Abstract
Despite investigative interest, the artificial derivation of pluripotent stem cells remains inefficient and incomplete reprogramming hinders its potential as a reliable tool in regenerative medicine. By contrast, fusion of terminally differentiated gametes at fertilization activates efficient epigenetic reprogramming to ensure totipotency of early embryos. Understanding the epigenetic mechanisms required for the transition from the fertilized egg to the embryo can improve efforts to reprogram differentiated cells to pluripotent/totipotent cells for therapeutic use. We review recent discoveries that are providing insight into the molecular mechanisms required for epigenetic reprogramming to totipotency in vivo.
Keywords: reprogramming, totipotency, epigenetic modification, embryonic genome activation, preimplantation development
Establishing the totipotency of early embryos
Investigations of iPSCs have provided insight into the establishment of pluripotency for potential clinical application. However, genetic and epigenetic abnormalities dampen enthusiasm for their use in regenerative medicine, and the establishment of totipotency presents a better opportunity for this purpose [1]. Unlike pluripotency, totipotency is defined as the ability of cells to differentiate into any cell type and, more stringently, to develop into a complete organism [2]. Although the molecular basis of this capacity remains largely unknown, four processes are designed to generate totipotent cells: (i) fusion of terminally differentiated, haploid sperm and egg into one-cell (1C) diploid zygotes (see Glossary) in vivo or by in vitro fertilization (IVF); (ii) somatic cell nuclear transfer (SCNT), which relies on the reprogramming activity of eggs; (iii) isolation of transient ESC/iPSC populations with a two-cell (2C)-like embryonic transcriptome [3]; and (iv) in vivo generation of iPSCs with totipotent features [4]. While the latter three are of investigative interest, SCNT suffers from inefficiency and transient ESC populations and, in vivo, produced iPSCs that lacked the ability to develop into an intact organism. Thus, cleavage stage embryos formed from fused gametes at fertilization provide the most physiological platform to investigate the signature of totipotency and devise strategies for reprogramming terminally differentiated cells.
Despite sharing major regulatory networks, totipotent and pluripotent cells exhibit significant differences such as core histone motility [5] and utilization of regulatory factors for gene expression [6]. Central to developing mouse embryos from differentiated gametes are epigenetic modifications to ensure expression or silencing of genes that define totipotency. Upon fertilization, haploid sperm penetrate into the cytoplasm of haploid eggs to form diploid 1C zygotes. This triggers the paternal genome to re-establish nucleosome structures with stored maternal histones, followed rapidly by the formation of the male and female pronuclei. There is minor activation of parental genomes at the 1C stage, but little translation from the de novo transcripts. This event has been described both as embryonic genome activation (EGA) and zygotic genome activation (ZGA). The former acronym will be used in this review.
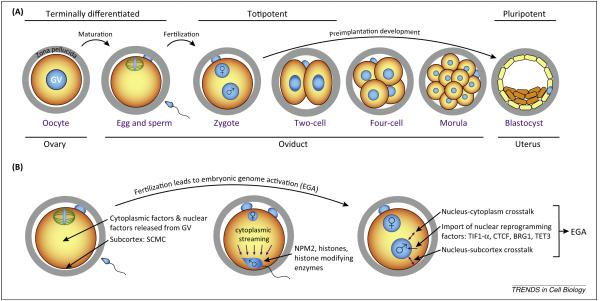
Initially, paternal and maternal chromatin have separate and asymmetric epigenetic profiles. Division of the 1C zygote forms 2C embryos with significantly increased transcription and each of the two blastomeres is totipotent [7]. Newly synthesized embryonic gene products gradually replace maternal factors as regulators of early development. Although a causal relationship has not been established, there is concomitant loss of totipotency of the embryo. For example, single four- and eight-cell (4C, 8C) blastomeres cannot form all the lineages of the embryo without aggregating with carrier blastomeres [8]. Ultimately, embryonic cells differentiate to form either the inner cell mass (ICM, origin of ESCs) or the trophectoderm (TE, precursor of the placenta) as the blastocyst prepares for implantation (Figure 1A).
Figure 1.
Mouse preimplantation development and maternal-to-embryonic transition. (A) After ovulation from the ovary into the oviduct, terminally differentiated mature eggs (surrounded by the extracellular zona pellucida) are fertilized by sperm to establish totipotent zygotes that divide during preimplantation development to become blastocysts prior to implantation in the uterus at embryonic day 4.5. (B) Upon fertilization, stored maternal factors activate the embryonic genome to trigger maternal-to-embryonic transition that results in formation of a totipotent zygote. Specifically, parental genomes are reorganized by chromatin remodeling factors with the aid of cytoplasmic streaming, and form pronuclei that import nuclear reprogramming factors and crosstalk with other cellular compartments. Abbreviations: BRG1, a chromatin remodeling protein; CTCF, CCCTC-binding factor; GV, germinal vesicle; NPM2, nucleoplasmin protein 2; SCMC, subcortical maternal complex; TET3, ten-eleven translocation protein 3; TIF-α, transcription intermediary factor 1 α.
