Abstract
The timely secretion of gonadal sex steroids is essential for the initiation of puberty, the post-pubertal maintenance of secondary sexual characteristics and the normal perinatal development of male external genitalia. Normal gonadal steroid production requires the actions of the pituitary-derived gonatrophins, LH and FSH. We report four human pedigrees with severe congenital gonadotrophin deficiency and pubertal failure in which all affected individuals are homozygous for loss-of-function mutations in TAC3 (encoding Neurokinin B) or its receptor TACR3 (encoding NK3R). Neurokinin B, a member of the substance P-related tachykinin family, is known to be highly expressed in hypothalamic neurons that also express kisspeptin1, a recently identified regulator of gonadotropin-releasing hormone secretion2. These findings implicate Neurokinin B as a critical central regulator of human gonadal function and suggest novel approaches to the pharmacological control of human reproduction and sex hormone-related diseases.
The hallmark of sexual maturity is secretion from the pituitary gland of the gonadotrophins luteinising hormone (LH) and follicle stimulating hormone (FSH) which act together, in gonadal tissue, to drive both sex hormone secretion and gametogenesis3. The production of LH and FSH from pituitary gonadotrope cells is largely controlled by the pulsatile delivery of gonadotrophin releasing hormone (GnRH) from a functionally interconnected group of secretory neurons whose cell bodies are located predominantly in the medial basal hypothalamus in humans3. This complex central neuroendocrine regulatory unit is susceptible to negative feedback regulation from the gonads by sex steroids3, however although other physiological regulators have been proposed, there is considerable uncertainty about the relative importance and species specificity of many of these inputs4. The gonadal endocrine system is active in utero and for the first few months of life before being centrally suppressed. The mechanisms subserving this neural suppression and the rekindling of the GnRH pulse generating system at the beginning of puberty are, however, an enduring enigma5.
Recently, through a combination of human and murine genetics, the hypothalamically-expressed peptide kisspeptin and its receptor GPR54 have been established to be necessary for normal GnRH secretion and thus gonadotrope function6,7. Human genetics has been a powerful contributor to the discovery of critical molecular elements of the central control of reproduction identifying a number of molecules which are essential for the embryonic migration of GnRH neurons to the hypothalamus from the olfactory placode (KAL1, NELF, FGFR1, PROK1, PROKR2). Humans with genetic defects in those molecules generally present clinically with failure to enter puberty and low circulating gonadotropin levels (hypogonadotropic hypogonadism (HH)) associated with impairment of olfactory sensation (MIM-308700,147950). Mutations either in the GnRH receptor or in GPR54 have been reported in humans with familial HH but normal olfaction (so called normosmic idiopathic HH (nIHH; MIM-146110)). However these genetic defects explain only a minority of cases of familial nIHH8 implying the existence of other, as yet unknown, key positive regulators of the central control of the reproductive axis.
