory. All patients were required to have a hemoglobin ≥ 10 g/dl. Of note, the majority of subjects included in the current study overlap with the individuals included in a previous study on depression, fatigue and inflammation in breast cancer patients (Torres et al., 2013).
All chemotherapy-treated patients received standard anthracycline- and/or taxane-based regimens. The time between the last cycle of chemotherapy and first assessment ranged from 3.7 to 18.0 weeks. Subjects underwent baseline fatigue assessments and peripheral blood sampling before radiation [after surgery and chemotherapy (if applicable)]. A subset of patients (N = 39) consented to undergo assessments again 6 months after completing radiation. Clinical and psychosocial variables were also collected.
2.1.1. Fatigue
Fatigue was assessed using the 20-item Multidimensional Fatigue Inventory (MFI) (Smets et al., 1995). The MFI measures general, physical, and mental fatigue as well as reduced activity and motivation. A clinically meaningful difference in MFI scores is 10 points based on studies of cancer patients (Purcell et al., 2010).
2.1.2. Biological samples
Peripheral blood samples were drawn into EDTA tubes between 8–11am (to reduce potential circadian effects) during the fatigue assessment. Plasma was separated and stored at −80 °C. Genomic DNA was isolated from buffy coat samples using Omega Bio-Tek chemistry and Kingfisher liquid handling device. DNA was quantified using Nanodrop and PicoGreen. A white blood cell count (WBC) with differential was performed to assess granulocyte and lymphocyte proportions in each sample.
For each subject, 485,513 CpG sites across the genome were interrogated using the HumanMethylation450 BeadChip (Illumina, San Diego, CA). 1ug of DNA was converted with sodium bisulfite, amplified, fragmented, and hybridized to the BeadChip per manufacturer’s instructions. One sample of male DNA was included on each BeadChip as a technical control and assessed for reproducibility using the Pearson correlation coefficient. CpGassoc (Barfield et al., 2012) was used to perform quality control and calculate β-values. Data points with probe detection p-values >0.001 were set to missing, and CpG sites with missing data for > 10% of samples were excluded from analysis. Samples with probe detection call rates <95% and those with an average intensity value of either <50% of the experiment-wide sample mean or <2000 arbitrary units (AU) were also excluded. 484,489 CpG sites remained eligible for analysis. For each individual sample and CpG site, the signals from methylated (M) and unmethylated (U) bead types were used to calculate a β-value [β = M/(U + M)] to approximate proportion of DNA methylated at a particular site. Samples from all subjects were included in the methylation analyses at baseline (N = 61). To determine the reproducibility and persistence of the findings, a subset of 39 of the original sample agreed to be evaluated again 6 months post radiation (range 162 to 239 days, mean 199.7 days, SD 18.8 days).
2.1.3. Inflammatory markers
Plasma concentrations of sTNFR2 and IL-6 were determined using sandwich ELISA according to manufacturer’s protocol (R&D Systems, Minneapolis, MN). All samples were assayed in duplicate. Quality controls (Randox Laboratories, Antrim, Northern Ireland) and plasma of low and high cytokine concentrations were included with every assay. The mean inter- and intra-assay coefficients of variation were ≤ 10%.
2.2. Statistical analysis
Chi-square tests were used to compare categorical variables between chemotherapy- and non-chemotherapy-treated patients. T-tests or Wilcoxon rank-sum tests were used to compare continuous variables with normal or non-normal distributions, respectively. To identify CpG sites that were differentially methylated in subjects receiving chemotherapy, MethLAB (Kilaru et al., 2012) was used to fit regression models that modeled β-values as a linear function of chemotherapy, adjusting for age, race, chip and row on chip, and proportions of granulocytes and lymphocytes. Secondary analyses were performed covarying for body mass index (BMI), smoking status, tumor stage and receptor status. The impact of additional covariates on DNA methylation was also evaluated including endocrine therapy, co-morbid medical conditions, concomitant medications (including those known to affect the immune system), and time since surgery. To adjust for the 484,489 tests performed, a strict Bonferroni criterion for significance was employed (p < 1.03 × 10−7).
To examine the relationship among chemotherapy, methylation status, inflammation and fatigue, outcome variables used were fatigue, as measured by MFI score, and log-transformed concentrations of sTNFR2 and IL-6, based on their association with fatigue in previous studies including our own (Bower et al., 2011; Liu et al., 2012; Torres et al., 2013). Each outcome was tested for association with chemotherapy and with methylation level (β-values) of CpG sites passing Bonferroni significance in the chemotherapy-methylation association tests. Each outcome variable was modeled as a linear function of (1) chemotherapy, adjusting for covariates and (2) chemotherapy and methylation level (β-value) at a specific CpG site, adjusting for covariates. Age, self-reported race, and proportions of granulocytes and lymphocytes were included in all initial models followed by secondary analyses including the clinical covariates noted above. To test whether the methylation status of CpG sites associated with chemotherapy (p < 1.03 × 10−7) mediated the association between chemotherapy and inflammation as well as fatigue, mediation analysis was performed using Sobel tests (Sobel, 1982). For each outcome, significance was determined using the Bonferroni–Holm (step-down) method to adjust for the multiple CpG sites tested (Holm, 1979). Finally, a follow-up analysis of the methylation status of CpG sites found to be significant at the baseline assessment and their relationship with inflammation and fatigue were repeated 6 months after radiation using the procedures described above. The Bonferroni– Holm method was used to determine significance of this follow- up analysis of reduced methylation.
3. Results
Sixty-one women agreed to participate in the study. No significant differences in clinical or treatment-related characteristics were found between patients who did and did not enroll in the protocol. Study cohort characteristics are summarized in Table 1. Chemotherapy-treated subjects (N = 22) were significantly younger and had more advanced stage tumors than non-chemotherapy- treated subjects (N = 39). There were no significant differences in race, BMI, smoking status or differential WBC. Half of the chemotherapy patients (N = 11) were treated with an anthracycline-based regimen (with the most common regimen being dose–dense adriamycin and cyclophosphamide for four cycles followed by paclitaxel for four cycles) (see Supplementary Table 1 for exact chemotherapy regimens of all subjects). The remaining patients (N = 11) were treated with non-antracycline-based regimens including docetaxol and cyclophosphamide for four cycles (N = 5 patients) and docetaxol, carboplatin, and trastuzumab for six cycles followed by one year of trastuzumab (N = 6).
Table 1.