This review focuses on investigations that define progressive changes in epigenetic reprogramming that affect chromatin dynamics and enable totipotency in the early mouse embryo. Future discoveries that guide chromatin reprogramming technologies will have profound influences on regenerative medicine. Personalized totipotent cells from terminally differentiated cells could provide pools of cells from which specific cell types could be established for the treatment degenerative diseases such as diabetes and Parkinsonism. Evolution has shaped the most efficient means of establishing totipotent cells from terminally differentiated gametes and understanding those molecular mechanisms should provide insight into how to recapitulate them for the therapeutic benefit of patients.
Stored maternal factors
During oogenesis, the volume of the germ cell increases dramatically and provides storage for maternal factors needed to compensate for the absence of transcription during meiotic maturation, ovulation, and early development. At fertilization, each gamete contributes a haploid genome, but the egg is the primary source for gene products (RNA, proteins) vital for the establishment of totipotency and EGA (Figure 1B). These factors are encoded by maternal effect genes, such as genes encoding nucleoplasmin (NPM) 2 [9] and the subcortical maternal complex (SCMC) [10-12]. Their genetic ablation in mice documents the essential roles of both nuclear and cytoplasmic factors in establishing the totipotency of early embryonic cells [13]. The large cytoplasmic volume of the 1C zygote may complicate protein trafficking and recent reports emphasize the importance of actin scaffold and actin flow-driven streaming in supporting the integrity and stability of subcellular structures [14] and redistribution of cytosolic components during genome reprogramming [15].
Although progress has been made in understanding the complexity of reprogramming to totipotency, the paucity of biological materials in mice has hampered efforts to compile a complete inventory and identification of key regulators. Notably, because matured eggs are transcriptionally inactive, information from translation profiling [16] and a detailed exploration of the egg proteome [17] is valuable. Further identification of intra- and extra-cellular maternal factors and their functions will greatly facilitate our understanding of how to improve chromatin reprogramming [18-20]. Accumulating evidence also documents reprogramming activity in the cytoplasm [21] and potential crosstalk between the nucleus and cytoplasm (especially the subcortex) further extends the scope for functional reprogramming.
Re-establishment of paternal chromatin
The repackaging of the male genome during spermatogenesis erases most, but not all, chromatin marks. Upon fertilization, the sperm nucleus undergoes decondensation and recondensation. During this time, protamines are replaced by stored maternal histones and nucleosomes are formed with both inherited paternal and newly deposited maternal histones.
Sperm epigenome for intergenerational inheritance
During spermatogenesis, somatic histones are hyperacetylated for removal and are replaced by transition nuclear proteins (encoded by Tnp1, Tnp2). These are replaced, in turn, by protamines (encoded by Prm1, Prm2), which are arginine and disulfide bond enriched nuclear proteins that condense the haploid male genome. During this histone-to-protamine transition, a few regions of the male genome (approximately 1% in mouse and 10% in human) escape histone removal and retain nucleosome structures.
The low efficiency of SCNT [21] and the aberrant epigenome in cloned embryos [22] illustrate the significance of a prepatterned sperm epigenome for correct reprogramming by maternal factors. DNA methylome profiling as well as ChIP-seq analyses in mouse and human sperm revealed the retention of histone variants and epigenetic memory with related DNA and histone modifications [23-28] (Figure 2A). While differences were observed between mouse and human, the extensive, but not deterministic, interrelationships with developmental regulators in early embryos suggest paternal transmission of epigenetic information to cleavage stage embryos. More nuanced experimental designs are required to elucidate the extent by which epigenetic marks in gametes are inherited by early embryos to determine their molecular mechanisms and their importance to development.
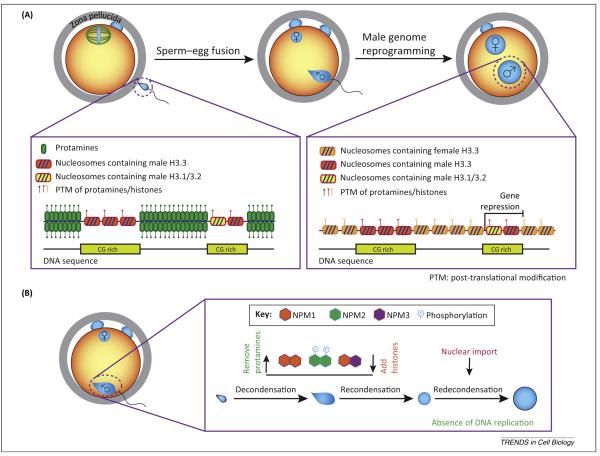
Figure 2.
