Abstract
Highly conserved Inhibitor of DNA Binding (ID1-4) genes encode multi-functional proteins whose transcriptional activity is based on dominant negative inhibition of basic helix-loop-helix (bHLH) transcription factors. Initial animal models indicated a degree of compensatory overlap between ID genes such that deletion of multiple ID genes was required to generate easily recognizable phenotypes. More recently, new model systems have revealed alterations in mice harboring deletions in single ID genes suggesting complex gene and tissue specific functions for members of the ID gene family. Because ID genes are highly expressed during development and their function is associated with a primitive, proliferative cellular phenotype there has been significant interest in understanding their potential roles in neoplasia. Indeed, numerous studies indicate an oncogenic function for ID1, 2 and 3. In contrast, the inhibitor of differentiation 4 (ID4) presents a paradigm shift in context of well-established role of ID1, 2 and 3 in development and cancer. Apart from some degree of functional redundancy such as HLH dependent interactions with bHLH protein E2A, many of the functions of ID4 are distinct from ID1, 2 and 3: ID4 proteins a) regulate distinct developmental processes and tissue expression in the adult, b) promotes stem cell survival, differentiation and/or timing of differentiation, c) epigenetic inactivation/loss of expression in several advanced stage cancers and d) increased expression in some cancers such as those arising in the breast and ovary. Thus, in spite of sharing the conserved HLH domain, ID4 defies the established model of ID protein function and expression. The underlying molecular mechanism responsible for the unique role of ID4 as compared to other ID proteins still remains largely un-explored. This review will focus on the current understanding of ID4 in context of development and cancer.
Keywords: ID4, bHLH, Development, Cancer Angiogenesis, Senescence, Proliferation, Androgen Independence, cancer, epigenetics, differentiation
1. Introduction
ID proteins (ID1, ID2, ID3 and ID4) are dominant negative transcriptional regulators of basic Helix Loop Helix (bHLH) transcription factors that lack the basic DNA binding domain but have intact HLH domain [1]. Thus ID proteins can interact with bHLH proteins, but the heterodimer fails to bind DNA and activate E-Box dependent transcription of target genes. ID proteins regulate the function of various ubiquitously expressed and tissue specific bHLH transcription factors as well as many non-bHLH proteins with different affinities [2-4] in complex transcriptional networks. In this review we focus on the current understanding of ID4 in development and cancer. The global perspective on ID proteins has been expertly reviewed elsewhere [1, 5-8].
The de-regulation of ID proteins contributes to developmental defects and neoplastic progression. Based on significant sequence homology of ID protein sub-types within the HLH domain, a degree of functional redundancy is expected at least in their interaction with bHLH transcription factors. However, recent studies suggest that each of the ID sub-type also participates/regulates unique biological activities which is evident from non-compensatory phenotypes in ID specific knockout models and preferred protein interactions, both bHLH and non-bHLH. In this context Inhibitor of differentiation/DNA-binding 4 (ID4) has emerged as an outlier in terms of expression and function. Evidence from animal models indicates that phenotypic changes and molecular pathways regulated by ID1, ID2 and ID3, in general are not similar to those regulated by ID4. The opposing function of ID4 (focus of this review) versus IDs 1, 2 and 3 also suggest that the core function of ID proteins as dominant negative bHLH transcriptional regulators may be just a fraction of their overall function. The majority of ID subtype specific function could involve unknown and perhaps yet undefined interactions with sequence specific bHLH or non-bHLH proteins resulting in non-overlapping biological endpoints.
2. ID4 sequence and structural properties
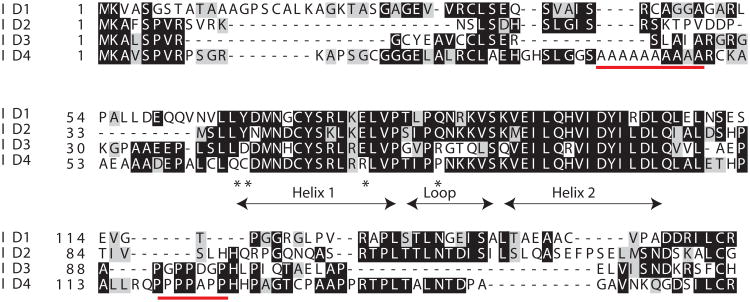
ID4 is located on a 4Mb region on chromosome 6p22.3 and consists of three exons [9]. Exons 1 and 2 code for the protein whereas exon 3 exists only as a 3′ un-translated region. ID4 is the longest protein within the ID family with 161 residues (Fig. 1) and shares the core HLH domain but divergent N- and C- terminal domains [2] compared with other ID proteins (Fig. 1). At least four amino acids are unique in the ID4 HLH domain: glutamine, cysteine and arginine in helix 1 and proline in the loop region (Fig. 1, indicated by asterisks). In general all ID proteins have between 3 (ID1), 4 (ID2) and 10 (ID3) amino acids changes respectively (7-24%) as compared to the conserved HLH domain sequence (41 amino acid stretch) [10]. The subtle changes in the core HLH domain but highly divergent N-terminal (Alanine rich) and C-terminal (Proline rich) domains (Fig. 1, indicated by red lines) could result in unique function of ID4 as opposed to the other ID protein family members. Therefore, ID4 can be considered as a remote homologue of ID1, 2 and 3.
Figure 1.
Protein sequence alignment of the four known human ID proteins (CLUSTAL W). The red lines indicate the alanine and proline rich regions at the N- and C-terminal of ID4. The conserved Helix-loop-Helix region and the amino acids that are unique to ID4 HLH domain is indicated (Asterisk).
The origins of ID proteins can be traced back to the D. melanogaster extramacrochaetae (emc) gene (reviewed in [11]). As orthologs of emc, the four ID proteins are paralogs that appeared through gene duplication in vertebrates.
The poly-alanine rich N-terminal tract in ID4 appears to be rapidly evolving. Similar to the poly-alanine tracts of HOXA13 [12] and class III POU [13] transcription factors that appeared only in mammals, the poly-alanine tract in ID4 also first appeared in primitive mammals such as opossum (marsupials) but is absent in lower vertebrates such as alligators, xenopus, zebra fish and birds (Gallus, Finch) (Fig. 2). Thus the poly-alanine tract in ID4 evolved independently as compared to its paralogs ID1, 2 and 3, hence conserved for functional reasons. The poly-alanine tract in ID4 is consistent with similar tracts which are preferentially found in many transcription factors and lies outside, usually towards the N-terminal of the functional domains (such as N-terminal to HLH domain in ID4). This tract generally acts as a flexible spacer element located between the functional domain of a protein and therefore essential to protein conformation, protein–protein interactions and/or DNA binding [14]. The length of the poly-alanine tract in ID4 (n=10) is also within threshold for normal function, whereas an increase in the length beyond this threshold results in human diseases (≤10) [15]. In fact, structural studies suggest that none of the N- and C- terminal fragments of any of the ID proteins adopts a helical conformation, except the N-terminal 27-64 fragment of ID4 [10], a motif that is dictated by the alanine residues 39 and 48 (Fig. 1). Thus the alanine rich N-terminal domain could be a functionally important domain in ID4. In this context, the unique function of ID4 suggests the potential for positive selective pressure for developing and maintaining the divergent Id4 locus during evolution.
Figure 2.

Alignment of ID4 protein sequences from various species. The respective ID4 sequences were obtained from NCBI and sequences were aligned using MAFFT v7 using default parameters. The phylogenetic analysis was also performed within MAFFT using NJ/UPGMA phylogeny.
The over-representation of proline within the C-terminal end may also impart additional structural and functional features unique to ID4. Proline is a strong promoter of intrinsic disorder [16, 17] and thus likely supports the internal disorder at the ID4 C-terminal domain. Indeed, the overall percent disorder (predicted by PONDR: Predictor of Naturally Disordered Regions [18]) in ID4 is highest among all the ID proteins: ID4-70.2%, ID2-59.7%, ID3-35.29% and ID1-28.6%. The low complexity proline rich region in ID4 suggests that ID4 like other internally disordered proteins also lacks a defined 3-D structure at the C-terminal that favors protein-protein interactions by presenting a larger interaction surface allowing multiple binding partners [17, 19]. Hence structural divergence through acquisition of new functionally relevant domains in ID4 suggests a potential novel role in development and differentiation as compared to IDs 1, 2 and 3.
3. The Role of ID4 during development and differentiation
ID proteins are expressed by essentially all cell lineages at some point during embryonic development. Consensus suggests that ID expression is highest in undifferentiated, proliferating populations and subsequently down-regulated as these cells exit from cell cycle and terminally differentiate [7].
By in situ hybridization on mouse development post-gastrulation, ID1, 2 and 3 expression is observed in multiple tissues whereas the expression of ID4 is non-overlapping and restricted to neuronal tissues and in the ventral portion of the epithelium in developing stomach [20]. In adult human tissues, ID4 expression is observed in brain, thyroid, testis and pancreas [21]. ID4 is also required for normal mammary [22] and prostate gland development [23].