Sample characteristics.
| Variable | No chemotherapy (N = 39) | Chemotherapy (N = 22) | p-Value |
|---|---|---|---|
| Age, Years | 58.7 ± 9.7 | 52.3 ± 9.5 | 0.02 |
| Race | |||
| White | 25 (64%) | 11 (50%) | 0.29 |
| African American | 14 (36%) | 11 (50%) | |
| BMI | 29.2 ± 6.8 | 27.7 ± 4.3 | 0.35 |
| Smoking History | |||
| Never | 33 (85%) | 16 (73%) | |
| Former | 6 (15%) | 3 (14%) | 0.06 |
| Current | 0 (0%) | 3 (14%) | |
| Leukocyte Subpopulation | |||
| Granulocytes | 58.7 ± 6.7 | 60.3 ± 10.2 | 0.47 |
| Lymphocytes | 31.4 ± 7.2 | 30.0 ± 13.7 | 0.61 |
| Monocytes | 7.8 ± 1.7 | 8.2 ± 2.8 | 0.45 |
| Eosinophils | 2.7 ± 2.3 | 2.8 ± 2.6 | 0.93 |
| Basophils | 0.63 ± 0.54 | 0.61 ± 0.61 | 0.90 |
| Stage | |||
| 0 | 16 (41%) | 0 (0%) | |
| I | 17 (44%) | 6 (27%) | <0.001 |
| II | 6 (15%) | 13 (59%) | |
| III | 0 (0%) | 3 (14%) | |
| Endocrine Treatment | 27 (69%) | 12 (54%) | 0.25 |
| BL MFI (SD) | 38.6 (10.1) | 56.3 (18.2) | <0.001 |
| BL sTNFR2 (pg/ml)(SD) | 2.8 (0.7) | 4.0 (2.3) | 0.01 |
| BL IL-6 (pg/ml)(SD) | 2.5 (2.4) | 4.1 (3.0) | 0.01 |
| 6 Month follow-up | |||
| No chemotherapy (N = 26) | Chemotherapy (N = 13) | ||
| 6mo MFI (SD) | 23.6 (18.8) | 23.4 (29.3) | 0.97 |
| 6mo sTNFR2 (pg/ml)(SD) | 3.5 (1.0) | 3.7 (1.6) | 0.63 |
| 6mo IL-6 (pg/ml)(SD) | 2.3 (1.7) | 2.7 (2.2) | 0.55 |
BL – baseline; BMI – body mass index; IL-6 – interleukin-6; MFI – Multidimensional Fatigue Inventory; mo – month; SD – standard deviation; sTNFR2 – soluble tumor necrosis factor 2.
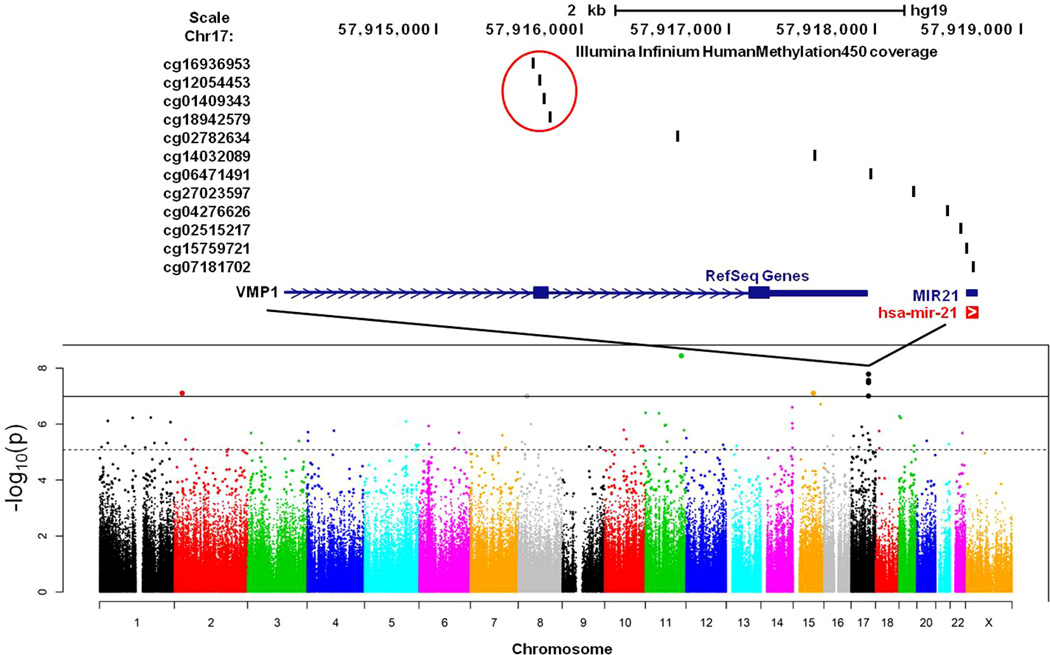
At baseline, among the 485,513 CpG sites tested, the methylation status of eight CpG sites were associated with chemotherapy treatment following Bonferroni adjustment for multiple tests (p < 1.03 × 10−7) (Figs. 2 and 3; Table 2). Significant CpG sites included cg26077811 in the body of ubiquitin carboxyl-terminal hydrolase 2 (USP2), cg25446789 in the body of dystrobrevin beta (DTNB), cg05438378 in the body of SMAD family member 3 (SMAD3), one CpG site (cg13518625) in an uncharacterized, non-genic region of chromosome 8, and four sites in exon 11 of trans-membrane protein 49 (TMEM49; Figs. 2 and 3). For each of these eight CpG sites, methylation was significantly lower in subjects who had been treated with chemotherapy; the four sites in TMEM49 demonstrated the largest difference in methylation between chemotherapy and non-chemotherapy-treated subjects (Fig. 3; Table 2). Of note, there were no significant associations between methylation and (1) time since chemotherapy or (2) type of chemotherapy categorized as anthracycline-based (yes/no) or treatment with trastuzumab (yes/no); this was true if all CpG sites across the genome were considered or just the 8 CpG sites that were found to differ between chemotherapy versus non-chemotherapy-treated patients.
Fig. 2.
Manhattan plot depicting the association of all CpG sites with chemotherapy.
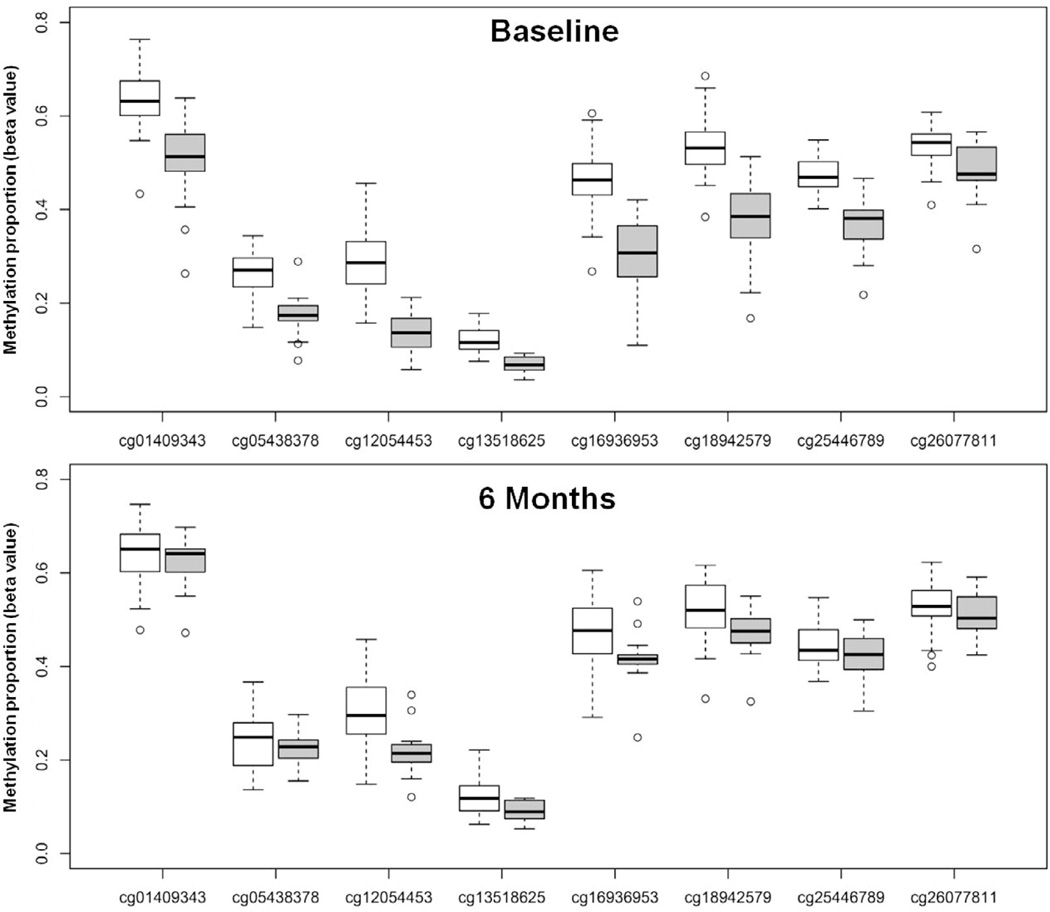
Fig. 3.