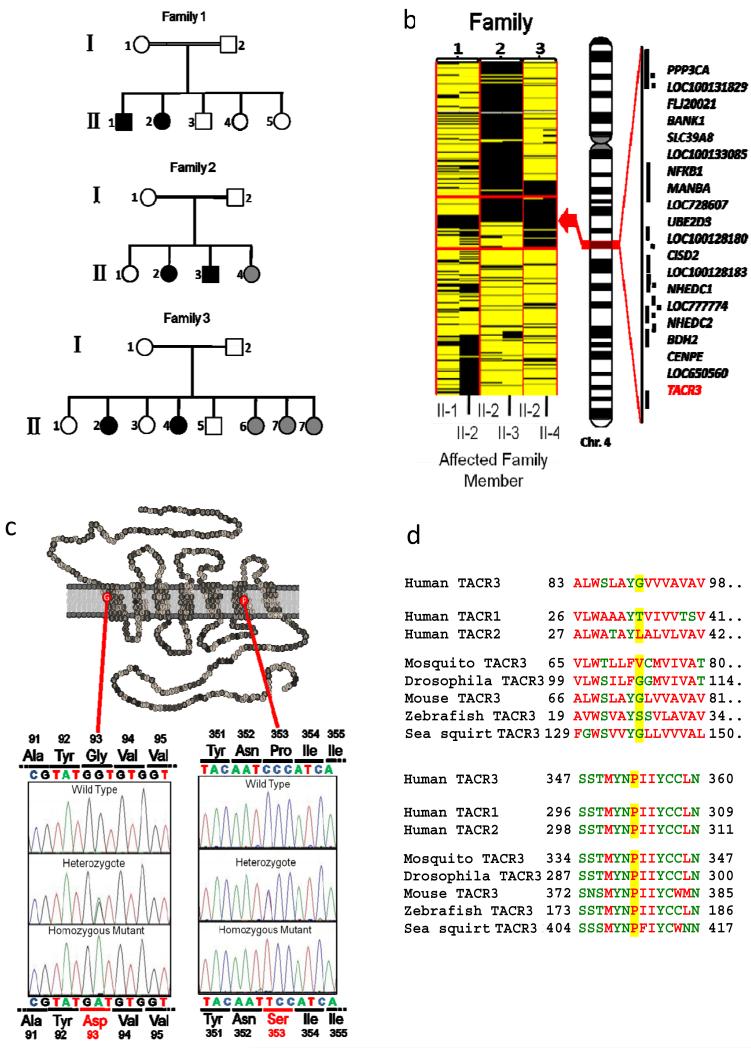
Nine consanguineous Turkish families with multiple members affected by nIHH were subjected to genome-wide SNP analysis, and within each family regions of homozygosity common to all affected, but not found in unaffected individuals, were identified9. In three families (Fig 1A) homozygosity at a locus on the long arm of chromosome 4 cosegregated with nIHH. Alignment of all 6 affected members of the three families refined the critical interval to a 2.74 MB genomic region extending from 102.20 to 104.94 MB encompassing 20 known or predicted genes (Fig 1B). Of those genes TACR3 (NM-001059) was prioritized for further analysis. TACR3 encodes NK3R, the receptor for Neurokinin B, a tachykinin peptide which is known to be highly expressed in hypothalamic neurons that also express kisspeptin1. Synthetic NK3R agonists have also been reported to modulate reproductive function but the site, and indeed direction, of such actions have been unclear10,11. Homozygous non-synonymous mutations in the coding sequence of TACR3 were found in all affected subjects (and no unaffected subjects) in all three families (supplementary note and figure). In family 1 all affected individuals were homozygous for a G to A transversion at cDNA nucleotide 278. This mutation leads to the substitution of glycine at residue 93 for aspartic acid (G93D). G93 lies in the first transmembrane domain of the G protein-coupled NK3R (Fig 1C), and, although it is not highly conserved (Fig 1D), introduction of a negatively charged side change in this very hydrophobic domain would likely be highly thermodynamically unfavourable. Indeed, only 1 of 739 members of the rhodopsin receptor superfamily has a charged side chain at this position. In families 2 and 3 a C to T transition at cDNA nucleotide 1057 was found, producing a proline to serine change at residue 353 (P353S), which lies within the sixth transmembrane domain (Fig 1C). P353 is exquisitely conserved (Fig 1D): 735 out of 739 members of the rhodopsin receptor superfamily have a proline at this position. Neither of these mutations were found in 100 Turkish controls, in the 6 other families from the original genome wide SNP-analysis, nor 50 patients with nIHH from a variety of ethnic groups.
Figure 1.
A. Families used to identify critical interval on chromosome 4 harbouring genetic defect causing nIHH B. Critical region on chromosome 4 showing homozygosity (black) in all affected individuals, with known or predicted genes in region indicated. C. Location of pathogenic mutation in NK3R D. evolutionary conservation of mutated residues.