ID4 is highly expressed in osteoblasts [24], adipocytes [25], prostate epithelial cells [23] neurons [26], testicular Sertoli cells [27] and during differentiation in glial cells [28] supporting its role as a pro-differentiation factor. ID4 is also expressed in germ cells at various stages of development: spermatogonial stem cells [29], spermatocytes, pachytene spermatocytes and spermatids [30].
Unexpectedly, overexpression of ID4 in oligodendrocyte progenitor cells (OPC) prevents differentiation associated with a decrease in the endogenous expression of all myelin genes. Conversely, OPCs lacking ID4 display precocious differentiation and increased apoptosis [31] suggesting that ID4 is required for the development of oligodendrocytes. A progressive decline in ID4 transcription is also a part of the intracellular timer that helps determine when oligodendrocyte precursor cells withdraw from the cell cycle and differentiate [32]. This unique phenotype supports the role of ID4 as inhibitor of differentiation, the classical function of ID proteins.
Immuno-cytochemical studies have shown that ID4 is localized to the nucleus in OPCs [32] and spermatids, but remains cytoplasmic in spermatocytes [30].
Collectively, the studies suggest that ID4 can act as pro- or anti-differentiation factor in a cell specific manner. The integration of various cellular events such as response to specific ligands and possibly cell specific interacting proteins could eventually determine whether ID4 regulates proliferation and/or differentiation and cellular localization.
4. ID4 knockout mouse models
4.1 Global ID4 knockout Model
Two different global ID4 knockout (ID4-/-) mouse models exist. In the first ID4-/- model (strain 129X1/Svj,) developed by Mark Israel's group (ID4tm1Mais in MGI database [33]), exons 1 and 2 were replaced by GFP and neomycin-resistance genes via homologous recombination in JM-1 ES line [26]. The second ID4-/- model (strain 129P2/OlaHsd) was developed by Fred Sablitzky's group (ID4tm1Fsky in MGI database) [34]. In this model the sequence encoding HLH domain and most of the C-terminus of the gene was replaced by the lacZ-neo cassette. The cassette was inserted in-frame, allowing the expression of a fusion gene encoding the N-terminal 65 amino acids of ID4 fused to beta-galactosidase.
Both the ID4-/- mouse models displayed essentially similar phenotypes in the brain (decreased brain size, abnormal fat cell morphology) and mammary gland. The ID4tm1Fsky model appears to have a more severe phenotype as a result of associated embryonic lethality (50% die in utero or neonatally). The surviving homozygous mutant mice lose weight rapidly, probably due to a defect in abnormal adipose tissue development and osteoporosis with only 20% surviving through adulthood. A similar phenotype in the ID4tm1Mais model was not reported.
4.1.1 ID4 and neuronal development (ID4tm1Mais and ID4tm1Fsky)
During development, depletion of ID4 in mice also revealed its essential role in normal brain development and function, where it is required for neural stem cell proliferation and differentiation [26], lateral expansion of the proliferative zone in the developing cortex, and hippocampus and proliferation in the ventricular zones [26, 34]. In the absence of ID4, neural precursor cells proliferate more slowly than their wild type counterpart, highlighting the essential regulatory role of ID4 during neural stem cell proliferation and fate determination. Though not investigated, the cognitive abilities of ID4-/- mice could provide some interesting clinical correlates.
4.1.2. ID4 and mammary gland morphogenesis (ID4tm1Mais and ID4tm1Fsky)
In mouse mammary gland, Id4 is expressed in cap cells, basal cells and in a subset of luminal epithelial cells where it promotes ductal elongation and branching morphogenesis [22]. Targeted ID4 deletion impairs ductal expansion and branching morphogenesis as well as cell proliferation induced by estrogen and/or progesterone. ID4 also maintained the survival of normal mammary cells as well as cultured mammary tumor cells [22].
4.1.3. ID4 and spermatogenesis (ID4tm1Mais)
In mice lacking ID4 expression, quantitatively normal spermatogenesis was found to be impaired due to progressive loss of the undifferentiated spermatogonial population during adulthood. ID4 expression was observed in spermatogonial stem cells (SSCs) in the mouse germline where it was shown to regulate self-renewal. Specifically, ID4 is expressed by a sub-population of type A single spermatogonia in mouse male germ line [29]. GDNF, a key growth factor driving self-renewal was found to up-regulate ID4 expression in isolated SSC-enriched fractions [29].
4.1.4. ID4 and Osteoporosis (ID4tm1Fsky)
A drastic reduction in osteoblast differentiation with a corresponding increase in differentiation toward adipocytes was observed in ID4-/- mice. ID4 promotes osteoblast differentiation by releasing Hes1 from Hes1-Hey2 complexes. Subsequently, Hes1 increases the stability and transcriptional activity of Runx2, a key molecule of osteoblast differentiation, which results in an enhanced osteoblast-specific gene expression [24].
4.1.5. ID4 and prostate cancer (ID4tm1Mais)
A large cohort of studies demonstrating a strong association between the loss of ID4 expression in many cancers suggests that ID4-/- mouse could present with multiple cancers at some point in the lifetime. To date, the cancer phenotype has been well established only in the prostate of ID4-/- mice. ID4 is highly expressed in wild type mouse prostate as well as the normal human prostate. We have shown that ID4 is a key regulator in the normal development of various androgen dependent organs of the male genital tract in general and specifically of the prostate. Prostates from ID4-/- mice have smaller size, decreased branching morphogenesis and decreased differentiated luminal cells as demonstrated by almost complete loss of NKX3.1 expression, a marker of differentiated luminal epithelial cells [23]. A similar defect in branching morphogenesis in the developing mammary gland of ID4-/- [22] therefore appears to be a consistent mechanism. The presence of PIN lesions, the earliest stage of prostate cancer [23] in 6 weeks old ID4-/- mouse prostate suggested that loss of ID4 may be an initiating event in prostate cancer development. ID4 had no effect on androgen receptor (AR) expression as well as its translocation to the nucleus. Interestingly, the expression of androgen dependent genes such as the homeobox NKX3.1 decreased; which was in part due to loss of AR binding on its respective response element. In contrast, the expression of other AR target genes such as probasin and Myc increased, suggesting that ID4 may differentially regulate AR binding to its corresponding binding site. Furthermore, expression of Pten, a known tumor suppressor, is also decreased in ID4-/- mice [23].
Clearly, tissue specific ID4 knockouts need to be developed in order to fully understand its role in development, differentiation and disease. The existing global ID4 knockouts should also be thoroughly analyzed since some of the effects could be subtle and may not result in easily identifiable large scale developmental phenotypes such as those observed in brain and reproductive tracts.
4.2 Tissue Specific ID4 Knockout models
Recently, the entire reading frame of Id4 (exons 1 and 2) flanked by LoxP sites was created [35]. This model is a significant step forward in developing tissue specific Id4 knockouts using targeted expression of Cre recombinase. The cross between MMTV- Cre which directs the expression primarily to secretory epithelium of virgin and lactating mammary gland, the salivary gland, seminal vesicle, skin, erythrocytes, B cells and T cells or K-14-Cre (KRT14-Cre) with expression primarily in ectoderm and its derivatives was used to understand the role of ID4 knockout on mammary gland development and ovarian function. The mammary gland phenotype in this model was essentially similar to that of the global Id4 KO model discussed above, that is delay in ductal morphogenesis, in part due to increase in the expression of estrogen receptor-α (ERα), progesterone receptor (PR) and FOXA1 [35]. In the ovary, Id4 is expressed in the granulosa cells of secondary and antral follicles. Consequently, lack of Id4 resulted in a significant increase in the numbers of secondary and antral follicles as well as atretic (degenerating) follicles. The authors attributed the ovarian phenotype in Id4-/- mice due to decreased estrogen biosynthesis [35].
5. ID4 and Cancer
ID4 appears to act primarily as a tumor suppressor in most cancers as opposed to ID1, ID2 and ID3 which acts as tumor promoters or supporting oncogenes [1, 36, 37]. In a small subset of cancers, ID4 also acts as a tumor promoter (Fig. 3). Beyond correlative studies showing ID4 expression in tumors, the underlying molecular mechanism remains largely unexplored.
Figure 3.
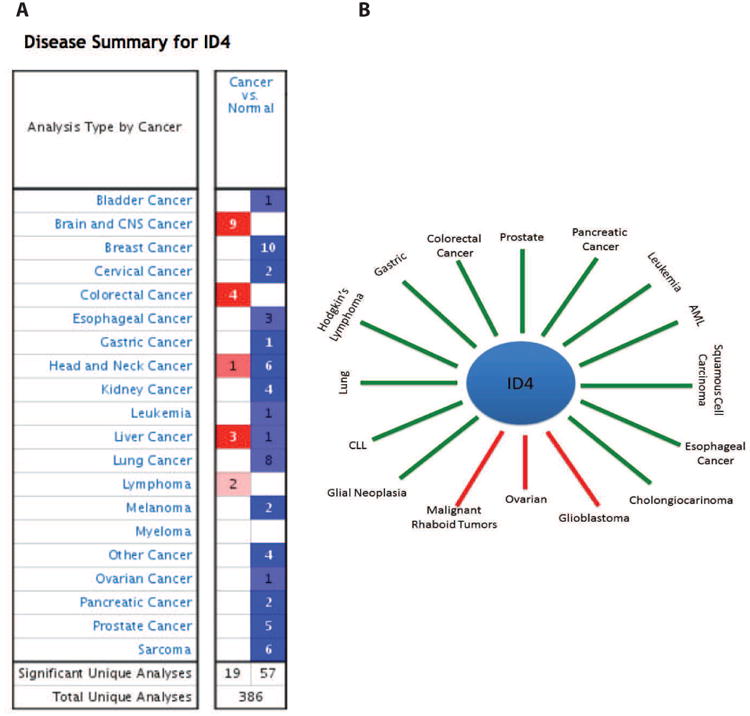
ID4 expression in cancers. A: Id4 expression profile in various cancers from Oncomine database. The blue and red boxes represent cancers in which ID4 expression is decreased or increased respectively as compared to normal counterparts. B: The ID4 expression in various cancers from literature mining (see text for references). The green lines points to cancers in which ID4 is down-regulated whereas red lines points to cancers where ID4 is up-regulated.