Distribution of methylation proportions in chemotherapy and non-chemotherapy-treated breast cancer patients at baseline and 6 months after radiation treatment.
Table 2.
Methylation differences (Δβ) between patients who received chemotherapy and those who did not, adjusted for age, race, and cell type proportions (N = 61).
| CpG site | Gene | Location | Δ β | p-Value |
|---|---|---|---|---|
| cg26077811 | USP2 | chr11:119232263 | −.074 | 3.65 × 10−9 |
| cg18942579 | TMEM49 | chr17:57915773 | −.161 | 1.65 × 10−8 |
| cg12054453 | TMEM49 | chr17:57915717 | −.154 | 2.75 × 10−8 |
| cg16936953 | TMEM49 | chr17:57915665 | −.168 | 3.26 × 10−8 |
| cg05438378 | SMAD3 | chr15:67383736 | −.089 | 7.78 × 10−8 |
| cg25446789 | DTNB | chr2:25810393 | −.085 | 7.84 × 10−8 |
| cg01409343 | TMEM49 | chr17:57915740 | −.138 | 9.88 × 10−8 |
| cg13518625 | chr8:29522838 | −.051 | 9.98 × 10−8 |
chr – chromosome; DTNB – dystrobrevin beta; SMAD3 - SMAD family member 3; TMEM49 – transmembrane protein 49; USP2 - ubiquitin carboxyl-terminal hydrolase 2.
Secondary analyses were performed to further verify that differences in methylation associated with chemotherapy were not due to additional differences in patient and tumor characteristics. For the eight CpG sites in Table 2, each of the following covariates was added to the model: BMI, smoking status, tumor stage and tumor receptor status. In all cases, the estimated decrease in methylation attributed to chemotherapy was unaffected, and none of the additional covariates were significantly associated with methylation. In addition, no significant associations with genome-wide DNA methylation status or the methylation status of the 8 identified CpG sites were found for endocrine therapy, co-morbid medical conditions, and concomitant medications (including those known to affect the immune system). Of note, time since surgery (surgery to baseline) differed significantly between chemotherapy and non-chemotherapy-treated patients (79.5 SD 51.0 versus 54.0 SD 12.0 days; t = 3.0, df = 59, p < 0.005; driven by patients who received adjuvant chemotherapy). Time since surgery was significantly associated with significantly reduced methylation at 4 CpG sites after Bonferroni correction (p < 1.03 × 10−7), although none of these overlapped with the 8 identified CpG sites described above. However, controlling for chemotherapy treatment as an additional covariate eliminated the association with time since surgery, suggesting that this variable was a surrogate marker for having received chemotherapy.
As shown in Table 1, chemotherapy-treated patients at baseline exhibited significantly higher plasma sTNFR2 concentrations (p < 0.001) and marginally increased plasma IL-6 (p = 0.069) than non-chemotherapy-treated patients after adjusting for age, race, and cell type proportions. Similar results were obtained when controlling for BMI, smoking status, tumor stage and receptor status. Chemotherapy-treated subjects also had significantly higher fatigue scores compared to non-chemotherapy-treated patients (p < 0.001). Similar results were obtained when controlling for BMI, smoking status, tumor stage and receptor status. No differences in fatigue (MFI scores) were found as a function of chemotherapy regimen (anthracycline-based, yes versus no: mean 54.3 SD 17.5 versus 57.9 SD 19.4, respectively, t = 0.46, df = 20, p = 0.65; or trastuzumab treatment, yes versus no: mean 58.2 SD 12.7 versus 55.6 SD 20.2, respectively, t = 0.29, df = 20, p = 0.77). sTNFR2 and IL-6 concentrations were positively correlated with fatigue in bivariate analyses (r = 0.31; p < 0.01 and r = 0.35; p = 0.015, respectively) and multivariate analyses controlling for BMI, smoking status, tumor stage and receptor status (both p < 0.05).