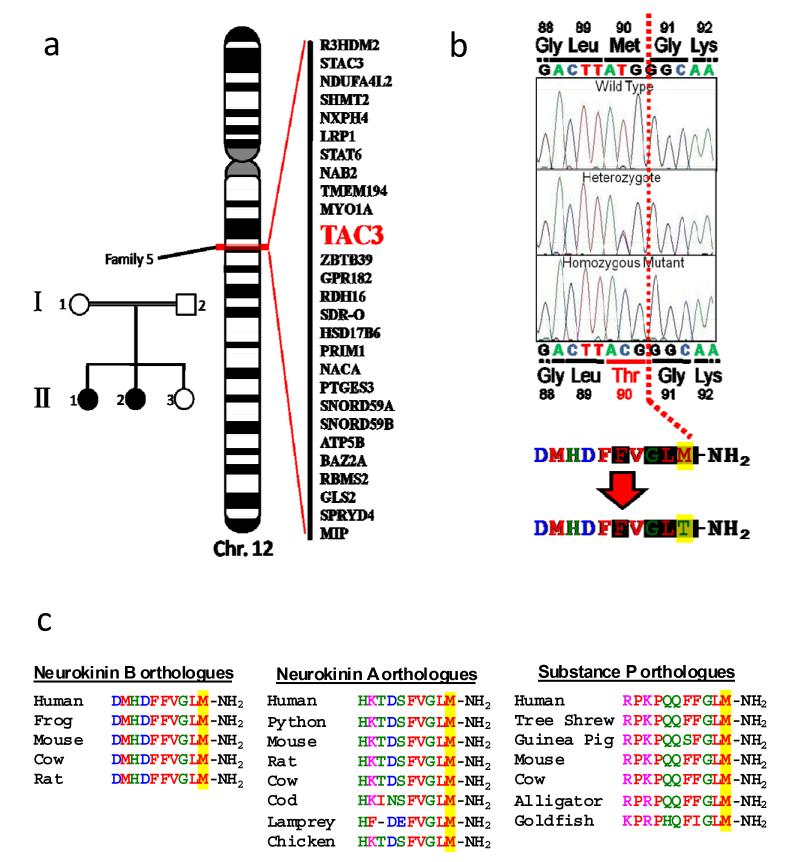
In one multiplex family in which TACR3 mutations had been excluded (Fig 2A) 2 autozygous regions were identified which were common to the two affected family members but not to their unaffected sister. Although these regions - from 125.7 to 130.4 MB on chromosome 4 and 54.5 to 57.6 MB on chromosome 12 - encompassed 21 and 102 known or predicted genes respectively, the only obvious biologically plausible candidate was TAC3 (NM-0013251), on chromosome 12 (Fig 2A), encoding neurokinin B, the preferred ligand for the neurokinin-3 receptor. On sequencing TAC3 we identified a homozygous T to C transversion at cDNA nucleotide 269 in both affected siblings, leading to substitution of methionine by threonine at residue 90 (M90T) of the prohormone (Fig 2B). The parents and unaffected sister were heterozygous for the mutation, which was not detected in 100 ethnically-matched controls. Met90 is the C terminal amino acid of the mature neurokinin B decapeptide, and falls within the canonical tachykinin motif, Phe-X-Gly-Leu-Met-NH2, which is universally conserved among known tachykinins12 (e.g. Fig 2C). In vivo NKB is known to be post-translationally modified by C-terminal amidation, which has been shown to be necessary for full activity of the closely related neurokinin A peptide13.
Figure 2.
A. Family used to identify region on chromosome 12 harbouring genetic defect causing nIHH with detail of subset of genes in critical interval B. Location of pathogenic mutation in TAC3. Shaded amino acids represent the tachykinin signature motif. C. Conservation of mutated residue among selected paralogues and orthologues.