5.1: ID4 as a tumor promoter
The role of ID4 as a tumor promoter is based on the evidence (experimental and meta-analysis) that increased ID4 expression is observed in a small subset of cancers (Fig. 3).
Breast Cancer
A clear picture relating ID4 expression with breast cancer has not emerged. Epigenetic silencing of ID4 is observed in Columnar Cell lesions (CCL) and Ductal Carcinoma in situ (DCIS)/invasive carcinoma [38] and majority of sporadic breast cancers [39]. ID4 expression is also absent in ERα positive atypical ductal hyperplasia, ductal carcinoma in situ and invasive carcinomas but present in normal ERa negative mammary epithelial cells [40] suggesting that ERa could negatively regulate ID4 [39, 40]. Increased ID4 expression is observed in basal celllike breast cancer [41], triple negative breast cancer (TNBC) (including 4T1 mouse mammary cancer cell line [42, 43]) but not in non-TNBC. In meta-analysis, no significant association was found between ID4 and breast cancer [44].
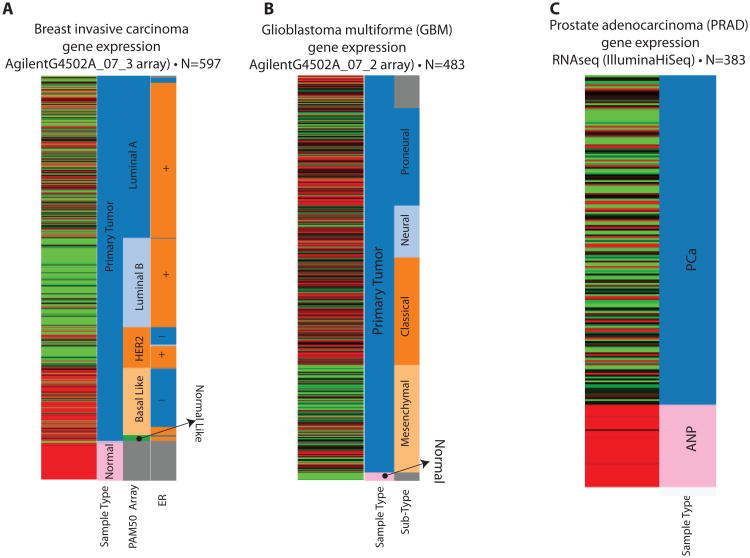
The ID4 gene expression profile in The Cancer Genome Atlas (TCGA) dataset is consistent with results discussed above. ID4 is highly expressed in the normal breast tissue and in Basal cell like cancers (ERα-ve) (Fig. 4A). Interestingly, ID4 expression was found to be lower in Luminal B type cancers as compared to Luminal A type (Fig. 4A). Both Luminal A and B type cancers are ER+ve, but Luminal B type cancer tend to be of higher histological grade than Luminal A type [45].
Figure 4.
ID4 expression profile from The Cancer Genome Atlas database. The respective cancers (A; Breast Cancer, B: GBM and C: Prostate cancer), the gene expression analysis platform and number of samples (N) used for analysis is indicated above each panel. The cluster is represented by decreased (green), increased (red) or no change (Black) in ID4 expression. The bars towards the right of each cluster represent sample type and sub type. The PAM50 array represents major molecular subtypes in breast cancer [111]. The Estrogen receptor status (ER) is indicated by + (present) and (-) absent. ANP: Adjacent normal Prostate, PCa: Prostate Cancer.
Other Cancers
Increased ID4 expression is observed in primary serous ovarian cancer cancer [46]. ID4 is also highly expressed in Glioblastoma [47]. In glioblastoma multiforme (GBM), high ID4 expression was also observed in TCGA data sets, specifically in classical, neuronal and pro-neural types (Fig. 4B).
A tumor-penetrating nano-complex (TPN) comprised of siRNA complexed with a tandem tumor-penetrating and membrane-trans-locating peptide, which enables the specific delivery of siRNA deep into the tumor parenchyma was employed in vivo to evaluate the role of ID4 as an oncogene [46] in tumors, where ID4 expression is elevated such as those of the ovary. Treatment of ovarian tumor-bearing mice with ID4-specific TPN suppressed growth of established tumors and significantly improved survival suggesting that targeting ID4 expression is a viable therapeutic strategy in cancers that over-express ID4.
5.2. ID4 DNA Alterations
ID4 is consistently co-amplified with E2F3 in aggressive bladder cancers [48]. On the contrary, genomic loss of ID4 is observed in Hodgkin's Lymphoma [49].
Cytogenetic abnormalities resulting from chromosomal 6 translocations are also very common in hematologic malignancies. ID4 was demonstrated in one case of B-cell lineage acute lymphoblastic leukemia (ALL) with t(6;14) (p22;q32) [50] indicating that ID4 may act as an oncogene in some human leukemia cases through its ability to sequester specific B-cell transcription factors. In general, ID4 expression is down-regulated in ALL [51], hence the translocation observed in one clinical case could be an isolated incidence in the backdrop of other genomic alterations.
5.3. ID4 as a tumor suppressor
Epigenetic silencing of ID4 in many cancers has prompted investigators to classify it as a tumor suppressor [40, 49, 52-57] (Fig. 3). ID4 is epigenetically silenced in: Leukemia [53], AML [58], CLL [59], ALL [51], glial neoplasia [60], squamous cell carcinoma [61], gastric cancer [62], pancreatic cancer [63], colorectal [64], lymphoma [65], cholangiocarcinoma [55], esophageal [66] and lung cancer [54]. Meta-analysis in Oncomine database [67] also suggested that ID4 expression is decreased in majority of cancers (Fig. 3).
5.3.1. ID4 and Prostate Cancer
Previous studies from our laboratory strongly support the role of ID4 as a tumor suppressor in prostate cancer (PCa) [52, 68]. Immuno-histochemical analysis of normal and malignant human prostate tissue microarray shows that ID4 is down regulated specifically in prostate cancer but not in normal adjacent prostate glands [68] (see also TCGA PRAD dataset in Fig. 4C). Decrease in ID4 expression is stage dependent with majority of high grade tumor samples showing negligible ID4 expression while high ID4 expression is observed in the normal prostate tissue [68]. We have shown that ID4 is expressed in PCa cell line LNCaP (low tumorigenic, androgen sensitive [69]), low in PC3, but not in C81 (androgen insensitive and more tumorigenic LNCaP derivative [70]), and DU145 cells [68]. Furthermore, similar to other cancers, the decrease in ID4 expression in prostate cancer tissue and cell lines was attributed to promoter hypermethylation [52, 68]. Our study was also validated by Vinarskaja et al who confirmed ID4 promoter hypermethylation and down-regulation in prostate cancer [71].
Silencing of ID4 in LNCaP prostate cancer cells (LNCaP(-)ID4) results in a castration resistant phenotype, partly due to gain of de novo steroidogenesis [72]. The LNCaP(-)ID4 cells also form tumors in castrated mice as compared to LNCaP cells [72] suggesting that ID4 is required to maintain the tumor suppressive function of AR whereas loss of ID4 results in tumor promoter activity of AR. These results could explain the Id4-/- mice prostate phenotype where AR is expressed at levels similar to wild type mice but associated with PIN lesions [23]. At the mechanistic level, ID4 may regulate the expression or function of specific but yet unknown AR co-regulators that may determine the final outcome of AR function.
5.4. ID4 in prognosis and survival
Inactivation of ID4 is also associated with poor differentiation and unfavorable prognosis in colorectal carcinoma [57]. In breast cancer loss of ID4 [73] is associated with recurrence free survival, and increased tumor relapse [56]. AML patients with myelodysplastic syndrome (MDS) exhibited a significantly higher frequency of ID4 methylation with shorter survival [74].
In chronic myeloid leukemia, methylation of the ID4 promoter increases as the disease progresses from ‘chronic phase’ to ‘accelerated phase’ to ‘blast crises’ [73]. In a separate study ID4 was found methylated in 86% of acute leukemia patients and 100% in leukemia-relapse patients [75]. ID4 expression was reduced by promoter methylation in 95% of B-cell lymphoma samples and 100% of follicular lymphoma samples.