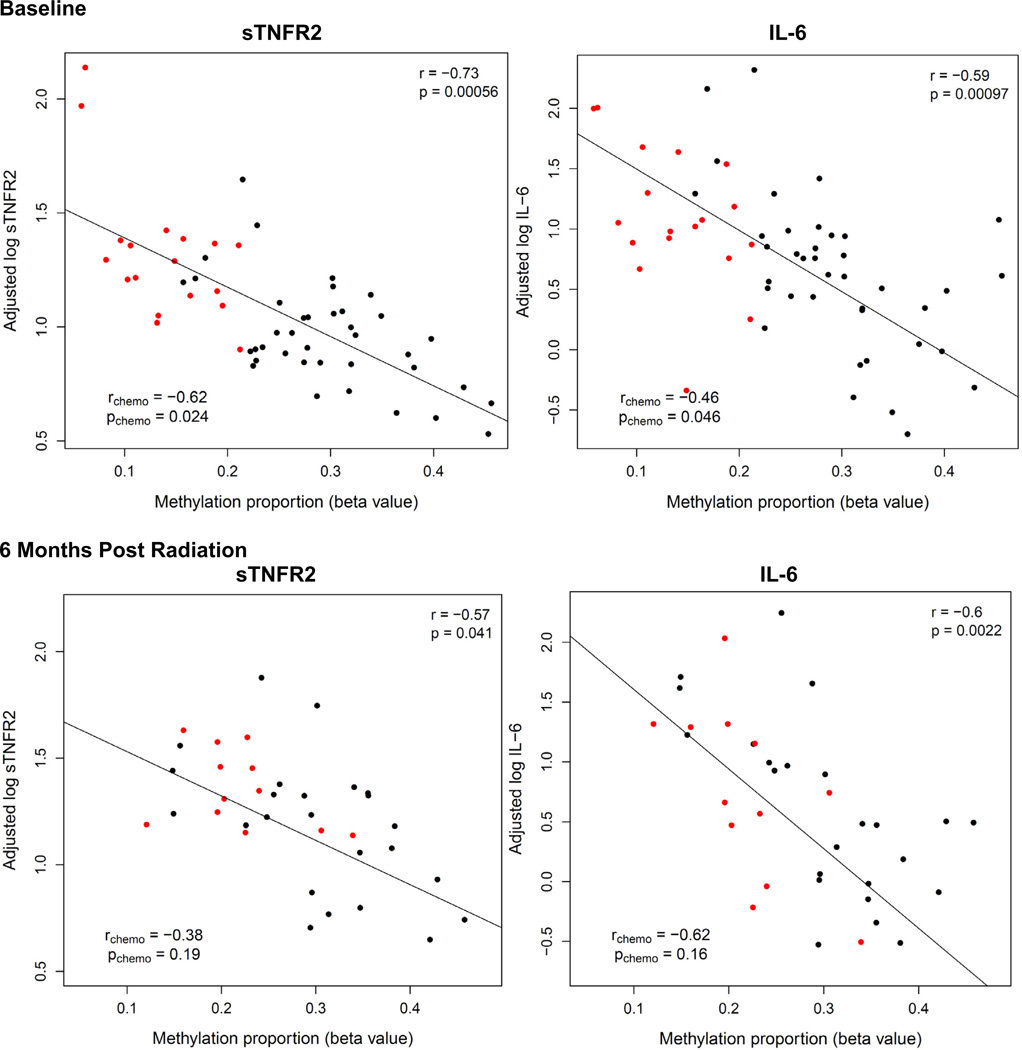
Methylation status at each of the 8 identified CpG sites associated with chemotherapy treatment at baseline was significantly correlated with sTNFR2 in the sample as a whole (N = 61), and all but one CpG site (cg13518625) correlated with sTNFR2 in patients treated with chemotherapy (N = 22) (see Table 3, Fig. 4 and Supplementary Fig. 1). Methylation status at 7 of the 8 chemotherapy-associated CpG sites significantly correlated with IL-6 in the sample as a whole, and in the patients treated with chemotherapy, similar correlations coefficients for the majority of the CpG sites were found, although they reached statistical significance for only 3 of the 8 CpG sites (see Table 3, Fig. 4 and Supplementary Fig. 2). No correlations were found between baseline methylation status and fatigue. To examine whether methylation changes at the eight significant CpG sites mediated the relationship among chemotherapy, inflammation, and fatigue, Sobel tests for mediation were performed (Sobel, 1982). Methylation at each of the identified CpG sites was found to mediate the relationship between chemotherapy and sTNFR2 and IL-6 concentrations (Table 3). For example, lower methylation status of the TMEM49 and USP2 CpG sites was associated with increased plasma sTNFR2 and IL-6 (Fig. 4; Supplementary Figs. 1 and 2). In contrast, methylation status of the identified CpG sites did not appear to mediate the relationship between chemotherapy and fatigue (Table 3). However, as expected, sTNFR2 (but not IL-6) concentrations mediated the relationship between chemotherapy and fatigue (p = 0.03).
Table 3.
Mediation analysis of the relationship among chemotherapy, methylation, inflammatory markers, and fatigue at baseline (N = 61).
| Outcome modeled on: | Gene | Outcome = MFI score | Outcome = log sTNR2 level | Outcome = log IL-6 level | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pChemo | pCpG | pSobel test | pChemo | pCpG | pSobel test | pChemo | pCpG | pSobel test | ||
| Chemotherapy only | – | 5.9 × 10−5 | – | – | 6.0 × 10−4 | – | – | .069 | – | – |
| Chemo + cg26077811 | USP2 | .006 | .92 | .54 | .69 | .0053 | .0030 | .062 | 6.0 × 10−5 | 6.0 × 10−5 |
| Chemo + cg18942579 | TMEM49 | .026 | .38 | .19 | .88 | 1.5 × 10−4 | 1.6 × 10−4 | .75 | .030 | .016 |
| Chemo + cg12054453 | TMEM49 | .072 | .10 | .052 | .89 | 5.6 × 10−4 | 5.0 × 10−4 | .26 | 9.6 × 10−4 | 7.9 × 10−4 |
| Chemo + cg16936953 | TMEM49 | .031 | .29 | .14 | .93 | 1.1 × 10−4 | 1.4 × 10−4 | .38 | .0028 | .0019 |
| Chemo + cg05438378 | SMAD3 | .003 | .89 | .55 | .37 | .014 | .0084 | .43 | .0032 | .0023 |
| Chemo + cg25446789 | DTNB | .017 | .42 | .21 | .54 | .0030 | .0022 | .67 | .014 | .0084 |
| Chemo + cg01409343 | TMEM49 | .0091 | .64 | .32 | .62 | .0018 | .0014 | .98 | .066 | .034 |
| Chemo + cg13518625 | .017 | .41 | .21 | .31 | .023 | .013 | .86 | .033 | .018 | |
Multidimensional Fatigue Inventory (MFI) score and cytokine concentrations [log(sTNFR2) or (log (IL-6)] were each modeled as a linear function of chemotherapy and methylation β values, adjusting for age, race, and cell type proportions. p-Values from these regression models are denoted pChemo and pCpG for chemotherapy and methylation respectively, while pSobel test represents the p-value from a Sobel test of methylation as a mediator between chemotherapy and the indicated outcome variable. IL: interleukin; sTNFR2 – soluble tumor necrosis factor 2. DTNB – dystrobrevin beta; SMAD3 – SMAD family member 3; TMEM49 – transmembrane protein 49; USP2 – ubiquitin carboxylterminal hydrolase 2.