Male genome reorganization during protamine-to-histone exchange. (A) Structural changes of male chromatin before and after fertilization. The male genome in sperm is densely packaged by protamines with a few regions retaining histones (including canonical histones and histone variants). After the protamine-to-histone exchange, protamines are replaced by maternal histones. The retained paternal histones may help determine gene expression in the early embryo. (B) During reorganization, the male genome experiences decondensation, recondensation, and re-decondensation, with protamine-to-histone exchange possibly collectively mediated by nucleoplasmin (NPM) family proteins, including NPM1, NPM2, and NPM3. Abbreviation: PTM, post-translational modification.
Protamine-to-histone exchange
Following fertilization, the sperm genome is decondensed to remove protamines and repackaged with stored maternal histones in the absence of DNA replication. This is one of the most dramatic chromatin remodeling events to occur during development. Continuous time lapse imaging showed that after rapid decondensation, the asymmetric shape of homogeneously decondensed paternal chromatin recondenses into a symmetric ovoid, followed by rapid swelling of this spherical structure into a pronucleus and the immediate import of nuclear factors [29] (Figure 2B). Notably, these dramatic morphological changes in paternal chromatin, from continuation of meiosis to pronucleus formation, occur over 2 h in mice in the absence of nuclear structures. Therefore, the maternal environment provides the opportunity for cytoplasmic and nuclear factors to collaborate in the initial reprogramming of parental genomes to form pronuclei.
To study this chromatin remodeling process, cell free systems using egg extracts have been developed in multiple species. NPM2 was first identified in frogs as a chaperone necessary and sufficient for the removal of sperm nuclear basic proteins (intermediates between protamines and histones) and the addition of histones to form nucleosome cores [30,31]. In mice, the Npm2 ortholog was genetically ablated without affecting sperm nuclear decondensation during embryonic development [9]. This suggests that other nucleoplasmins (NPM1 and NPM3) compensate for the loss of NPM2, which is supported by in vitro studies showing cooperation of NPM1, NPM2, and NPM3 in chromatin disassembly and assembly [32] (Figure 2B). Comparative analysis with the histone-to-protamine exchange in spermatogenesis should provide further insight in the molecular mechanism of the protamine to histone exchange as these two remodeling processes may share similar mechanisms including protein degradation [33], transition protein incorporation, and histone variant replacement [34].
Chromatin reprogramming towards totipotency
The two separate pronuclei, harboring asymmetric epigenetic modifications, approximate each other during syngamy and form the 1C diploid zygote, which subsequently divides [35]. During cleavage stage embryogenesis, striking changes in epigenetic modifications (deposition of histone variants, re-establishment of histone marks, and DNA demethylation) occur throughout the genome.
Histone variant dynamics
Canonical histones (H2A, H2B, H3, H4) are expressed primarily in S phase and are incorporated into chromatin in a replication-coupled manner. In addition, variant histones share sequence homology and structural similarity with canonical histones, but harbor specialized functions (Figure 3A). These variants are expressed and incorporated into chromatin throughout the cell cycle, and preferentially replace canonical histones at specific genomic regions to form nucleosomes with unique biophysical characteristics. Histone variants play essential roles in chromatin reprogramming [20,36-38] and participate in early embryogenesis (Figure 3B) among other developmental processes.
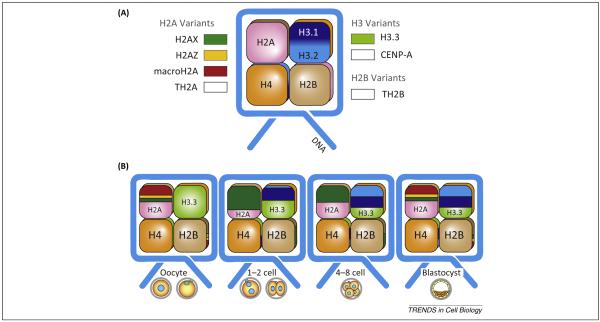
Figure 3.
Histone variants during preimplantation development. (A) Schematic representation of nucleosome structure and histone variants. (B) Changes in histone variant deposition during preimplantation development. Notably, there are significant contributions of H2A.X and H3.3 to totipotent 1C and 2C chromatin, which links the presence of these histones to totipotency. Abbreviation: CENP-A, centromeric histone 3.