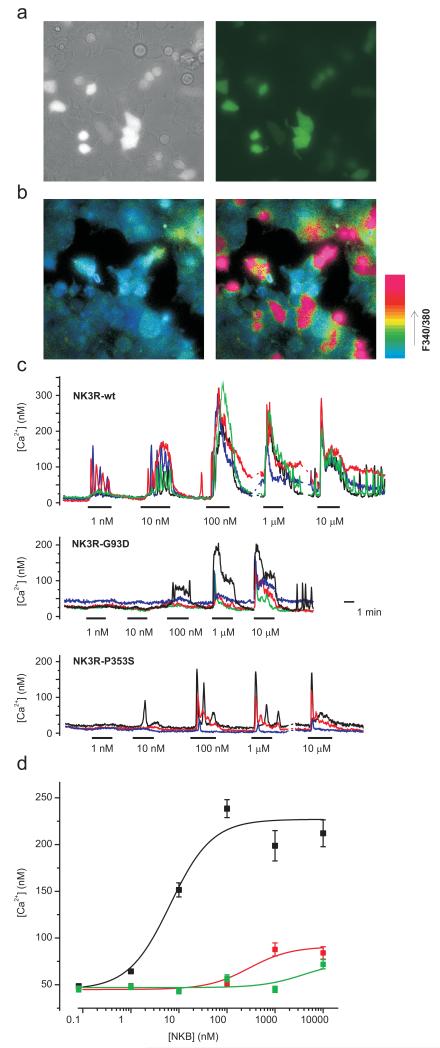
NK3R activation is linked to Gq proteins and its action results in an increase in intracellular calcium. To evaluate the functional consequences of the TACR3 mutations we expressed wild type and mutant TACR3 in HEK293 cells and assessed the ability of neurokinin B (NKB), to stimulate calcium flux. In transiently transfected cells, identified on the basis of coexpressed GFP, NKB elicited a robust increase in intracellular calcium with an EC50 of 6.6±1.3 nM, while no response to NKB was seen in untransfected cells (Fig 3). Both G93D and P353S mutant receptors showed severely attenuated ability to respond to stimulation, with EC50s for NKB of 290±146 nM and 4.0±2.1 μM respectively. Thus, both the missense TACR3 mutations found in all affected subjects in Families 1 through 3 show unequivocal evidence of impaired receptor signaling.
Figure 3.
Both missense mutations in NK3R result in loss of receptor function. A) Bright field (left) and GFP-fluorescent only (right) identifying TACR3 transfected HEK293 cells. B) Pseudocolour images of 340/380 nm ratio for the same optical field as in (A) before (left) and during (right) the application of 100 nM wt-NKB. Note that only GFP-positive cells respond. C) Representative Ca2+-responses of 3-5 wt- or mutant TACR3 transfected cells to application of different NKB doses as indicated. D) NKB dose responses for wt (black squares, n=93-206), G93D- (red squares, n=112-145) and P353S-mutant (green squares, n=156) TACR3. Peak [Ca2+]-responses recorded as in (C) in at least 5 dishes/construct were averaged over 30s and the data was fit with the logistic function [Ca2+] = (A1-A2)/(1+([NKB]/EC50))+A2 using origin software (Microcal). The resulting fit parameters were: wt-TACR3: A1=45±6 nM, A2=227±6 nM, EC50=6.6±1.3 nM; TACR3-G93D: A1=45±2 nM, A2=91±5 nM, EC50=290±146 nM; TACR3-P353S: A1=47±3 nM, A2=75 nM (fixed), EC50=4.0±2.1 μM. Error bars represent 1SE.
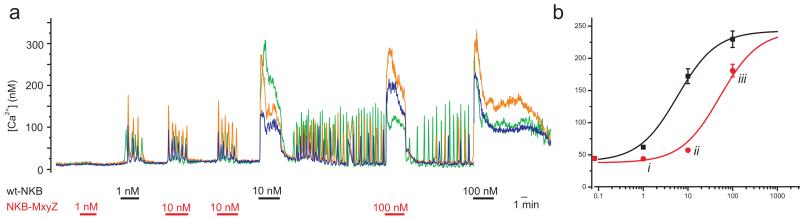
To assess the activity of the M90T mutant NKB found in Family 4, the mature mutant decapeptide was synthesized in both C terminal-amidated and non-amidated form, and the effects of each peptide on wild type receptor were assessed in the same heterologous expression system. The amidated mutant peptide (predicted to be the form that would occur in nature) had severely reduced activity, being around 10 fold less potent than amidated wild type NKB at stimulating the NK3R (EC50-M90T = 52±6 nM; Fig 4). As expected, the non-amidated mutant NKB had little detectable ability to elicit calcium flux (data not shown).