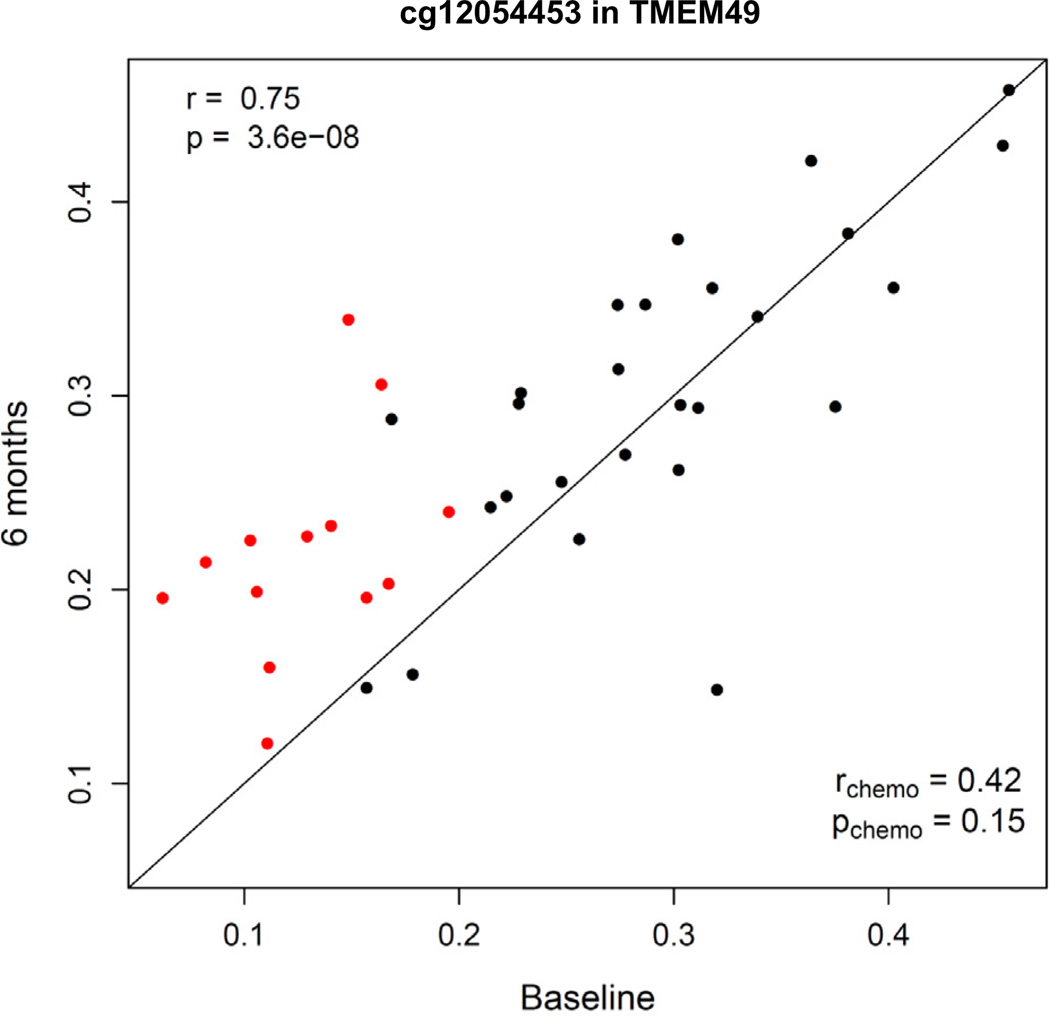
Fig. 4.
Relationship between plasma cytokine concentrations and methylation status of cg12054453 in exon 11 of TMEM49 at baseline (N = 61) and 6 months after radiation (N = 39).
To examine the reproducibility and persistence of the findings, a subset of 39 patients from the original sample was studied 6 months after completion of radiation treatment (see Methods and Fig. 1). The mean time interval from the end of radiation to the 6 month sampling point was 199.7 days SD 18.8 days, range 162–239 days. There was decay in the methylation differences between groups at follow-up, however after Holm-adjustment for 8 sites, 4 of the 8 CpG sites identified at baseline continued to exhibit reduced methylation in the subset of chemotherapy (N = 13) versus non-chemotherapy-treated (N = 26) patients (Fig. 3, Table 4). As noted in the methods section, a Bonferonni–Holm correction for 8 comparisons was used in this analysis, because it was a follow- up analysis of only the 8 CpG sites identified by the genome- wide analysis at baseline which involved 450,000 CpG sites and required a much more stringent correction: cut off of p < 1.03 × 10−7. Similar to baseline, methylation status at 6 months post radiation in these 4 CpG sites correlated with either sTNFR2 or IL-6 in the sample as a whole (Fig. 4, Supplementary Table 2 – see pCpG for log IL-6 and log sTNFR2). In chemotherapy-treated patients alone (n = 13), no significant correlations were found between methylation status and either sTNFR2 or IL-6, although similar correlation coefficients were found in chemotherapy-treated patients and the group as a whole in all but one of these 4 CpG sites. There was also a significant correlation between methylation levels in all 8 identified CpG sites at baseline and 6 months post radiation in the sample as a whole (Fig. 5, Supplementary Fig. 3). When chemotherapy patients were analyzed alone (N = 13), the correlation coefficients between methylation status at baseline and 6 months were similar, albeit lower, than in the group as a whole and did not reach statistical significance (Fig. 5, Supplementary Fig. 3).
Table 4.
Methylation differences (Δβ) between patients who received chemotherapy and those who did not at baseline and 6 months post radiation, adjusted for age, race, and cell type proportions (N = 39).
| CpG site | Gene | Baseline | 6 mos post-baseline | ||
|---|---|---|---|---|---|
| Δβ | p | Δβ | p | ||
| cg26077811 | USP2 | −.08 | 3.2 × 10−6 | −.028 | .045 |
| cg18942579 | TMEM49 | −.22 | 3.6 × 10−15 | −.060 | .0027 |
| cg12054453 | TMEM49 | −.19 | 3.9 × 10−9 | −.088 | 8.4 × 10−4 |
| cg16936953 | TMEM49 | −.21 | 1.0 × 10−10 | −.061 | .014 |
| cg05438378 | SMAD3 | −.10 | 3.4 × 10−6 | −.027 | .12 |
| cg25446789 | DTNB | −.09 | 1.9 × 10−7 | −.039 | .0095 |
| cg01409343 | TMEM49 | −.18 | 1.5 × 10−12 | −.042 | .031 |
| cg13518625 | −.06 | 3.4 × 10−6 | −.038 | .0012 | |
Fig. 5.
Relationship between methylation status of cg12054453 in exon 11 of TMEM49 at baseline and 6 months after radiation.
Changes in methylation (β-values) between baseline and 6 months post radiation were also inversely correlated with changes in either sTNFR2 or IL-6 over the same time period in all but one of the 8 identified CpG sites in the sample as a whole (p < 0.05; Supplementary Table 3). Thus, as methylation status increased or decreased from baseline to 6 months, plasma cytokine concentrations decreased or increased, respectively. In addition, Sobel tests confirmed that reduced methylation at 4 of the identified sites continued to mediate the relationship between chemotherapy and sTNFR2 (cg12054453 only) and/or IL-6 (4 sites, p < 0.05) 6 months post radiation (Supplementary Table 2 – see pSobel for log IL-6 and log sTNFR2). Controlling for time since radiation did not affect these results. Of note, no relationship was found between inflammatory mediators and fatigue at 6 months in this patient subset, and fatigue levels did not differ between chemotherapy and non-chemotherapy-treated patients at 6 months.
4. Discussion
The current data indicate that prior chemotherapy was associated with decreased DNA methylation in specific CpG sites which was associated with increased inflammatory markers up to 6 months after initial evaluation. It is well known that chemotherapy induces epigenetic changes in a variety of cancer cell lines and malignant tumors, including breast carcinomas, and epigenetic changes are believed to be one mechanism responsible for the development of cancer treatment resistance and poor outcome (Baker et al., 2005; Calcagno et al., 2008; Chekhun et al., 2007; Ogawa et al., 2012). This is the first study, however, to identify differentially methylated CpG sites in DNA extracted from noncancerous mononuclear cells circulating in the peripheral blood of chemotherapy versus non-chemotherapy-treated breast cancer patients that are associated with inflammatory markers. In fact, only 8 of the 485,513 CpG sites interrogated were associated with prior chemotherapy treatment. The results of this study support the hypothesis that chemotherapy may promote changes in DNA methylation, not only in cancer cells but in normal cells, and that these differentially methylated genes may contribute to an inflammatory state. Although not directly related to fatigue, these methylation changes through their effects on inflammatory mediators may set the stage for the development of co-morbidities related to inflammation including cancer-related fatigue in vulnerable individuals.