5.5. ID4 and angiogenesis
Glioblastoma derived tumor cell expressing elevated levels of ID4 produce enlarged xenografts in immunosuppressed mice that were better vascularized than corresponding control tumors suggesting a novel pro-angiogenic function to ID4 (mediated by Matrix GLA protein (MGP)) [76]. In glial neoplasms down regulation of ID4 is associated with inhibition of angiogenesis [60]. In breast cancer cells increased ID4 expression may also promote angiogenesis through stabilization of GRO-α and IL-8 mRNA [77]) (Table I). Collectively, these studies suggest that ID4 supports angiogenesis in cancers in which it is over-expressed.
Table I. Downstream targets of ID4.
| Downstream Targets |
|---|
| Jagged 1 |
| BRCA1 |
| P38 MAPK |
| Sox2 |
| CDKN1A |
| p21/p27 |
| GRO-α |
| IL-8 |
| miRNA-9 |
| ERα |
| FOXA1 |
5.6. ID4 and Chemo-resistance
High ID4 expression was shown to promote chemo resistance to anticancer drugs in glioma stem cells (GSCs) by suppressing microRNA-9 which in turn led to an increase in the expression of SOX2 (Table I). SOX2 is a non bHLH transcription factor known to play key role in development and maintenance of GSCs and is repressed by microRNA-9. The resulting ID4 mediated increased expression of SOX2 further induced ABC transporters such as ABCC3 and ABCC6 through direct transcriptional regulation resulting in chemo resistance of GSCs [78].
It is evident from the above studies that ID4 expression and association with various disease modalities even in same cancer types such as breast and glioblastoma is often conflicting. The analytical tools such as specific ID4 antibodies, RT-PCR strategy (given that ID4 gene is highly CG rich) and specific CpG islands in the promoter itself needs to be revaluated and validated. This is apparent from a prostate cancer study which demonstrated a positive correlation between ID4 overexpression with the Gleason score and metastatic progression [79]. Use of highly specific ID4 antibodies, TCGA prostate adenocarcinoma (Fig. 4C) and Oncomine databases (Fig. 3) clearly presented an inverse correlation between ID4 expression and prostate cancer. Most of the ID4 promoter methylation studies on cancers where ID4 is silenced have focused on the proximal promoter (surrounding the transcriptional start site) hence comparable and confirms epigenetic inactivation. Cancer heterogeneity may also contribute equally to inconsistent results. It is likely that ID4 expression is limited to a sub-set of cancers (e.g Luminal Type A vs. Luminal Type B breast cancers, Fig. 4A), particularly in context of heterogeneity observed in many cancers. Decreased angiogenesis with better prognosis and increased survival in a subset of glioblastoma with low ID4 [60, 80] whereas high ID4 particularly in cells which are in close proximity to the vasculature [76, 81] clearly supports an association between ID4 expression and cancer subtypes. A strong relationship between ID4 expression and cancer subtypes, particularly in GBM and breast is also evident from TCGA datasets (Fig. 4A and B). A clear discrimination between cancer subtypes (Breast and Glioblastoma), apparent from TCGA datasets (Fig. 4A and B) suggests that ID4 expression is a strong predictor of associated disease modalities.
The contrasting expression of ID4 in various cancers (and sub-types) is suggestive of a complex molecular mechanism which could involve tissue/stage/sub-type specific interaction proteins, post-transcriptional modifications and regulatory mechanisms.
6. Regulation of ID4 gene expression
To date no mutations have been reported in the ID4 gene [65]. Thus regulation ID4 expression appears to be the primary mechanism which determines the function of ID4.
6.1. Transcriptional regulation
At least two functional elements located downstream of the TATA box have been identified in the ID4 promoter. The first, a positive regulatory element contains a consensus E-Box that binds the bHLH leucine zipper upstream stimulatory factor (USF). The second, a negative regulatory element is a GA motif recognized by Sp1 and Sp3 transcription factors [82].
Interestingly, the predicted ER response element [83] could result in the down-regulation of ID4 expression in ER positive breast cancer cells treated with estradiol [40]. Increased ID4 expression in TNBC [84] therefore could in part be due to loss of ER. In sporadic breast tumors, ID4 is also inversely correlated with ER mRNA expression [39]. In several tumor specimens, ID4 mRNA expression was lowest in samples expressing high levels of ER mRNA whereas treatment of ER positive breast cancer cells with estradiol resulted in decreased expression of ID4 [85].
Our studies demonstrated that ID4 is regulated by androgens in normal prostate epithelial cells and less metastatic prostate cancer cell line LNCaP [52]. Ectopic ID4 expression also induced re-expression of androgen receptor and its downstream transcriptional target PSA in otherwise androgen receptor negative prostate cancer cell line DU145 [52].
AR-mediated prostate tumorigenesis in mice expressing mutant androgen receptor (E231G) demonstrated negligible ID4 expression together with other signature genes to predict biochemical relapse in androgen receptor dependent prostate cancer [86]. Taken together, these studies suggest that ID4 exists in positive feedback loop with AR but in a negative feedback loop with ER. However, evidence that ER or AR binds directly to ID4 promoter is lacking.
ID4 is also the downstream target of BMP4 in neural progenitor cells and apparently mediates the inhibitory effects on oligodendroglial differentiation [87, 88] whereas ID4 dependent osteoblast differentiation is independent of BMP4 [24], suggesting cell type specific regulation (Table II).
Table II. Transcription regulation of ID4.
| Transcriptional Regulation |
|---|
| Epigenetic: Cdc42/PRMT5/YY1/EZH2/DNMT1 |
| Mutant p53 |
| BMP 2/4 |
| USF |
| Sp1/Sp3 |
| Hormonal: AR/ER |
6.2. Regulation of ID4 expression by microRNA
Recent studies have demonstrated that ID4 expression is also regulated by miRNAs. ID4 is repressed by miR342 (breast cancer cell line MDA-MB-231 [89], miR335 (breast cancer cell line MCF7 [90] and senescence associated (SA) miR485-5p (fibroblast cell lines [91]).
6.3. Epigenetic Regulation
Gene silencing through promoter hypermethylation is the most widely accepted mechanism involved in the regulation of ID4 expression. ID4 promoter is CpG rich that spans the proximal promoter through entire exon 1, thus making this region highly attractive for epigenetic modifications. Several studies have not only extensively characterized the down regulation of ID4 expression through promoter hypermethylation in many cancers and cell lines, but have also linked ID4 expression to clinico-pathological variables, such as stage, tumor grade, age, cancer recurrence and poor prognosis such as in lung, bladder, breast and colorectal cancer [56, 57] as discussed above. Demethylation of the breast cancer cell lines (BT20, MCF7 and T47D) by 5-Aza-2′-Deoxycytidine (AZA) and Trichostatin A [56] and prostate cancer cell line DU145 by AZA [52] resulted in clear re-expression of ID4 further supporting hypermethylation as the primary mechanism involved in ID4 down-regulation in cancers.
Studies in colorectal carcinoma demonstrated that ID4 promoter is regulated by Cdc42 [64], a small GTPase of the Rho family. High Cdc42 expression in many cancers [92] including colorectal carcinoma, may suggest a role of cdc42 in ID4 promoter methylation. How a protein involved in promoting cell cycle regulates epigenetic re-programming of the ID4 promoter remains to be addressed (Table II).
PRMT5, a type II protein arginine methyl-transferase catalyzes mono- and di-methylation of arginine residues in histones and results in promoter DNA hypermethylation [93]. PRMT5 was shown to occupy the CpG rich islands in ID4 promoter resulting in its promoter hypermethylation during glial cell and oligodendrocyte differentiation [28]. In cancer cells, high PRMT5 expression may tend to support its role in epigenetic silencing of ID4. However, in prostate cancer cells PRMT5 expression is primarily cytoplasmic and promotes growth. In contrast, PRMT5 is nuclear in benign prostate epithelial cells where it inhibits growth [94]. Thus PRMT5 localization (predominantly cytoplasmic) in prostate cancer does not correlate with its role in ID4 methylation or association with CpG islands, which one would expect to be in the nucleus. The transcription factor YY1 was also shown to inhibit ID4 transcription by recruiting histone deacetylase-1 to its promoter [95] suggesting that histone acetylation may also play a role in regulating ID4 expression (Table II).
EZH2 (enhancer of Zeste 2), part of the Polycomb repressor complex 2 (PRC2) is involved in epigenetic re-programming in both normal and disease states including cancer. EZH2 is specifically involved in covalent modification of histone tails, specifically tri-methylation (me3) of lysine 27 (K27) on histone 3 (H3) (H3K27me3), a repressive mark found on many gene promoters that are silenced [96]. EZH2, as part of the PRC2 complex also recruits DNMTs (DNA methyl transferases) that in turn promotes DNA methylation at CpG islands (CGI) thus connecting the two key epigenetic repression systems [97]. Increased EZH2 expression is also observed in many cancers [98] including prostate cancer [99]. We recently demonstrated that ID4 is an EZH2 target gene in prostate cancer [100]. Assembly of PRC2 complex initiated by increased recruitment of EZH2 on ID4 promoter increases the repressive H3K27me3 histone modification and recruits DNMT1/3a resulting in ID4 promoter hyper-methylation in DU145 prostate cancer cell lines and prostate cancer tissue (Table II).