In addition to two canonical isoforms (H3.1 and H3.2), histone H3 has three variants in mammals: H3.3, centromeric CENP-A, and testis-specific H3.4. The most investigated variant is H3.3, which incorporates into chromatin in both a replication-independent and a replication-coupled manner. H3.3 differs from canonical H3 in only four amino acids, which are essential for interacting with the chaperones HIRA and Daxx/ATRX. Early immunofluorescence and ChIP-seq assays document enrichment of H3.3 in transcriptionally active regions where many regulatory elements depend on its chaperone HIRA [39,40]. H3.3 also localizes to telomeres where its presence depends on ATRX [40]. Following mouse fertilization, HIRA deposits maternal H3.3 onto paternal chromatin during the protamine-to-histone exchange [41-43] to establish new paternal nucleosomes [44] and activate pluripotency genes [45]. H3.3 is transiently removed from maternal chromatin during this stage, indicating erasure of maternal epigenetic memory [42]. H3.3 occupies both euchromatin (noncondensed) and heterochromatin (condensed) up to the four-cell stage, after which its distribution is restricted to euchromatin [42]. H3.3 is required to establish double-stranded RNA (dsRNA)-dependent pericentrometric heterochromatin in early embryos, an activity dependent on H3.3 lysine 27 [46]. In addition, H3.3 maintains a decondensed chromatin state in early mouse embryos by antagonizing linker H1, an activity dependent on H3.3 lysine 36 [47].
Histone H2A has the most diverse variants among core histones in mammals. Notable H2A variants include H2A.X, H2A.Z, and macroH2A. H2A.X is abundant in cleavage stage chromatin, and its C-terminal amino acids determine its specific association in the chromatin of early embryos [48]. H2A.Z is required for pluripotency [49] and macroH2A inhibits epigenetic reprogramming to pluripotency [50]. H2A.Z and macroH2A are highly expressed in matured egg chromatin, and are later silenced after fertilization [48], suggesting that H2A.X is the major H2A variant in totipotent early embryos.
To decipher the effect of histone variants on totipotency, additional investigation is needed to uncover: (i) the relationship between sperm-retained histone variants and newly deposited histones in paternal chromatin after fertilization; (ii) the binding profiles of individual histone variants in early embryos; and (iii) the presence and possible function of homotypic and heterotypic nucleosomes containing one or more histone variants in early embryos [51,52].
Asymmetric histone modifications
Real-time imaging has provided a dynamic view of histone modifications during early development [53,54]. After protamine removal, the maturing paternal genome is repack-aged by newly synthesized and acetylated maternal histones. By contrast, maternal chromatin maintains its histone methylation pattern throughout early cleavage stages, leading to epigenetic asymmetry between paternal and maternal genomes (Figure 4A). This asymmetry correlates with asymmetric transcriptional activity and may account for the different reprogramming capacities of the two pronuclei [55].
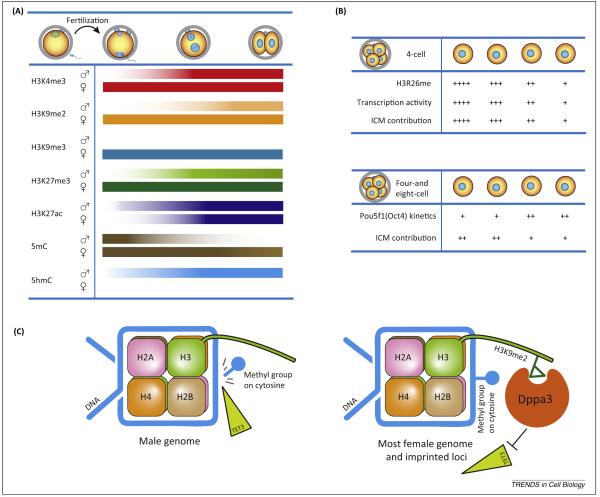
Figure 4.
Epigenetic changes during the maternal-to-embryonic transition. (A) Summary of histone and DNA modification dynamics. Acetylated histones are deposited on the male genome after fertilization and re-establish histone methylation patterns that correlate with reduced DNA methylation. Density of color reflects extent of modification. (B) Epigenetic asymmetry among blastomeres is detected as early as four-cell (4C) embryos and this polarity predicts the contributions of the individual blastomere to the inner cell mass (ICM) or trophectoderm layer (TE). (C) TET enzymes, specifically TET3, mediates active DNA demethylation during maternal-to-embryonic transition by converting 5mC to 5hmC, but can be protected from demethylation by H3 lysine 9 di-methylation (H3K9me2)-recruited Dppa3.
In cleavage stage mouse embryogenesis, epigenetic rather than transcriptome asymmetry is associated with cell fate decision and can be detected as early as the 4C stage [56]. Using histone arginine methyltransferase activity, global asymmetric H3 arginine methylation patterns were detected in 4C mouse embryos [57] (Figure 4B). The blastomeres with high and low methylation patterns preferentially formed the ICM and TE, respectively. Similar molecular asymmetry is observed for POU5F1 (Oct4) binding sites as embryos progress from 4C to 8C stage and reflects different accessibility of POU5F1 to the genome [58] (Figure 4B). Nevertheless, it remains unclear how embryonic polarity is initially established and there is no evidence of asymmetric distribution of maternal RNA or protein in the fertilized egg. Therefore, other mechanisms including cell–cell contact [59] and cell position sensing [60] may play regulatory roles in this process. Understanding the mechanisms underlying early establishment of cell polarity should provide insight into developing strategies to inhibit differentiation and maintain totipotency during cell division.