Several mechanisms may account for the DNA methylation differences observed in chemotherapy-treated patients. As previously suggested, chemotherapy may directly alter the methylation status of the identified CpG sites. Alternatively, the observed methylation differences between chemotherapy versus non-chemotherapy-treated patients may result from the inflammatory response to chemotherapy-related tissue injury and cell death. Indeed, inflammatory markers including IL-6 have been shown to directly impact DNA methylation status in a variety of cell types including nonmalignant and malignant cells such as breast cancer cells (D’Anello et al., 2010; Papageorgis et al., 2010; Shanmugam and Sethi, 2012; Soltanpour et al., 2013). A third possibility is that the differences in methylation as a function of chemotherapy are not related to chemotherapy itself but are related to factors associated with chemotherapy treatment. However, all analyses adjusted for differences in age, race and differential WBC as well as BMI, smoking status, cancer stage, and tumor receptor status, and no effects on methylation status were found for type. In addition, time since chemotherapy, endocrine therapy, co-morbid medical conditions, and concomitant medications (including those known to affect the immune system).
Consistent with previous studies, increased sTNFR2 and IL-6 were associated with cancer-related fatigue at baseline. Moreover, as we have previously shown, increases in sTNFR2 and IL-6 were associated with prior chemotherapy exposure (Torres et al., 2013). Chemotherapy was associated with differential methylation of several CpG sites, which appeared to mediate the relationship between chemotherapy and increases in these inflammatory markers. Nevertheless, the epigenetic changes seen in chemotherapy patients did not mediate the relationship between chemotherapy and fatigue. Thus, there appears to be a more proximal and direct relationship between the observed DNA methylation changes and inflammation, while additional intervening factors may influence the relationship between inflammation and fatigue, thereby obscuring the relationship between DNA methylation and fatigue as a function of chemotherapy. One of the factors that may contribute to this observation is genetic vulnerability. For example, although clear relationships exist between the administration of interferon-alpha and the induction of inflammation, only a subset of IFN-alpha-treated subjects are vulnerable to symptoms of depression including fatigue. Recent data have identified several genetic factors that contribute to this susceptibility including polymorphisms in the genes for the serotonin transporter and indoleamine 2,3 dioxygenase (Bull et al., 2009; Lotrich, 2009; Smith et al., 2012). Taken together with our findings, these data suggest that there are additional factors including genetic vulnerability that may contribute to the relationship between inflammation and fatigue, and thus the relationship between DNA methylation and fatigue may only be revealed in individuals with genetic and/or environmentally-derived neural sensitivity to the behavioral effects of inflammatory states.
The biologic mechanisms linking the identified epigenetic changes to inflammation are currently unknown, and did not involve methylation changes within inflammatory cytokine genes or inflammatory transcription factors. Nevertheless, each of the identified CpG sites, excluding those in DTNB, has been implicated in the inflammatory response. For example, the largest chemotherapy-associated decrease in DNA methylation, involving 4 of the 8 identified CpG sites, was found in a region within TMEM49, which lies in close proximity to the reported promoter for the primary miRNA (pri-miR) transcript that gives rise to the mature oncomiR, miR-21, (Cai et al., 2004) a well-characterized oncogenic miRNA that is upregulated in many tumor types, including breast cancer (Cancer Genome Atlas Research Network, 2013; Kumarswamy et al., 2011; Ozsolak et al., 2008; Tsujiura et al., 2010; Xu et al., 2011). In cancer and non-cancer cell lines, miR-21 has been found to downregulate PTEN and PDCD4, which results in increased activation of nuclear factor kappa B (NF-kB) and IL-6. In turn, IL-6 has been shown to activate STAT3, which can bind to and regulate the promoter of miR-21, resulting in a positive feedback loop that drives further NF-kB activation and inflammation (Iliopoulos et al., 2010; Yasuda et al., 2010; Young et al., 2010). Interestingly, lower methylation of CpG sites in SMAD3, a transcription factor known to directly upregulate miR-21 (Zhong et al., 2011), was also found in the chemotherapy- versus non-chemotherapy treated patients. Finally, the epigenetic changes noted in USP may also contribute to NF-KB activation, based on a recent report demonstrating that USP2 is a modulator of TNF-alpha-induced NF-kB signaling (Metzig et al., 2011). Data on the expression of miRNA was not available for the current study sample. Therefore, we examined the relationship between methylation of the 4 sites in TMEM49 and the 1 site in SMAD3 and miR-21 expression in 276 breast cancer samples analyzed as part of The Cancer Genome Atlas project (http://cancergenome.nih.gov/). Interestingly, there was a significant inverse relationship between methylation of the sites in exon 11 of TMEM49 as well as an intron in SMAD3 and miR-21 expression (all p < 0.05; Supplementary Fig. 4). These data support a relationship between chemotherapy-associated epigenetic changes and inflammation possibly mediated by upregulation of miR-21 and NF-kB activity.
Limitations of the study warrant consideration. Patients were not assessed before chemotherapy and therefore, a cause and effect relationship between chemotherapy and the observed epigenetic changes cannot be definitively established. Nevertheless, other factors that contribute to the decision to administer chemotherapy were controlled for in the analyses and did not influence the findings. Moreover, the diversity of the sample (40% African American) suggests that the findings were not a function of race. In addition, all patients had breast conserving surgery prior to study entry, and therefore the potential influence of diverse surgical treatments on study outcomes were minimized. The relatively small number of subjects limits the power and generalizability of the findings. However, the unexpectedly large effect size of the methylation changes in this small sample reflects the potential significance of the findings. Indeed, among the 485,513 CpG sites tested, 8 were significantly associated with chemotherapy and four were in exon 11 of transmembrane protein 49 (TMEM49). Of note these findings were subjected to strict Bonferroni correction methods requiring a significance level of p < 1.03 × 10−7. Reduced methylation in 4 of the 8 identified CpG sites was also reproduced 6 months after radiation, limiting the likelihood of spurious results. Reduced methylation in all of the 8 CpG sites also continued to correlate with peripheral inflammatory markers, although due to the limited number of chemotherapy-treated patients at 6 months, correlations between methylation status and plasma inflammatory markers in chemotherapy-treated patients alone did not reach statistical significance. Furthermore, differences in absolute levels of inflammatory markers in chemotherapy- versus non-chemotherapy-treated patients were not apparent at 6 months, possibly reflecting the small number of chemotherapy patients and the decay in methylation changes at 6 months. This decay in the methylation changes in chemotherapy-treated patients 6 months post radiation suggests a dynamic (and potentially reversible), treatment- related effect (e.g. chemotherapy) on DNA methylation patterns that may contribute to individual variability in responses to cancer therapy. Other examples of reversible methylation changes in peripheral tissues have been observed after environmental exposures including exercise and dietary manipulations (Barres et al., 2012; Jacobsen et al., 2012). No correlation was found between inflammatory markers and fatigue 6 months post radiation, although this may be explained by the relatively small number of patients assessed at this time point (N = 39) as well as the relatively low and variable levels of fatigue. Finally, DNA samples were extracted from a mix of PBMCs, and although leukocyte subtypes (granulocytes and lymphocytes) were controlled for in the analysis, it remains possible that chemotherapy-induced changes in cell type distribution, including the percentages or numbers of specific lymphocyte subsets may account for the findings. Moreover, which cell type may be most sensitive to chemotherapy-associated epigenetic changes remains unknown. Monocyte and T cell lymphocyte lineages constitute the greatest fraction of PBMCs under normal conditions, and both of these cell types have been shown to produce and to respond to IL-6 and TNF and express sTNFR2 (Gehr et al., 1992; Khalaf et al., 2010; Suzuki and Mihara, 2012; Tartaglia et al., 1993).