Epigenetic inactivation in many but not all cancers suggests that ID4 could be highly regulated in a tissue specific manner. A basal transcriptional machinery consisting of a subset of bHLH (USF1) transcription factors together with the assembly of tissue specific positive (AR, BMP4) or negative (ER) regulatory complexes could determine optimal ID4 transcription. The binding of Sp1 to GC rich regions, which are targets of hyper-methylation, could be a rate limiting step in the assembly of such transcription factor complexes on ID4 promoter that could eventually determine ID4 transcriptional output.
Overwhelming evidence now suggests that ID4 is epigenetically silenced in many cancers. Hence therapeutic strategies which alter the cancer epigenome via inhibition of DNMT1 such as by 5-Aza-2′-Deoxycytidine (AZA) [101] are viable strategies. Alternatively, mechanisms involved in regulating ID4 levels at the transcriptional level such as those involving ER and AR could be exploited to elevate ID4 levels in breast and prostate cancer. A study recently demonstrated that treatment cells by arsenic trioxide (ATO) reversed the hyper-methylation of ID4 promoter and decreased proliferation of Raji cells [102]. The specificity of this treatment on ID4 promoter however remains to be investigated.
7. Mechanism of Action of ID4
All known ID4 interactions are mediated by the HLH domain (Table III). Protein interaction studies have demonstrated that ID4 sequesters OLIG1 and OLIG2, members of class B bHLH proteins which plays key roles in early oligodendrocyte specification and regulates oligodendrocyte differentiation [87, 88]. Interestingly OLIG1 and OLIG2 only interact with ID4 and to a lesser extent with ID2 but not with ID1 or ID3. These studies suggest that ID4 preferentially binds specific bHLH proteins either due to sequence specific domains in ID4 or in the corresponding interacting proteins [87].
Table III. ID4 protein Interactions (bHLH proteins are in bold).
| Protein Interactions |
|---|
| Olig1/Olig2 |
| wt and mut-p53/E2F1/p65 |
| CBP/p300 |
| Hes1 |
| E2A |
| MyoD |
| Twist1 |
Apart from interacting with OLIG1/2, ID4 also interacts with bHLH proteins HES1 to promote osteoblast differentiation [24] and Twist1 in GBM [80] resulting in decreased MMP2 mediated invasion of glioblastoma derived cells. (ID4 protein interaction network is summarized in Table III).
Functional studies demonstrated that ID4 down-regulates BRCA1 [41, 84, 103] and allows for anchorage-independent growth of breast (SKBr3) and ovarian (PA-1) cancer cells [103] which are ERa-ve. Interestingly, another study demonstrated that BRCA1 induces the expression of ID4 [85].
In ID4-/- mice, a decrease in mammary gland ductal expansion and branching morphogenesis was attributed to activation of p38MAPK [22]. Thus ID4 may regulate major stress activated pathways through down-regulation of p38MAPK.
A novel but unexpected mechanism of action of Id4 has recently emerged. In a ChIP study, ID4 was found to be part of the transcriptional complex that could potentially regulate ERα and Foxa1 (Table I). The lack of ID4 DNA binding domain in ID4 suggests that it most likely regulates transcription as part of a larger protein complex [35].
As opposed to the protein interaction network of ID1 and ID2, the regulatory network of ID4 remains to be fully explored. Given the inverse association between ID1 and ID4 in cancer, the ID4 regulatory network is expected to be unique and non-overlapping. The major question that needs to be addressed is whether ID4 negatively regulates ID1 dependent pathways. If in fact this happens to be case then the HLH independent mechanisms should be explored such as contributions of the ID4 non-HLH domains towards selective binding with bHLH and non-bHLH proteins. Moreover, the degree of overlapping functions between a subset of ID proteins such as ID2 and ID4 which both bind the non-bHLH protein Rb and bHLH proteins OLIG1/OLIG2 (with different affinities) needs to be closely examined to evaluate structure/function relationship. Recent data supporting the indirect interaction of ID4 with DNA as part of the larger transcriptional complex is also interesting and opens an entirely novel dimension to the function of ID4.
8. The role of ID4 in cell cycle and proliferation
Ectopic expression of ID4 blocks cell cycle and inhibits proliferation that is associated with increased expression of cyclin dependent kinase inhibitors p21 and p27 in prostate cancer cell line DU145 [52]. The ID4 dependent cell cycle arrest appears to be primarily at S-Phase in DU145 and PC3 prostate cancer cell lines and not at G1 as would be expected from increased p21 and p27 expression [52]. The block in S-phase appears to be due to a decrease in the expression of E2F1 that is required for transition through S-phase [104]. Loss of ID4 also promotes progression into S-phase in neuronal early cortical progenitor cells [26] and LNCaP prostate cancer cell lines. Epigenetic knockdown of ID4 through promoter hypermethylation in colon carcinoma by Cdc42, a member of Rho GTPases family may be required for cell cycle progression [64] (Table II). Nevertheless, these results suggest that unlike ID1, ID2 and ID3, the expression of ID4 is not associated with progression of cell cycle and points towards the role of ID4 in promoting differentiation in specific cell types since both these processes are largely mutually exclusive.
ID4 can also drive the malignant transformation of astrocytes via de-regulation of cell cycle and differentiation through the up-regulation of cyclin E and activation of Jagged-Notch1 signaling but only in primary murine Ink4a/Arf-/- astrocytes [81]. Increased ID4 expression [47] and deletion of the INK4a-ARF locus is also found in majority of human malignant gliomas [105]. These results suggest that the Ink4a/Arf could be an upstream regulator of ID4 in a subset of gliomas. Whether ID4 itself drives the expression of Ink4a/Arf or vice versa to block cell cycle could be an interesting avenue to investigate.
9. ID4-P53 Cross talk
The molecular mechanism by which mutant p53 may function has been reviewed extensively [106]. These pathways include alterations in the DNA-binding ability of mutant p53, interaction of mutant p53 with other proteins, including transcription factors or proteins not directly related to the regulation of gene expression. It is clear that the effects of mutant p53 are strongly context dependent, and interactions that promote activity in some circumstances may be inhibitory in others.
Fontemaggi and colleagues have extensively characterized the molecular interaction between ID4 and mutant p53 in promoting neo-angiogenesis in breast cancer [77]. In breast cancer cell lines, ID4 is a transcriptional target of gain-of-function p53 mutants R175H and R280K in SKBr3 and MDA-MB-231 but not in MCF7 breast cancer cells expressing wild type p53. The protein complex involving mutant p53–E2F1-p65 (NF-kB p65 subunit RelA) assembles on specific regions of the ID4 promoter and positively controls ID4 expression [77]. The study further demonstrated that mutant p53 R273H had the highest regulatory control over ID4 expression [77]. ID4 expression was also increased in response to Adriamycin and Cisplatin treatments on mutant p53/ID4 expressing cell lines. Studies involving the knockdown of mutant p53 abrogated this response, indicating that in response to DNA damage mutant p53 in complex with E2F1 and p300 bind to the CDE element closest to the transcriptional start site, on the ID4 promoter, to achieve “mutant p53 associated” chemo-resistance [107]. Thus direct interaction between mutant p53 and E2F1 on the ID4 promoter supports the oncogenic functions of mutant p53 in at least breast cancer.
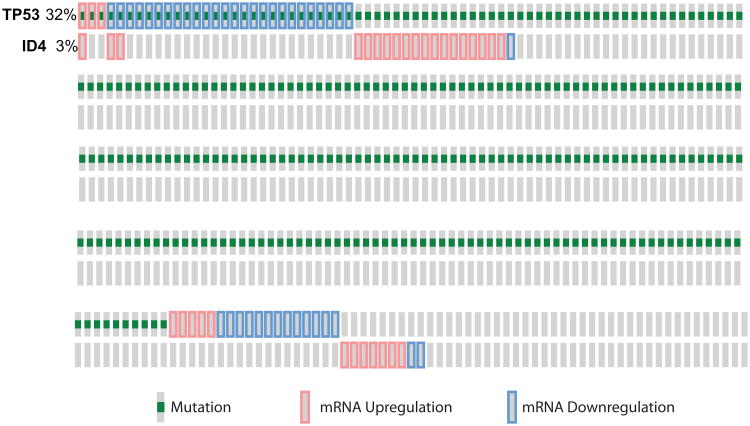
Surprisingly, a negative correlation was observed between mutant p53 and ID4 expression in meta-analysis of clinical studies [108]. Similarly, in the TCGA dataset (Breast Invasive Carcinoma, TCGA Provisional), p53 mutations were observed in 32% of cases (306 cases out of 956) whereas ID4 was altered in only 3% of the cases. Interestingly, ID4 expression was altered in only 6% of cases in which p53 was mutated. Hence a significant correlation was not observed between p53 mutations and ID4 expression (Fig. 5).
Figure 5. Breast Invasive Carcinoma (TCGA, Provisional)/All Complete Tumors: (956)/User-defined List/2genes.
Relation between p53 mutations and ID4 expression in breast cancer obtained from TCGA provisional dataset (cBIO Portal). A total of 956 samples were analyzed of which 306 (32% demonstrated p53 mutations (green) and 3% demonstrated ID4 alterations (red bars: up-regulated, blue bars: down-regulated). Only 20 samples with p53 mutations showed alterations in ID4 expression. The figure only shows the samples with p53 mutations and ID4 expression. The grey bars represent samples with no change in either p53 (mutations/expression) or ID4 (expression) (majority of these samples not shown). The expression is based on microarray analysis with Z-score of more than 2.0.