Despite progress in establishing essential roles of histone modifications during cleavage stage embryogenesis, experimental approaches are complicated by the redundancy of histone genes, nonhistone targets of histone modifying enzymes, and unclear mechanisms of dominant negative histone mutants.
Active DNA demethylation activity
Recent investigations of active demethylation have documented the importance of nucleotide(s) excision and repair. One important pathway is dependent on prior modification of methylated cytosine. Ten eleven translocation (TET) proteins convert 5-methylcytosine (5mC) to 5-hydroxy-methylcytosine (5hmC) [61], and further to 5-formylcytosine (5fC) and 5-carboxymethylcytosine (5caC) for excision [62,63]. 5hmC can be recognized by epigenetic regulators for gene regulation, indicating its role beyond just a demethylation intermediate [64].
The paternal genome, highly methylated during spermatogenesis, is actively demethylated beginning in the early 1C embryo [65,66]. The histone modification, H3 lysine 9 di-methylation, is specifically enriched in maternal chromatin and recruits maternal factor Dppa3/PGC7/Stella [67] (Figure 4C). Long considered to protect maternal DNA and imprinted loci from demethylation, recent sequence data documents active demethylation in maternal, as well as paternal, DNA [68,69]. High levels of 5hmC on the paternal genome are detected in the 1C zygote [70,71] which also undergoes replication-dependent loss [72] and, among the TET family of proteins, only TET3 is expressed at this point. In the absence of TET3, the paternal genome remains highly methylated in early embryos and severely reduces fecundity [73], which indicates a pivotal role of paternal DNA demethylation in chromatin reprogramming. Cullin-ring finger ligase-4 (CRL4) ubiquitin ligase has recently been reported to induce TET3 activity and play an essential role in female fecundity [74], further strengthening the importance of active DNA demethylation. The above observations have been recently expanded by genome-scale DNA methylation maps of mouse [68,69,75] and human [76,77] preimplantation, which revealed specific methylation patterns of repetitive elements and differentially methylated regions (DMRs) [75]. Real-time imaging of DNA methylation dynamics in live embryos will facilitate the identification of additional epigenetic regulators in early mouse embryos [78].
Higher-order chromatin structure
Adding further complexity to the investigation of genomic reprogramming is the role of higher-order chromatin architecture and localization within the nucleus as determinants of epigenetic modification and transcriptional regulation. For example, when early mouse embryos express a nuclear envelope protein fused to a zinc finger protein recognizing major satellite sequences, pericentric chromatin is tethered to the nuclear periphery, and heterochromatin establishment, as well as development, is disrupted [79]. A similar strategy was used to bridge enhancer–promoter interactions to manipulate endogenous gene expression in erythroid cells [80]. Therefore, it will be valuable to further characterize dynamic changes of higher-order chromatin architecture in early mouse embryos and define their relationship with transcriptional activity and nuclear localization.
In summary, although dynamic changes of epigenetic status during reprogramming to totipotency have been extensively investigated, the only genome-wide study reported is the DNA methylome [68,69,75-77]. The small amount of biological material available from embryos not only impedes genome-wide scans of histone variant occupancy and histone modifications, but also imposes limits on biochemical analyses of histone chaperones and histone modifying enzymes. For example, H3.3 is by far the pre-dominant H3 isoform packaging the male genome at early zygote stage, but whether H3.3 binding is concentrated at particular loci by specific chaperones is unknown. In addition, it is not clear whether individual histone variants are marked by modifications with variant-specific functions, which appears to be true for H3.3 lysine36 tri-methylation [81]. Therefore, novel strategies are required for detailed investigations on epigenome changes during transition from terminally differentiated haploid germ cells to early embryos to better understand the epigenetic reprogramming necessary for totipotency.