The notion that chemotherapy may leave an epigenetic imprint in non-malignant peripheral immune cells that is associated with inflammation represents a novel development in understanding how some breast cancer survivors may experience persistent inflammation months to years after treatment completion. Prospective studies are needed to determine which cancer therapies impart the greatest epigenetic burden, how long these changes persist, whether they are reversible and what mechanisms are involved.
Supplementary Material
Acknowledgments
Financial support
This work was supported in part by the National Cancer Institute (R21 CA155511); The Cooper Family Foundation Breast Cancer Initiative; and the Winship Cancer Institute of Emory University Kennedy Survivorship Pilot Grant Award. Research reported in this publication was also supported in part by the NIH/NCI under award number P30CA138292. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbi.2014.02.010.
Footnotes
Conflicts of interest
None.
Contributor Information
Alicia K. Smith, Email: alicia.smith@emory.edu.
Karen N. Conneely, Email: kconnee@emory.edu.
Thaddeus W.W. Pace, Email: twwpace@email.arizona.edu.
Donna Mister, Email: dmister@emory.edu.
Jennifer C. Felger, Email: jfelger@emory.edu.
Varun Kilaru, Email: vkilaru@emory.edu.
Mary J. Akel, Email: makel@emory.edu.
Paula M. Vertino, Email: pvertin@emory.edu.
Andrew H. Miller, Email: amill02@emory.edu.
Mylin A. Torres, Email: matorre@emory.edu.
References
- Ari F, Napieralski R, Ulukaya E, Dere E, Colling C, Honert K, Kruger A, Kiechle M, Schmitt M. Modulation of protein expression levels and DNA methylation status of breast cancer metastasis genes by anthracycline-based chemotherapy and the demethylating agent decitabine. Cell Biochem. Funct. 2011;29:651–659. doi: 10.1002/cbf.1801. [DOI] [PubMed] [Google Scholar]
- Avraham A, Uhlmann R, Shperber A, Birnbaum M, Sandbank J, Sella A, Sukumar S, Evron E. Serum DNA methylation for monitoring response to neoadjuvant chemotherapy in breast cancer patients. Int. J. Cancer. 2012;131:E1166–E1172. doi: 10.1002/ijc.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene. 2005;24:8061–8075. doi: 10.1038/sj.onc.1208955. [DOI] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28:1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Kwan L, Breen EC, Cole SW. Inflammation and behavioral symptoms after breast cancer treatment: do fatigue, depression, and sleep disturbance share a common underlying mechanism? J. Clin. Oncol. 2011;29:3517–3522. doi: 10.1200/JCO.2011.36.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Tao ML, Hu W, Belin TR, Sepah S, Cole S, Aziz N. Inflammatory biomarkers and fatigue during radiation therapy for breast and prostate cancer. Clin. Cancer Res. 2009;15:5534–5540. doi: 10.1158/1078-0432.CCR-08-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull SJ, Huezo-Diaz P, Binder EB, Cubells JF, Ranjith G, Maddock C, Miyazaki C, Alexander N, Hotopf M, Cleare AJ, Norris S, Cassidy E, Aitchison KJ, Miller AH, Pariante CM. Functional polymorphisms in the interleukin-6 and serotonin transporter genes, and depression and fatigue induced by interferon-alpha and ribavirin treatment. Mol. Psychiatry. 2009;14:1095–1104. doi: 10.1038/mp.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno AM, Fostel JM, To KK, Salcido CD, Martin SE, Chewning KJ, Wu CP, Varticovski L, Bates SE, Caplen NJ, Ambudkar SV. Single-step doxorubicin-selected cancer cells overexpress the ABCG2 drug transporter through epigenetic changes. Br. J. Cancer. 2008;98:1515–1524. doi: 10.1038/sj.bjc.6604334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekhun VF, Lukyanova NY, Kovalchuk O, Tryndyak VP, Pogribny IP. Epigenetic profiling of multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals novel hyper- and hypomethylated targets. Mol. Cancer Ther. 2007;6:1089–1098. doi: 10.1158/1535-7163.MCT-06-0663. [DOI] [PubMed] [Google Scholar]
- D’Anello L, Sansone P, Storci G, Mitrugno V, D’Uva G, Chieco P, Bonafe M. Epigenetic control of the basal-like gene expression profile via Interleukin-6 in breast cancer cells. Mol. Cancer. 2010;9:300. doi: 10.1186/1476-4598-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehr G, Gentz R, Brockhaus M, Loetscher H, Lesslauer W. Both tumor necrosis factor receptor types mediate proliferative signals in human mononuclear cell activation. J. Immunol. 1992;149:911–917. [PubMed] [Google Scholar]
- Geinitz H, Zimmermann FB, Stoll P, Thamm R, Kaffenberger W, Ansorg K, Keller M, Busch R, van Beuningen D, Molls M. Fatigue, serum cytokine levels, and blood cell counts during radiotherapy of patients with breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2001;51:691–698. doi: 10.1016/s0360-3016(01)01657-1. [DOI] [PubMed] [Google Scholar]
- Giese-Davis J, Collie K, Rancourt KM, Neri E, Kraemer HC, Spiegel D. Decrease in depression symptoms is associated with longer survival in patients with metastatic breast cancer: a secondary analysis. J. Clin. Oncol. 2011;29:413–420. doi: 10.1200/JCO.2010.28.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SC, Brons C, Bork-Jensen J, Ribel-Madsen R, Yang B, Lara E, Hall E, Calvanese V, Nilsson E, Jorgensen SW, Mandrup S, Ling C, Fernandez AF, Fraga MF, Poulsen P, Vaag A. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia. 2012;55:3341–3349. doi: 10.1007/s00125-012-2717-8. [DOI] [PubMed] [Google Scholar]