ID4 functions as a tumor suppressor in ER+ve breast tumors where it is frequently inactivated by promoter hypermethylation; however, ID4 displays oncogenic activity in ER-ve breast cancer cells SKBr3 and MDA-MB-237 that express mutant p53 [107]. These results suggest that ER may have a role in regulating ID4 dependent expression of mutant p53. The corresponding estrogen receptor levels in the meta-analyses discussed above were not reported but could determine the outcome of the ID4-mutant p53 cross-talk. These conflicting results between cell lines and clinical studies suggest that the ID4 regulatory mechanism could be controlled by a complex mechanism that still remains to be investigated.
Our studies have demonstrated that ID4 promotes transcriptional activity of wild type p53 in a CBP/P300 dependent acetylation at K320 and K373 residues [69]. Interestingly, ID4 also restores the wild type transcriptional activity of mutant p53 in DU145 cells through a similar mechanism as reflected by the activation of p53 dependent processes such as increased apoptosis, senescence and attenuation of cell cycle in ID4 over-expressing DU145 cells [104] in spite of mutations in Rb and p16 [109, 110]. Moreover, EMSA and ChIP analysis demonstrated that ID4 promotes binding of mutant p53 to the consensus wild type p53 DNA response element and enrichment to p53 binding sites on p21, BAX and PUMA promoters. Co-elution of ID4 as part of the p53-CBP/p300 complex with p53 antibody and co-elution of p53 with ID4 antibody suggests that ID4 can recruit CBP/P300 on wt- and mutant p53 to promote acetylation [69]. Whether ID4 can also restore the wild type transcriptional activity of various p53 hot spot mutants remains to be established.
10. Conclusions
It is evident that ID4 shows divergence from the other ID family members during development, differentiation and diseases such as cancer. The mouse knockout models have clearly established the essential role of ID4 in development that is not compensated by other ID proteins. Overall, the role of ID4 in cancer appears to be that of a tumor suppressor that is largely based on its epigenetic silencing in majority of cancers. The expression of ID4 in a limited cancer types (GBM) is associated with favorable prognosis further supporting the role of ID4 as a putative tumor suppressor. How ID4 acts as a tumor suppressor is an open question and should be the major focus of future studies. The other major question that remains to be addressed is the mechanism by which ID4 acts as a tumor promoter in limited cancer sub-types such as TNBC, basal cell carcinoma of the breast and ovarian cancer. Interaction of ID4 with bHLH proteins such as OLIG and TWIST and cross talk with BRCA1, p38MAPK and Notch signaling pathway may provide a significant insight into the mechanism of action of ID4 that needs to be further explored. The lack of a DNA binding domain and unique sequence of ID4 (Alanine and Proline rich domains that support protein interactions) also suggest that ID4 may act as a major hub in protein-protein interaction networks, possibly as a putative co-chaperone. These interactions may help unravel the complex biology of ID4 in development and cancer. The strong anti-cancer effect of ID4 in prostate cancer suggests that mechanisms to up-regulate ID4 expression in cancers is a therapeutic strategy and needs to be further explored. Specifically, how ER and AR pathways can be used to regulate ID4 expression in hormone related cancers such as breast and prostate respectively appears to be an attractive therapeutic approach.
Acknowledgments
The work was supported by NIH/NCI CA128914 (JC) and in part by NIH/NCRR/RCMI G12MD007590 (CCRTD). The authors wish to thank Prof. Deborah Core (PhD), Dept. of English, Eastern Kentucky University, Richmond, KY for reviewing of the manuscript.
Abbreviations
- ID
Inhibitor of DNA Binding
- bHLH
basic Helix Loop Helix
- emc
extramacrochaetae
- OPC
oligodendrocyte progenitors cells
- CCL
Columnar Cell lesions
- DCIS
Ductal Carcinoma in situ
- ER
Estrogen Receptor
- TNBC
triple negative breast cancer
- ALL
acute lymphoblastic leukemia
- PCa
Prostate Cancer
- MDS
myelodysplastic syndrome
- MGP
Matrix GLA protein
- GSCs
glioma stem cells
- USF
upstream stimulatory factor
- AR
androgen receptor
- AZA
5-Aza-2′-Deoxycytidine
- EZH2
enhancer of Zeste 2
- PRC2
Polycomb repressor complex 2
- DNMTs
DNA methyl transferases
- SSCs
spermatogonial stem cells
- PIN
Prostatic Intraepithelial Neoplasia
- KO
Knock out
- ATO
arsenic trioxide
- TPN
tumor-penetrating nano-complex
Footnotes
Authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 2.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabellini C, Del Bufalo D, Zupi G. Involvement of RB gene family in tumor angiogenesis. Oncogene. 2006;25:5326–5332. doi: 10.1038/sj.onc.1209631. [DOI] [PubMed] [Google Scholar]
- 4.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 6.Ling F, Kang B, Sun XH. Chapter Five - Id Proteins: Small Molecules, Mighty Regulators. In: Reshma T, editor. Current Topics in Developmental Biology. Vol. 110. Academic Press; 2014. pp. 189–216. [DOI] [PubMed] [Google Scholar]
- 7.Coppe JP, Smith AP, Desprez PY. Id proteins in epithelial cells. Exp Cell Res. 2003;285:131–145. doi: 10.1016/s0014-4827(03)00014-4. [DOI] [PubMed] [Google Scholar]
- 8.Hasskarl J, Munger K. Id proteins--tumor markers or oncogenes? Cancer Biol Ther. 2002;1:91–96. doi: 10.4161/cbt.50. [DOI] [PubMed] [Google Scholar]
- 9.Pagliuca A, Bartoli PC, Saccone S, Della Valle G, Lania L. Molecular cloning of ID4, a novel dominant negative helix-loop-helix human gene on chromosome 6p21.3-p22. Genomics. 1995;27:200–203. doi: 10.1006/geno.1995.1026. [DOI] [PubMed] [Google Scholar]
- 10.Kiewitz SD, Cabrele C. Synthesis and conformational properties of protein fragments based on the Id family of DNA-binding and cell-differentiation inhibitors. Biopolymers. 2005;80:762–774. doi: 10.1002/bip.20287. [DOI] [PubMed] [Google Scholar]
- 11.Campuzano S. Emc, a negative HLH regulator with multiple functions in Drosophila development. Oncogene. 2001;20:8299–8307. doi: 10.1038/sj.onc.1205162. [DOI] [PubMed] [Google Scholar]
- 12.Mortlock DP, Sateesh P, Innis JW. Evolution of N-terminal sequences of the vertebrate HOXA13 protein. Mammalian genome: official journal of the International Mammalian Genome Society. 2000;11:151–158. doi: 10.1007/s003350010029. [DOI] [PubMed] [Google Scholar]
- 13.Sumiyama K, Washio-Watanabe K, Saitou N, Hayakawa T, Ueda S. Class III POU genes: generation of homopolymeric amino acid repeats under GC pressure in mammals. Journal of molecular evolution. 1996;43:170–178. doi: 10.1007/BF02338824. [DOI] [PubMed] [Google Scholar]
- 14.Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S. Polyalanine expansions in human. Human molecular genetics. 2004;13 Spec No 2:R235–243. doi: 10.1093/hmg/ddh251. [DOI] [PubMed] [Google Scholar]
- 15.Pelassa I, Cora D, Cesano F, Monje FJ, Montarolo PG, Fiumara F. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Human molecular genetics. 2014;23:3402–3420. doi: 10.1093/hmg/ddu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. Journal of Molecular Graphics and Modelling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 17.Kini RM, Evans HJ. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun. 1995;212:1115–1124. doi: 10.1006/bbrc.1995.2084. [DOI] [PubMed] [Google Scholar]
- 18.Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. Neural Networks,1997, International Conference on. 1997;1:90–95 vol.91. [Google Scholar]
- 19.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic Disorder Is a Common Feature of Hub Proteins from Four Eukaryotic Interactomes. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 1996;207:235–252. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Rigolet M, Rich T, Gross-Morand MS, Molina-Gomes D, Viegas-Pequignot E, Junien C. cDNA cloning, tissue distribution and chromosomal localization of the human ID4 gene. DNA Res. 1998;5:309–313. doi: 10.1093/dnares/5.5.309. [DOI] [PubMed] [Google Scholar]