Embryonic genome activation and totipotency
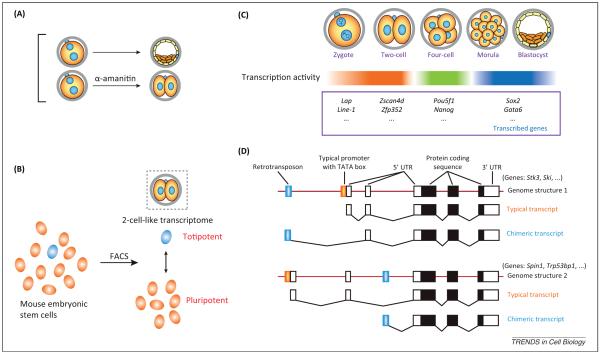
Following fertilization, maternal factors reprogram parental genomes to restore totipotency to the early embryo and ensure embryonic gene activation. EGA is first detected in mice at the late 1C stage, with higher activity in the male than the female pronucleus, and becomes robust at the 2C stage. For example, pericentric satellites, which are essential for chromocenter formation in early embryos, are activated with parental asymmetry in 1C zygotes and dramatically up-regulated at the 2C stage [82]. Blocking EGA with α-amanitin, an inhibitor of RNA polymerase II elongation, results in embryonic arrest at the 2C stage, which correlates EGA with the establishment of totipotency (Figure 5A). A recent study described transient ESCs/iPSCs populations with a transcriptome that closely resembles that observed in the blastomeres of totipotent, 2C embryos [3] (Figure 5B). Thus, totipotency occasionally can be regained in pluripotent cells and may result from fluctuating expression of pluripotent regulators that overcome genome-wide epigenetic barriers to totipotency. Notably, EGA is characterized by more efficient utilization of TATA-less promoters [83], activation of repetitive elements (especially retrotransposons silenced in most cell types) [84], uncoupling of transcription and translation in 1C zygotes [85], and activation of enhancers for transcription in 2C embryos [86].
Figure 5.
Embryonic genome activation as a hallmark of totipotency. (A) Preimplantation development is blocked at the two-cell (2C) stage by treatment with α-amanitin that inhibits transcription elongation. (B) A transient subpopulation of mouse embryonic stem cells has been identified as totipotent with 2C-like transcriptome, which correlates embryonic genome activation with totipotency. (C) Transcriptome profiling documents distinct patterns of maternal transcript degradation and waves of transcription activations during preimplantation development. (D) Retrotransposons are activated during the preimplantation stage and provide alternative first exons to activate downstream embryonic genes, producing chimeric transcripts that are absent in other cell types. Abbreviation: FACS, fluorescence-activated cell sorting.
Ultra-large scale and single-cell transcriptome profiling, coupled with nano-CAGE analysis, have documented waves of de novo transcription in mouse and human pre-implantation embryos that are triggered by activating functional gene expression modules in stage-specific patterns [87-89] (Figure 5C). Specifically, single-cell analyses used to capture expression of transcription factors and epigenetic modifiers during preimplantation development describe a complex transcriptional network and asymmetric expression pattern among individual cells [90,91]. Despite progress, it has been difficult to identify the earliest de novo transcripts because of a large, residual maternal RNA pool in early mouse embryos. One approach is to inhibit transcription with α-amanitin and compare the transcriptome of treated and untreated embryos [92,93], although conclusions drawn from these studies may be complicated by possible drug induced effects on subsequent development.
Retrotransposons and the Zscan4 family of transcription factors are well-known EGA genes. Expression of retrotransposons makes a significant contribution to the transcriptome of early mouse embryos. At least one function is to act as alternative promoters for the activation of protein-coding genes by generating chimeric transcripts with retrotransposon–gene junctions [84] (Figure 5D). The most active LINE-1 retrotransposon provides a stimulatory auto-enhancing loop, indicating that maternal retrotransposon transcripts robustly activate retrotransposons in the genome after fertilization [89]. These retrotransposon-activated EGA genes can be reactivated in pluripotent ESCs in the absence of lysine-specific demethylase LSD1, implying that histone-modifying enzymes silences EGA genes during totipotency-to-pluripotency transition [94]. Expression of the Zscan4 gene family is temporally and spatially restricted, and is essential for early mouse development, with one member, Zscan4d, identified as a 2C specific gene. This gene family plays important roles in genome stability and maintenance of telomeres [95]. Zscan4 also reactivates early embryonic genes during iPSCs generation, documenting a hierarchy of activated embryonic genes [96].
Concluding remarks
Recent investigations document that the totipotency of early embryos is specified by unique epigenetic status including histone variants, histone modifications, and patterns of DNA methylation. These structural features of chromatin may facilitate access of reprogramming factors/cofactors to specific genomic regions and promote formation of higher-order chromatin structure necessary to activate or repress the genes needed for totipotency. Detailed understanding of this reprogramming process should provide insight into fundamental mechanisms that may translate into improved reprogramming of differentiated adult cells for therapeutic use in regenerative medicine. The field is still in its infancy and there is much to be learned about interacting genetic and epigenetic hierarchies, transcriptome dynamics and the role of maternal and embryonic regulators (Box 1). Further detailed molecular analyses and screening will necessitate technological advances to improve micromanipulation of early mouse embryos and overcome the lack of available biological materials. The use of nanofluidics [97] and rapid progress in micro-technologies hold promise for genome-sequencing [98], methylome analyses [99], allele specific histone modification assays [100], chromosome conformation mapping [101], and nucleosome mapping [102] of individual embryonic cells. By focusing these technologies on the physiological reprogramming that occurs in the transition from terminally differentiated gametes to the totipotency of the early embryo, greater insight is anticipated in establishing protocols for converting somatic cells to totipotency for use in regenerative medicine and control of degenerative diseases that afflict patients worldwide.