- Kesler S, Janelsins M, Koovakkattu D, Palesh O, Mustian K, Morrow G, Dhabhar FS. Reduced hippocampal volume and verbal memory performance associated with interleukin-6 and tumor necrosis factor-alpha levels in chemotherapy-treated breast cancer survivors. Brain Behav. Immun. 2012 doi: 10.1016/j.bbi.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf H, Jass J, Olsson PE. Differential cytokine regulation by NF-kappaB and AP-1 in Jurkat T-cells. BMC Immunol. 2010;11:26. doi: 10.1186/1471-2172-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru V, Barfield RT, Schroeder JW, Smith AK, Conneely KN. MethLAB: a graphical user interface package for the analysis of array-based DNA methylation data. Epigenetics. 2012;7:225–229. doi: 10.4161/epi.7.3.19284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mills PJ, Rissling M, Fiorentino L, Natarajan L, Dimsdale JE, Sadler GR, Parker BA, Ancoli-Israel S. Fatigue and sleep quality are associated with changes in inflammatory markers in breast cancer patients undergoing chemotherapy. Brain Behav. Immun. 2012;26:706–713. doi: 10.1016/j.bbi.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotrich FE. Major depression during interferon-alpha treatment: vulnerability and prevention. Dialogues Clin. Neurosci. 2009;11:417–425. doi: 10.31887/DCNS.2009.11.4/felotrich. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzig M, Nickles D, Falschlehner C, Lehmann-Koch J, Straub BK, Roth W, Boutros M. An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int. J. Cancer. 2011;129:607–618. doi: 10.1002/ijc.26124. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Liggett TE, Melnikov AA, Monitto CL, Kusuke D, Shiga K, Kobayashi T, Horii A, Chatterjee A, Levenson VV, Koch WM, Sidransky D, Chang X. Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle. 2012;11:1656–1663. doi: 10.4161/cc.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgis P, Lambert AW, Ozturk S, Gao F, Pan H, Manne U, Alekseyev YO, Thiagalingam A, Abdolmaleky HM, Lenburg M, Thiagalingam S. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res. 2010;70:968–978. doi: 10.1158/0008-5472.CAN-09-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell A, Fleming J, Bennett S, Burmeister B, Haines T. Determining the minimal clinically important difference criteria for the Multidimensional Fatigue Inventory in a radiotherapy population. Support. Care Cancer. 2010;18:307–315. doi: 10.1007/s00520-009-0653-z. [DOI] [PubMed] [Google Scholar]
- Raison CL, Borisov AS, Majer M, Drake DF, Pagnoni G, Woolwine BJ, Vogt GJ, Massung B, Miller AH. Activation of central nervous system inflammatory pathways by interferon-alpha: relationship to monoamines and depression. Biol. Psychiatry. 2009;65:296–303. doi: 10.1016/j.biopsych.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam MK, Sethi G. Role of epigenetics in inflammation-associated diseases. Subcell. Biochem. 2012;61:627–657. doi: 10.1007/978-94-007-4525-4_27. [DOI] [PubMed] [Google Scholar]
- Shapiro CL, Recht A. Side effects of adjuvant treatment of breast cancer. N. Engl. J. Med. 2001;344:1997–2008. doi: 10.1056/NEJM200106283442607. [DOI] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Parshad R, Gupta SD, Ralhan R. DNA methylation of circulating DNA: a marker for monitoring efficacy of neoadjuvant chemotherapy in breast cancer patients. Tumour Biol. 2012;33:1837–1843. doi: 10.1007/s13277-012-0443-y. [DOI] [PubMed] [Google Scholar]
- Smets EM, Garssen B, Bonke B, De Haes JC. The Multidimensional Fatigue Inventory (MFI) psychometric qualities of an instrument to assess fatigue. J. Psychosom. Res. 1995;39:315–325. doi: 10.1016/0022-3999(94)00125-o. [DOI] [PubMed] [Google Scholar]
- Smith AK, Simon JS, Gustafson EL, Noviello S, Cubells JF, Epstein MP, Devlin DJ, Qiu P, Albrecht JK, Brass CA, Sulkowski MS, McHutchinson JG, Miller AH. Association of a polymorphism in the indoleamine- 2,3-dioxygenase gene and interferon-alpha-induced depression in patients with chronic hepatitis C. Mol. Psychiatry. 2012;17:781–789. doi: 10.1038/mp.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel ME. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol. Methodol. 1982;13:290–312. [Google Scholar]
- Soltanpour MS, Amirizadeh N, Zaker F, Oodi A, Nikougoftar M, Kazemi A. MRNA expression and promoter DNA methylation status of CDKi p21 and p57 genes in ex vivo expanded CD34(+) cells following co-culture with mesenchymal stromal cells and growth factors. Hematology. 2013;18:30–38. doi: 10.1179/1607845412Y.0000000030. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Mihara M. Adiponectin induces CCL20 expression synergistically with IL-6 and TNF-alpha in THP-1 macrophages. Cytokine. 2012;58:344–350. doi: 10.1016/j.cyto.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Swisher EM, Gonzalez RM, Taniguchi T, Garcia RL, Walsh T, Goff BA, Welcsh P. Methylation and protein expression of DNA repair genes: association with chemotherapy exposure and survival in sporadic ovarian and peritoneal carcinomas. Mol. Cancer. 2009;8:48. doi: 10.1186/1476-4598-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia LA, Goeddel DV, Reynolds C, Figari IS, Weber RF, Fendly BM, Palladino MA., Jr Stimulation of human T-cell proliferation by specific activation of the 75-kDa tumor necrosis factor receptor. J. Immunol. 1993;151:4637–4641. [PubMed] [Google Scholar]
- Tevaarwerk AJ, Lee JW, Sesto ME, Buhr KA, Cleeland CS, Manola J, Wagner LI, Chang VT, Fisch MJ. Employment outcomes among survivors of common cancers: the Symptom Outcomes and Practice Patterns (SOAPP) study. J. Cancer Surviv. 2013 doi: 10.1007/s11764-012-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Pace TW, Liu T, Felger JC, Mister D, Doho GH, Kohn JN, Barsevick AM, Long Q, Miller AH. Predictors of depression in breast cancer patients treated with radiation: role of prior chemotherapy and nuclear factor kappa B. Cancer. 2013;119:1951–1959. doi: 10.1002/cncr.28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Williams LA, Krishnan S, Liao Z, Liu P, Mao L, Shi Q, Mobley GM, Woodruff JF, Cleeland CS. Serum sTNF-R1, IL-6, and the development of fatigue in patients with gastrointestinal cancer undergoing chemoradiation therapy. Brain Behav. Immun. 2012;26:699–705. doi: 10.1016/j.bbi.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratten C, Kilmurray J, Nash S, Seldon M, Hamilton CS, O’Brien PC, Denham JW. Fatigue during breast radiotherapy and its relationship to biological factors. Int. J. Radiat. Oncol. Biol. Phys. 2004;59:160–167. doi: 10.1016/j.ijrobp.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Schmid T, Rubsamen D, Colburn NH, Irie K, Murakami A. Downregulation of programmed cell death 4 by inflammatory conditions contributes to the generation of the tumor promoting microenvironment. Mol. Carcinog. 2010;49:837–848. doi: 10.1002/mc.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MR, Santhanam AN, Yoshikawa N, Colburn NH. Have tumor suppressor PDCD4 and its counteragent oncogenic miR-21 gone rogue? Mol. Interv. 2010;10:76–79. doi: 10.1124/mi.10.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 2011;22:1668–1681. doi: 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.