- 22.Dong J, Huang S, Caikovski M, Ji S, McGrath A, Custorio MG, Creighton CJ, Maliakkal P, Bogoslovskaia E, Du Z, Zhang X, Lewis MT, Sablitzky F, Brisken C, Li Y. ID4 regulates mammary gland development by suppressing p38MAPK activity. Development. 2011;138:5247–5256. doi: 10.1242/dev.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma P, Knowell AE, Chinaranagari S, Komaragiri S, Nagappan P, Patel D, Havrda MC, Chaudhary J. Id4 deficiency attenuates prostate development and promotes PIN-like lesions by regulating androgen receptor activity and expression of NKX3.1 and PTEN. Mol Cancer. 2013;12:67. doi: 10.1186/1476-4598-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokuzawa Y, Yagi K, Yamashita Y, Nakachi Y, Nikaido I, Bono H, Ninomiya Y, Kanesaki-Yatsuka Y, Akita M, Motegi H, Wakana S, Noda T, Sablitzky F, Arai S, Kurokawa R, Fukuda T, Katagiri T, Schonbach C, Suda T, Mizuno Y, Okazaki Y. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS genetics. 2010;6:e1001019. doi: 10.1371/journal.pgen.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murad JM, Place CS, Ran C, Hekmatyar SK, Watson NP, Kauppinen RA, Israel MA. Inhibitor of DNA binding 4 (ID4) regulation of adipocyte differentiation and adipose tissue formation in mice. J Biol Chem. 2010;285:24164–24173. doi: 10.1074/jbc.M110.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhary J, Johnson J, Kim G, Skinner MK. Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology. 2001;142:1727–1736. doi: 10.1210/endo.142.5.8134. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Vogel G, Yu Z, Almazan G, Richard S. Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J Biol Chem. 2011;286:44424–44432. doi: 10.1074/jbc.M111.277046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85:347–356. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sablitzky F, Moore A, Bromley M, Deed RW, Newton JS, Norton JD. Stage- and subcellular-specific expression of Id proteins in male germ and Sertoli cells implicates distinctive regulatory roles for Id proteins during meiosis, spermatogenesis, and Sertoli cell function. Cell Growth Differ. 1998;9:1015–1024. [PubMed] [Google Scholar]
- 31.Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- 32.Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. Embo J. 2000;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE G. Mouse Genome Database. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedford L, Walker R, Kondo T, van Cruchten I, King ER, Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev Biol. 2005;280:386–395. doi: 10.1016/j.ydbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Best SA, Hutt KJ, Fu NY, Vaillant F, Liew SH, Hartley L, Scott CL, Lindeman GJ, Visvader JE. Dual roles for Id4 in the regulation of estrogen signaling in the mammary gland and ovary. Development. 2014;141:3159–3164. doi: 10.1242/dev.108498. [DOI] [PubMed] [Google Scholar]
- 36.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 37.G J, Wice BM. Forced expression of Id-1 in the adult mouse small intestinal epithelium is associated with development of adenomas. J Biol Chem. 1998;273:25310–25319. doi: 10.1074/jbc.273.39.25310. [DOI] [PubMed] [Google Scholar]
- 38.Verschuur-Maes AH, de Bruin PC, van Diest PJ. Epigenetic progression of columnar cell lesions of the breast to invasive breast cancer. Breast Cancer Res Treat. 2012;136:705–715. doi: 10.1007/s10549-012-2301-4. [DOI] [PubMed] [Google Scholar]
- 39.Roldan G, Delgado L, Muse IM. Tumoral Expression of BRCA1, Estrogen Receptor Alpha and ID4 Protein in Patients with Sporadic Breast Cancer. Cancer Biol Ther. 2006;5:505–510. doi: 10.4161/cbt.5.5.2597. [DOI] [PubMed] [Google Scholar]
- 40.de Candia P, Akram M, Benezra R, Brogi E. Id4 messenger RNA and estrogen receptor expression: inverse correlation in human normal breast epithelium and carcinoma. Hum Pathol. 2006;37:1032–1041. doi: 10.1016/j.humpath.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 42.Park SJ, Kim RJ, Nam JS. Inhibitor of DNA-binding 4 contributes to the maintenance and expansion of cancer stem cells in 4T1 mouse mammary cancer cell line. Laboratory animal research. 2011;27:333–338. doi: 10.5625/lar.2011.27.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, Vargas-Roig LM, Roque M. Methylation profile of triple-negative breast carcinomas. Oncogenesis. 2012;1:e17. doi: 10.1038/oncsis.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S, Kon M, DeLisi C. Pathway-based classification of cancer subtypes. Biology direct. 2012;7:21. doi: 10.1186/1745-6150-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23:S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 46.Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, Hirsch MS, Mesirov JP, Drapkin R, Root DE, Lo J, Fogal V, Ruoslahti E, Hahn WC, Bhatia SN. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Science translational medicine. 2012;4:147ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng W, Rushing EJ, Hartmann DP, Azumi N. Increased inhibitor of differentiation 4 (id4) expression in glioblastoma: a tissue microarray study. Journal of Cancer. 2010;1:1–5. doi: 10.7150/jca.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol Cancer. 2005;4:16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slovak ML, Bedell V, Hsu YH, Estrine DB, Nowak NJ, Delioukina ML, Weiss LM, Smith DD, Forman SJ. Molecular karyotypes of Hodgkin and Reed-Sternberg cells at disease onset reveal distinct copy number alterations in chemosensitive versus refractory Hodgkin lymphoma. Clin Cancer Res. 2011;17:3443–3454. doi: 10.1158/1078-0432.CCR-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellido M, Aventin A, Lasa A, Estivill C, Carnicer MJ, Pons C, Matias-Guiu X, Bordes R, Baiget M, Sierra J, Nomdedeu JF. Id4 is deregulated by a t(6;14)(p22;q32) chromosomal translocation in a B-cell lineage acute lymphoblastic leukemia. Haematologica. 2003;88:994–1001. [PubMed] [Google Scholar]
- 51.Hu HB, Hu Q. ID4 methylation patterns in childhood T line and B line lymphocytic leukemia. Zhongguo dang dai er ke za zhi = Chinese journal of contemporary pediatrics. 2010;12:940–942. [PubMed] [Google Scholar]
- 52.Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:173. doi: 10.1186/1471-2407-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, Liu S, Smith LT, Lee S, Rassenti L, Marcucci G, Byrd J, Caligiuri MA, Plass C. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- 54.Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sanchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. Journal of translational medicine. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhm KO, Lee ES, Lee YM, Kim HS, Park YN, Park SH. Aberrant promoter CpG islands methylation of tumor suppressor genes in cholangiocarcinoma. Oncology research. 2008;17:151–157. doi: 10.3727/096504008785114110. [DOI] [PubMed] [Google Scholar]
- 56.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, Knuchel R, Dahl E. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer. 2008;8:154. doi: 10.1186/1471-2407-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10:7475–7483. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 58.Jiang D, Hong Q, Shen Y, Xu Y, Zhu H, Li Y, Xu C, Ouyang G, Duan S. The diagnostic value of DNA methylation in leukemia: a systematic review and meta-analysis. PLoS One. 2014;9:e96822. doi: 10.1371/journal.pone.0096822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen SS, Claus R, Lucas DM, Yu L, Qian J, Ruppert AS, West DA, Williams KE, Johnson AJ, Sablitzky F, Plass C, Byrd JC. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117:862–871. doi: 10.1182/blood-2010-05-284638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martini M, Cenci T, D'Alessandris GQ, Cesarini V, Cocomazzi A, Ricci-Vitiani L, De Maria R, Pallini R, Larocca LM. Epigenetic silencing of Id4 identifies a glioblastoma subgroup with a better prognosis as a consequence of an inhibition of angiogenesis. Cancer. 2013;119:1004–1012. doi: 10.1002/cncr.27821. [DOI] [PubMed] [Google Scholar]
- 61.Ruchusatsawat K, Wongpiyabovorn J, Protjaroen P, Chaipipat M, Shuangshoti S, Thorner PS, Mutirangura A. Parakeratosis in skin is associated with loss of inhibitor of differentiation 4 via promoter methylation. Hum Pathol. 2011;42:1878–1887. doi: 10.1016/j.humpath.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Chan AS, Tsui WY, Chen X, Chu KM, Chan TL, Li R, So S, Yuen ST, Leung SY. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003;22:6946–6953. doi: 10.1038/sj.onc.1206799. [DOI] [PubMed] [Google Scholar]
- 63.Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res. 2011;17:4341–4354. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomez Del Pulgar T, Valdes-Mora F, Bandres E, Perez-Palacios R, Espina C, Cejas P, Garcia-Cabezas MA, Nistal M, Casado E, Gonzalez-Baron M, Garcia-Foncillas J, Lacal JC. Cdc42 is highly expressed in colorectal adenocarcinoma and downregulates ID4 through an epigenetic mechanism. Int J Oncol. 2008;33:185–193. [PubMed] [Google Scholar]
- 65.Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta T, Saito H. Frequent DNA methylation but not mutation of the ID4 gene in malignant lymphoma. Journal of clinical and experimental hematopathology : JCEH. 2007;47:15–18. doi: 10.3960/jslrt.47.15. [DOI] [PubMed] [Google Scholar]
- 66.Smith E, De Young NJ, Pavey SJ, Hayward NK, Nancarrow DJ, Whiteman DC, Smithers BM, Ruszkiewicz AR, Clouston AD, Gotley DC, Devitt PG, Jamieson GG, Drew PA. Similarity of aberrant DNA methylation in Barrett's esophagus and esophageal adenocarcinoma. Mol Cancer. 2008;7:75. doi: 10.1186/1476-4598-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Neoplasia. ONCOMINE: a cancer microarray database and integrated data-mining platform. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma P, Chinaranagari S, Patel D, Carey J, Chaudhary J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Medicine. 2012;1:n/a–n/a. doi: 10.1002/cam4.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knowell AE, Patel D, Morton DJ, Sharma P, Glymph S, Chaudhary J. Id4 dependent acetylation restores mutant-p53 transcriptional activity. Mol Cancer. 2013;12:161. doi: 10.1186/1476-4598-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin MF, Meng TC, Rao PS, Chang C, Schonthal AH, Lin FF. Expression of human prostatic acid phosphatase correlates with androgen-stimulated cell proliferation in prostate cancer cell lines. J Biol Chem. 1998;273:5939–5947. doi: 10.1074/jbc.273.10.5939. [DOI] [PubMed] [Google Scholar]