Box 1. Outstanding questions and important future investigations.
Determine triggering events for the activation of the totipotent embryonic genome.
Identify genome wide profiles of the epigenome in cleavage stage embryos.
Elucidate epigenetic asymmetry between parental genomes and among blastomeres.
Analyze regulatory network of maternal effect genes affecting cleavage stage development.
Define key reprogramming factors in the nucleus and cytoplasm required for totipotency.
Establish efficient protocols to generate gametes in vitro or create totipotent cell lines to provide sufficient material for biochemical identification of required reprogramming factors.
Acknowledgments
We thank Dr M.E. Torres-Padilla for her critical reading of the manuscript and useful suggestions. We apologize to authors whose work could not be cited because of space constraints. This research was supported by the Intramural Research Program of the National Institutes of Health, NIDDK (ZIA-DK015603).
Glossary
- Active DNA demethylation
an enzymatic process to remove DNA methylation, in contrast to passive DNA demethylation carried out by serial dilutions during cell divisions without DNA methylation activity
- Blastocyst
the 128-cell blastocyst has two distinct cell lineages and a fluid-filled cavity called blastocoel. The outer trophectoderm layer (TE) is the precursor of the placenta and the inner cell mass (ICM) forms the fetus and is the source of embryonic stem cells
- ChIP-seq
ChIP with high-throughput sequencing. It is a widely used genome-wide approach to determine DNA-binding sites of proteins of interest. Cleavage stage embryogenesis: after fertilization, the 1C embryo undergoes three cell divisions to form eight blastomeres prior to compacting into a morula
- Embryonic genome activation (EGA)/zygotic genome activation (ZGA)
after fertilization, embryonic genes are transcriptionally activated by maternal regulators to initiate developmental programs in the early embryo
- Gametes
Mammalian male gametes are small, motile sperm while female gametes are relatively large, nonmotile eggs surrounded by a proteinaceous zona pellucida
- Higher-order chromatin structure
following the formation of nucleosomes, nuclear DNA is further organized topologically into higher-order structures by forming genome-wide physical networks in a nonrandom manner
- Histone variant
noncanonical histones with amino acid sequences distinct from canonical histones. They usually have specific functions. Unlike canonical histones, they can be incorporated into chromatin outside of S-phase and their genes contain introns
- Nano-CAGE
identification of promoter location and gene expression by capturing and quantifying 5′ ends of transcripts using small amounts of total RNA. Combined nano-CAGE and RNA-seq facilitates genome-wide transcriptome studies
- Oogenesis/spermatogenesis
during gametogenesis, oocytes are established by oogenesis in the ovary and sperm by spermatogenesis in the testis. Each gamete undergoes reductive divisions during meiosis to form haploid genomes
- Preimplantation development
embryonic development from 1C zygotes to blastocysts. After this early development, blastocysts hatch from the surrounding zona pellucida and implant on the uterine wall
- Reprogramming
a process that occurs in vivo during primordial germ cell development and again during cleavage stage embryogenesis to erase or rewrite epigenetic chromatin marks
- Subcortex
an actin-enriched cytoplasmic layer at the inner aspect of the plasma membrane. The maternal subcortical maternal complex (SCMC) is enriched in the subcortex of eggs and is required for progression beyond two-cell (2C) development
- Syngamy
fusion of gametes at the start embryogenesis. During syngamy, parental pronuclei migrate towards each other and interdigitate their nuclear membranes which breakdown so that chromosomes can congress on the mitotic spindle before the first cleavage division
- Zygote
diploid one-cell (1C) embryo produced at fertilization and is the earliest stage of embryo that develops into a complete organism
References
- 1.Ma H, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condic ML. Totipotency: what it is and what it is not. Stem Cells Dev. 2014;23:796–812. doi: 10.1089/scd.2013.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abad M, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 5.Boskovic A, et al. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G, et al. Establishment of totipotency does not depend on Oct4A. Nat. Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarkowski AK. Experiments on the development of isolated blastomers of mouse eggs. Nature. 1959;184:1286–1287. doi: 10.1038/1841286a0. [DOI] [PubMed] [Google Scholar]
- 8.Rossant J. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J. Embryol. Exp. Morphol. 1976;36:283–290. [PubMed] [Google Scholar]
- 9.Burns KH, et al. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 10.Li L, et al. A subcortical maternal complex essential for pre-implantation mouse embryogenesis. Dev. Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu XJ, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat. Commun. 2014;5:4887. doi: 10.1038/ncomms5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, et al. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 2013;15:1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajduk A, et al. Rhythmic actomyosin-driven contractions induced by sperm entry predict mammalian embryo viability. Nat. Commun. 2011;2:417. doi: 10.1038/ncomms1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, et al. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, et al. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Munoz E, et al. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345:822–825. doi: 10.1126/science.1254745. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa M, et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 20.Shinagawa T, et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14:217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Kang E, et al. Nuclear reprogramming by interphase cytoplasm of two-cell mouse embryos. Nature. 2014;509:101–104. doi: 10.1038/nature13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan MM, et al. Mouse ooplasm confers context-specific reprogramming capacity. Nat. Genet. 2012;44:978–980. doi: 10.1038/ng.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 24.Carone BR, et al. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erkek S, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 27.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samans B, et al. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev. Cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Adenot PG, et al. Dynamics of paternal chromatin changes in live one-cell mouse embryo after natural fertilization. Mol. Reprod. Dev. 1991;28:23–34. doi: 10.1002/mrd.1080280105. [DOI] [PubMed] [Google Scholar]