Figure 4.
The missense mutations in NKB results in severe loss of agonist function at the NK3R A) Representative Ca2+-responses of 3 wt-TACR3 transfected cells to application of different NKB and NKB-M90T doses as indicated. B) NKB dose responses for NKB (black squares), and NKB-M90T (red circles). Peak [Ca2+]-responses recorded as in (A) in 5 dishes (n=113 cells) were averaged over 30s and the data was fit with the logistic function [Ca2+] = (A1-A2)/(1+([NKB]/EC50))+A2 using origin software (Microcal). The resulting fit parameters were: NKB: A1=40±8 nM, A2=242±11 nM, EC50=5.8±1.5 nM; NKB-M90T: A1=38±4 nM, A2=242 nM (fixed), EC50=52±6 nM. Error bars represent 1SE; responses to similar doses of the two peptides were compared using a paired t-test; i p=3*10−12, ii p=3*10−20, iii p=6*10−7.
The phenotypes of subjects homozygous for either TAC3 or TACR3 mutations provide strong evidence that NKB, acting through its receptor NK3R, is essential for the central hormonal control of human reproduction (supplementary note and supplementary table 1). All subjects who are in the second decade of life or older show unequivocal evidence of failure of pubertal progression with low circulating sex steroid levels and pre-pubertal levels of circulating gonadotropins. Furthermore, all affected males have micropenis, strongly suggesting failure of normal intrauterine and perinatal activation of the reproductive axis14. The only homozygote (out of 9) in any of the four families who does not yet exhibit a clear phenotype is a 5 year-old female in whom HH would not be expected to have become overt.
NK3R is widely expressed, particularly in the central nervous system15, and previous studies in rodents utilising pharmacological or genetic manipulation have suggested potential roles in a wide range of CNS functions16-17. Additionally, polymorphisms in the TACR3 locus have been weakly genetically associated with a range of human neurobehavioural phenotypes18. In this regard, it is notable that patients with homozygous loss of function mutations in TACR3 lack any other clear neurobehavioural or other phenotypic abnormalities outside the reproductive axis. However, both affected subjects with TAC3 mutations do have mild learning disability (supplementary note) suggesting that NKB action, possibly through receptors other than NK3R, may have effects on higher cognitive function.
The precise mechanism whereby NKB and the NK3R exert their effects on the control of reproduction remains to be established. It is most likely, however, that this effect is exerted at the level of hypothalamic GnRH release. NK3R is expressed on rodent GnRH-expressing neurons19,20. Axons of neurons expressing NKB are closely anatomically apposed to those of GnRH neurons within the median eminence of the hypothalamus19,21, and NKB immunoreactive varicosities have been reported to in direct contact with GnRH immunoreactive axons22. NKB expression is highest in the arcuate nucleus, where it colocalises with estrogen receptor alpha (Era) and dynorphin23, implicated in progesterone feedback to GnRH secretion24. Nevertheless NK3R and NKB are also expressed in peripheral organs including ovaries and uterus25, so additional peripheral effects cannot be formally excluded. However there is a paucity of pharmacological or murine genetic evidence directly implicating NKB or NK3R in gonadal function. Mice with genetic ablation of Tac2 (the murine orthologue of TAC3) have not been described and studies of Tacr3-/- mice have not reported a reproductive phenotype17,26,27. Notably, central administration of a potent NK3R agonist to ovariectomized, oestrogen-primed rats suppressed LH production rather than stimulating it10. It thus remains possible that there is true divergence in the reproductive role of NKB/NK3R between humans and rodents.