- 71.Vinarskaja A, Goering W, Ingenwerth M, Schulz WA. ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World journal of urology. 2012;30:319–325. doi: 10.1007/s00345-011-0750-8. [DOI] [PubMed] [Google Scholar]
- 72.Patel D, Knowell AE, Korang-Yeboah M, Sharma P, Joshi J, Glymph S, Chinaranagari S, Nagappan P, Palaniappan R, Bowen NJ, Chaudhary J. Inhibitor of differentiation 4 (ID4) inactivation promotes de novo steroidogenesis and castration-resistant prostate cancer. Mol Endocrinol. 2014;28:1239–1253. doi: 10.1210/me.2014-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang XR, Kang HY, Cen J, Li YH, Wang LL, Yu L. Methylation status of id4 gene promoter in patients with chronic myeloid leukemia. Zhongguo shi yan xue ye xue za zhi/Zhongguo bing li sheng li xue hui = Journal of experimental hematology/Chinese Association of Pathophysiology. 2010;18:1402–1404. [PubMed] [Google Scholar]
- 74.Wang H, Wang XQ, Xu XP, Lin GW. ID4 methylation predicts high risk of leukemic transformation in patients with myelodysplastic syndrome. Leukemia research. 2010;34:598–604. doi: 10.1016/j.leukres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 75.Uhm KO, Lee ES, Lee YM, Park JS, Kim SJ, Kim BS, Kim HS, Park SH. Differential methylation pattern of ID4, SFRP1, and SHP1 between acute myeloid leukemia and chronic myeloid leukemia. Journal of Korean medical science. 2009;24:493–497. doi: 10.3346/jkms.2009.24.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuzontkoski PM, Mulligan-Kehoe MJ, Harris BT, Israel MA. Inhibitor of DNA binding-4 promotes angiogenesis and growth of glioblastoma multiforme by elevating matrix GLA levels. Oncogene. 2010;29:3793–3802. doi: 10.1038/onc.2010.147. [DOI] [PubMed] [Google Scholar]
- 77.Fontemaggi G, Dell'Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, Del Bufalo D, Strano S, Blandino G. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nature structural & molecular biology. 2009;16:1086–1093. doi: 10.1038/nsmb.1669. [DOI] [PubMed] [Google Scholar]
- 78.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 79.Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Id proteins expression in prostate cancer: high-level expression of Id-4 in primary prostate cancer is associated with development of metastases. Mod Pathol. 2006;19:931–941. doi: 10.1038/modpathol.3800602. [DOI] [PubMed] [Google Scholar]
- 80.Rahme GJ, Israel MA. Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene. 2014 doi: 10.1038/onc.2013.531. [DOI] [PubMed] [Google Scholar]
- 81.Jeon HM, Jin X, Lee JS, Oh SY, Sohn YW, Park HJ, Joo KM, Park WY, Nam DH, DePinho RA, Chin L, Kim H. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes & development. 2008;22:2028–2033. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pagliuca A, Cannada-Bartoli P, Lania L. A role for Sp and helix-loop-helix transcription factors in the regulation of the human Id4 gene promoter activity. J Biol Chem. 1998;273:7668–7674. doi: 10.1074/jbc.273.13.7668. [DOI] [PubMed] [Google Scholar]
- 83.Lu XC, Chi XH, Lou FD, Zhu HL, Fan H, Li SX, Yu L. Bioinformatics scan analysis for predicting drug targeted modulation on ID4 gene expression. Zhongguo shi yan xue ye xue za zhi/Zhongguo bing li sheng li xue hui = Journal of experimental hematology/Chinese Association of Pathophysiology. 2007;15:594–598. [PubMed] [Google Scholar]
- 84.Wen YH, Ho A, Patil S, Akram M, Catalano J, Eaton A, Norton L, Benezra R, Brogi E. Id4 protein is highly expressed in triple-negative breast carcinomas: possible implications for BRCA1 downregulation. Breast Cancer Res Treat. 2012;135:93–102. doi: 10.1007/s10549-012-2070-0. [DOI] [PubMed] [Google Scholar]
- 85.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King MC. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci U S A. 2002;99:7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson VC, Day TK, Bianco-Miotto T, Selth LA, Han G, Thomas M, Buchanan G, Scher HI, Nelson CC, Greenberg NM, Butler LM, Tilley WD. A gene signature identified using a mouse model of androgen receptor-dependent prostate cancer predicts biochemical relapse in human disease. Int J Cancer. 2012;131:662–672. doi: 10.1002/ijc.26414. [DOI] [PubMed] [Google Scholar]
- 87.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 88.Srikanth M, Kim J, Das S, Kessler JA. BMP Signaling Induces Astrocytic Differentiation of Clinically Derived Oligodendroglioma Propagating Cells. Molecular Cancer Research. 2014;12:283–294. doi: 10.1158/1541-7786.MCR-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crippa E, Lusa L, De Cecco L, Marchesi E, Calin GA, Radice P, Manoukian S, Peissel B, Daidone MG, Gariboldi M, Pierotti MA. miR-342 Regulates BRCA1 Expression through Modulation of ID4 in Breast Cancer. PLoS One. 2014;9:e87039. doi: 10.1371/journal.pone.0087039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heyn H, Engelmann M, Schreek S, Ahrens P, Lehmann U, Kreipe H, Schlegelberger B, Beger C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer. 2011;129:2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 91.Napolitano M, Comegna M, Succoio M, Leggiero E, Pastore L, Faraonio R, Cimino F, Passaro F. Comparative Analysis of Gene Expression Data Reveals Novel Targets of Senescence-Associated microRNAs. PLoS One. 2014;9:e98669. doi: 10.1371/journal.pone.0098669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arias-Romero LE, Chernoff J. Targeting Cdc42 in cancer. Expert Opinion on Therapeutic Targets. 2013;17:1263–1273. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature structural & molecular biology. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu Z, Li Y, Lee P, Liu T, Wan C, Wang Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One. 2012;7:e44033. doi: 10.1371/journal.pone.0044033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Current Opinion in Genetics & Development. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 98.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Y, Yu J. EZH2, an epigenetic driver of prostate cancer. Protein Cell. 2013;4:331–341. doi: 10.1007/s13238-013-2093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chinaranagari S, Sharma P, Chaudhary J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget. 2014 doi: 10.18632/oncotarget.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang LF, Huang S, Huang C, Li CR, Li DJ. Effect of 5-Aza-CdR on biological activity and inhibitor of DNA binding 4 gene expression in human erythroleukemia cell line K562. Zhongguo shi yan xue ye xue za zhi/Zhongguo bing li sheng li xue hui = Journal of experimental hematology/Chinese Association of Pathophysiology. 2011;19:1388–1392. [PubMed] [Google Scholar]
- 102.Qu F, Zhao CH, Diao YQ, Zhu XL, Chen J, Li M, Liu CP, Jiang L, Jin J. Methylation of Id4 gene and inhibitive effect of arsenic trioxide on it in Raji cells. Zhonghua xue ye xue za zhi = Zhonghua xueyexue zazhi. 2010;31:821–825. [PubMed] [Google Scholar]
- 103.Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci U S A. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carey JP, Knowell AE, Chinaranagari S, Chaudhary J. Id4 promotes senescence and sensitivity to doxorubicin-induced apoptosis in DU145 prostate cancer cells. Anticancer Res. 2013;33:4271–4278. [PMC free article] [PubMed] [Google Scholar]
- 105.Liggett WH, Sidransky D. Role of the p16 tumor suppressor gene in cancer. Journal of Clinical Oncology. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 106.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 107.Dell'Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G. ID4: a new player in the cancer arena. Oncotarget. 2010;1:48–58. doi: 10.18632/oncotarget.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coradini D, Fornili M, Ambrogi F, Boracchi P, Biganzoli E. TP53 mutation, epithelial-mesenchymal transition, and stemlike features in breast cancer subtypes. Journal of biomedicine & biotechnology. 2012;2012:254085. doi: 10.1155/2012/254085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bookstein R, Rio P, Madreperla SA, Hong F, Allred C, Grizzle WE, Lee WH. Promoter deletion and loss of retinoblastoma gene expression in human prostate carcinoma. Proc Natl Acad Sci U S A. 1990;87:7762–7766. doi: 10.1073/pnas.87.19.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tamimi Y, Bringuier PP, Smit F, van Bokhoven A, Debruyne FM, Schalken JA. p16 mutations/deletions are not frequent events in prostate cancer. Br J Cancer. 1996;74:120–122. doi: 10.1038/bjc.1996.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]