- 30.Philpott A, et al. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- 31.Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- 32.Okuwaki M, et al. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res. 2012;40:4861–4878. doi: 10.1093/nar/gks162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian MX, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montellier E, et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013;27:1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamboni L, et al. First cleavage division of the mouse zygot. An ultrastructural study. Biol. Reprod. 1972;7:170–193. doi: 10.1093/biolreprod/7.2.170. [DOI] [PubMed] [Google Scholar]
- 36.Jullien J, et al. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5:17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaspar-Maia A, et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nashun B, et al. Dramatic replacement of histone variants during genome remodeling in nuclear-transferred embryos. Epigenetics. 2011;6:1489–1497. doi: 10.4161/epi.6.12.18206. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Padilla ME, et al. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama T, et al. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Heijden GW, et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Inoue A, Zhang Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014;21:609–616. doi: 10.1038/nsmb.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen D, et al. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7325–7330. doi: 10.1073/pnas.1406389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santenard A, et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CJ, et al. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nashun B, et al. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137:3785–3794. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- 49.Hu G, et al. H2A.Z. facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasque V, et al. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J. Cell Sci. 2012;125:6094–6104. doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber CM, et al. H2A.Z. nucleosomes enriched over active genes are homotypic. Nat. Struct. Mol. Biol. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin C, et al. H3.3/H2A.Z. double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki K, et al. Real-time imaging of histone H4 hyperacetylation in living cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi-Takanaka Y, et al. Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. J. Cell Biol. 2009;187:781–790. doi: 10.1083/jcb.200904137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, et al. Asymmetric reprogramming capacity of parental pronuclei in mouse zygotes. Cell Rep. 2014;6:1008–1016. doi: 10.1016/j.celrep.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 56.VerMilyea MD, et al. Transcriptome asymmetry within mouse zygotes but not between early embryonic sister blastomeres. EMBO J. 2011;30:1841–1851. doi: 10.1038/emboj.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres-Padilla ME, et al. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plachta N, et al. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 59.Fierro-Gonzalez JC, et al. Cadherin-dependent filopodia control preimplantation embryo compaction. Nat. Cell Biol. 2013;15:1424–1433. doi: 10.1038/ncb2875. [DOI] [PubMed] [Google Scholar]
- 60.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 61.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 66.Mayer W, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 67.Nakamura T, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 68.Wang L, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.08.003. http://dx.doi.org/10.1016/j.stem.2014.08.003. [DOI] [PubMed]
- 70.Iqbal K, et al. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 72.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 74.Yu C, et al. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science. 2013;342:1518–1521. doi: 10.1126/science.1244587. [DOI] [PubMed] [Google Scholar]
- 75.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo H, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 77.Smith ZD, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okada Y, et al. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jachowicz JW, et al. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev. 2013;27:2427–2432. doi: 10.1101/gad.224550.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Probst AV, et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Davis W, Jr, Schultz RM. Developmental change in TATA-box utilization during preimplantation mouse development. Dev. Biol. 2000;218:275–283. doi: 10.1006/dbio.1999.9486. [DOI] [PubMed] [Google Scholar]
- 84.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 85.Nothias JY, et al. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- 86.Wiekowski M, et al. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev. Biol. 1991;147:403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- 87.Park SJ, et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27:2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fadloun A, et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 90.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 91.Burton A, et al. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013;5:687–701. doi: 10.1016/j.celrep.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 92.Hamatani T, et al. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 93.Posfai E, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26:920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zalzman M, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirata T, et al. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Murphy PJ, et al. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7772–7777. doi: 10.1073/pnas.1218495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 99.Lorthongpanich C, et al. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341:1110–1112. doi: 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- 100.Gomez D, et al. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Small EC, et al. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2462–E2471. doi: 10.1073/pnas.1400517111. [DOI] [PMC free article] [PubMed] [Google Scholar]