In summary, we have identified loss-of-function mutations in either neurokinin B or its receptor in four out of nine multiplex families affected by hypogonadotrophic hypogonadism. These findings establish that NKB action via the NK3R is necessary for the central neuroendocrine control of human reproduction. These families represents the first examples of inherited defects of tachykinin signalling in any human disorder and suggest that the NKB/NK3R system may provide a novel avenue for the pharmacological manipulation of human fertility and the treatment of sex steroid-related diseases, such as cancer of the breast and prostate.
Methods Summary
The ethics committee of the Cukurova University Faculty of Medicine approved this study, and full informed consent was obtained for each participant. nIHH was defined by conventional criteria (supplementary methods). Normal karyotypes were confirmed and mutations in GNRHR, GNRH1, GPR54, KISS1, KAL1, NELF, PROK2, PROK2R and FGFR1 genes were ruled out by sequencing. Whole genome SNP analysis used 250K NspI Affymetrix SNP microarrays (Affymetrix, CA, USA) and data were analyzed using AutoSNPa software9. PCR-amplified exons and splice junctions of TAC3 or TACR3 were sequenced on an ABI PRISM 3130 autosequencer (Applied Biosystems, Foster City, CA). For primer sequences and annealing temperatures see supplementary Table 2.
Wild type or mutant TACR3 in pIRES2-AcGFP1 (Clontech) were transfected into HEK293A cells (QBiogene) using polyethylenimine (Sigma) before seeding cells into poly-L-lysine-coated glass-bottomed dishes 16 hours later. At 24 hours cells were loaded with Fura2-AM for 30 min. Experiments were performed on an inverted fluorescence microscope (Eclipse TE2000, Nikon, UK) with a 40x oil-immersion objective. Fura2 was excited at 340, 360 and 380 nm, and GFP at 475 nm, using a 75W xenon arc lamp and a monochromator (Cairn Research, Faversham, UK) and MetaFluor software (Molecular Devices, UK). Emission, filtered at 510/80 and 535/25 for Fura2 and GFP respectively, was recorded with a QuantEM CCD camera (Photometrics, Roper Scientific, UK). NKB (Sigma) or custom made mutant NKB with or without C terminal amidation (New England Peptides) were perfused in bath solution at ~1 ml/min with a chamber volume of ~0.2 ml. 340/380 nm ratio was calculated from images taken at 100 ms excitation at each wavelength at 0.5 Hz. Free Ca2+-concentrations were calculated for individual cells after background subtraction using the equation of Grynkiewicz et al (1985) assuming a KD of 224 nM. Minimal and maximal signals were recorded in the presence of 5 mM ionomycin in 5 mM EGTA/0 mM Ca2+ and 5 mM Ca2+ respectively at the end of the experiment. Further detail is given in the supplementary methods.
Supplementary Material
Acknowledgements
This work was supported by grants to A.K.T., N.O.M., and B.Y. from The Scientific and Technological Research Council of Turkey (TÜBİTAK: The Support Programme for Scientific and Technological Research Projects (1001), Grant no: 106S276), by the Wellcome Trust (R.K.S.: Intermediate Clinical Fellowship 080952/Z/06/Z; S.O.: Programme Grant 078986/Z/06/Z; F.R.: Senior Basic Science Fellowship 084210/Z/07/Z) and the U.K. NIHR Cambridge Biomedical Research Centre. F.R. has also received support from St. John’s College, Cambridge.
Footnotes
List of URLs. AutoSNPa is available at http://dna.leeds.ac.uk/autosnpa/. The PRINTS-S database of protein family fingerprints is at http://www.bioinf.man.ac.uk/dbbrowser/PRINTS.
Supplementary Information is linked to the online version of this manuscript at www.nature.com/nature
References
- 1.Goodman RL, et al. Endocrinology. 2007;148(12):5752. doi: 10.1210/en.2007-0961. [DOI] [PubMed] [Google Scholar]
- 2.Gianetti E, Seminara S. Reproduction. 2008;136(3):295. doi: 10.1530/REP-08-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plant TM. J Neuroendocrinol. 2008;20(6):719. doi: 10.1111/j.1365-2826.2008.01708.x. [DOI] [PubMed] [Google Scholar]
- 4.Herbison AE. In: Knobil and Neill’s Physiology of Reproduction. Neill JD, editor. Elsevier; Amsterdam: 2006. pp. 1415–1482. [Google Scholar]
- 5.Herbison AE. Hormone research. 2007;68(Suppl 5):75. doi: 10.1159/000110583. [DOI] [PubMed] [Google Scholar]
- 6.Seminara SB, et al. N Engl J Med. 2003;349(17):1614. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 7.de Roux N, et al. Proc Natl Acad Sci U S A. 2003;100(19):10972. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley WF, Jr., Pitteloud N, Seminara S. Trans Am Clin Climatol Assoc. 2008;119:29. [PMC free article] [PubMed] [Google Scholar]
- 9.Carr IM, et al. Hum Mutat. 2006;27(10):1041. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 10.Sandoval-Guzman T, Rance NE. Brain Res. 2004;1026(2):307. doi: 10.1016/j.brainres.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Rance NE. Peptides. 2008 [Google Scholar]
- 12.Almeida TA, et al. Curr Med Chem. 2004;11(15):2045. doi: 10.2174/0929867043364748. [DOI] [PubMed] [Google Scholar]
- 13.Patacchini R, et al. J Pharmacol Exp Ther. 1993;264(1):17. [PubMed] [Google Scholar]
- 14.Grumbach MM. J Clin Endocrinol Metab. 2005;90(5):3122. doi: 10.1210/jc.2004-2465. [DOI] [PubMed] [Google Scholar]
- 15.Pinto FM, et al. Eur J Pharmacol. 2004;494(2-3):233. doi: 10.1016/j.ejphar.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Hasenohrl RU, et al. Neuropeptides. 2000;34(5):272. doi: 10.1054/npep.2000.0824. [DOI] [PubMed] [Google Scholar]
- 17.Siuciak JA, et al. Psychopharmacology (Berl) 2007;194(2):185. doi: 10.1007/s00213-007-0828-6. [DOI] [PubMed] [Google Scholar]
- 18.Saito S, et al. Neuroreport. 2008;19(4):471. doi: 10.1097/WNR.0b013e3282f600b4. [DOI] [PubMed] [Google Scholar]; Foroud T, et al. Alcohol Clin Exp Res. 2008;32(6):1023. doi: 10.1111/j.1530-0277.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krajewski SJ, et al. J Comp Neurol. 2005;489(3):372. doi: 10.1002/cne.20626. [DOI] [PubMed] [Google Scholar]
- 20.Todman MG, Han SK, Herbison AE. Neuroscience. 2005;132(3):703. doi: 10.1016/j.neuroscience.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 21.Goubillon ML, et al. Endocrinology. 2000;141(11):4218. doi: 10.1210/endo.141.11.7743. [DOI] [PubMed] [Google Scholar]
- 22.Ciofi P, Leroy D, Tramu G. Neuroscience. 2006;141(4):1731. doi: 10.1016/j.neuroscience.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 23.Burke MC, Letts PA, Krajewski SJ, Rance NE. J Comp Neurol. 2006;498(5):712. doi: 10.1002/cne.21086. [DOI] [PubMed] [Google Scholar]
- 24.Goodman RL, et al. Endocrinology. 2004;145(6):2959. doi: 10.1210/en.2003-1305. [DOI] [PubMed] [Google Scholar]
- 25.Patak E, et al. Br J Pharmacol. 2003;139(3):523. doi: 10.1038/sj.bjp.0705279. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loffler S, et al. Regul Pept. 2004;122(2):131. doi: 10.1016/j.regpep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Nordquist RE, et al. Psychopharmacology (Berl) 2008;198(2):211. doi: 10.1007/s00213-008-1119-6. [DOI] [PubMed] [Google Scholar]
- 27.Kung TT, et al. Pharmacol Res. 2004;50(6):611. doi: 10.1016/j.phrs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Rosenfield RL. In: Pediatric endocrinology: A practical clinical guide. Radovick S, MacGillivray MH, editors. Humana Press; 2003. p. 451. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.