Abstract
Previous studies have shown that an attenuated West Nile virus (WNV) nonstructural (NS) 4B-P38G mutant induces stronger innate and adaptive immune responses than wild-type WNV in mice, which has important applications to vaccine development. To investigate the mechanism of immunogenicity, we characterized WNV NS4B-P38G mutant infection in two human cell lines--THP-1 cells and THP-1 macrophages. Although the NS4B-P38G mutant produced more viral RNA than the parental WNV NY99 in both cell types, there was no detectable infectious virus in the supernatant of either cell type. Nonetheless, the attenuated mutant boosted higher innate cytokine responses than virulent parental WNV NY99 in these cells. The NS4B-P38G mutant infection of THP-1 cells led to more diverse and robust innate cytokine responses than that seen in THP-1 macrophages, which were mediated by toll-like receptor (TLR)7 and retinoic acid-inducible gene 1(RIG-I) signaling pathways. Overall, these results suggest that a defective viral life cycle during NS4B-P38G mutant infection in human monocytic and macrophage cells leads to more potent cell intrinsic innate cytokine responses.
Keywords: West Nile virus, NS4B Protein, Immune Response, Cytokine
1. Introduction
West Nile virus (WNV), is a mosquito-borne flavivirus that has been a serious public health problem in North America for more than a decade [1]. WNV infection of the central nervous system (CNS, neuroinvasive disease) commonly presents as encephalitis, meningitis, or acute flaccid paralysis. The overall mortality rate among people who develop acute WNV neuroinvasive disease is about 10%, although it increases significantly in the elderly and immunocompromised [2-3]. About 20-50% of WNV convalescent patients develop persistent sequelae years after their acute illness; symptoms included muscle weakness and pain, fatigue, headaches, memory loss, and ataxia [4-8]. At present, no vaccines nor therapeutics for treatment or prevention of WNV infection have been approved for human use.
The WNV genome is a single-stranded, positive-sense RNA molecule, which is translated into a single polypeptide and co- and post-translationally processed into 10 proteins – three structural proteins (envelope (E), membrane, nucleocapsid) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) [9-10]. The nonstructural (NS) proteins are known to be associated with evasion of host innate immunity and are important determinants in flaviviral pathogenesis [11-14]. The NS4B P38 residue is conserved in most vector-borne flaviviruses. An attenuated mutation with a P38G substitution in NS4B protein was recently made by utilizing site-directed mutagenesis of a WNV New York 1999 strain (NY99) infectious clone [15]. Full-genome sequencing of the NS4B-P38G mutant revealed two other compensatory mutations -- NS4B-T116I and NS3-N480H. Although the NS4B-P38G mutant showed enhanced vector competence in Culex tarsalis mosquitoes than the wild-type parental control strain [16], studies in animal models have indicated that the NS4B-P38G mutant has several features that may make it an important mutation for inclusion in live attenuated vaccine candidates. First, it is highly attenuated in mice compared to the wild-type WNV NY99. Second, it induces stronger innate and adaptive immune responses. Lastly, mice immunized with the NS4B-P38G mutant were all protected from subsequent lethal wild-type WNV infection [17]. We previously reported that myeloid differentiation factor 88- dependent innate signaling pathways contribute to a robust, cell-mediated immune response in mice [18]; and we postulated that WNV NS4B-P38G mutant could have a similar impact on host immunity in humans. In this study, we characterized the NS4B-P38G mutant infection and immunity in two human cell lines (THP-1 cells and THP-1 macrophages) as a first model to investigate the utility of the NS4BP38G mutant as a potential vaccine candidate in humans.
2. Materials and methods
2.1. Cell lines and WNV infection
Human monocytic leukemia cells (THP-1) were propagated in RPMI medium 1640 with l-glutamine and 25 mM HEPES buffer (Invitrogen, Carlsbad, CA) supplemented with 1 mM sodium pyruvate (Sigma, St. Louis, MO), and 10% fetal bovine serum (FBS, Sigma). In some experiments, THP-1 cells were treated with phorbol 12-myristate 13-acetate (PMA, Sigma) to differentiate into macrophage cells as described previously [19]. Two WNV strains were used: the parental strain WNV NY99 [10], which had been passaged once in Vero cells and twice in C6/36 cells. The WNV NS4B-P38G mutant was produced by utilizing site-directed mutagenesis and then passaged twice in Vero cells [15]. Supernatants and cells were collected for measurement of viral load and cytokine production.
2.2. Plaque assay
Vero cells were seeded in 6-well plates in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% FBS 24h before infection. Serial dilutions of culture supernatants were added and incubated for 1h. Subsequently, MEM containing 1% low-melting-point agarose were added, and the plates were incubated for 4 days. A second overlay of 4 ml 1% agarose-medium containing 0.055% neutral red (Sigma) was then added to visualize plaques. Virus concentrations were determined as PFU/ml.
2.3 Quantitative PCR (Q-PCR) for viral load and cytokine production
WNV-infected samples were re-suspended in Trizol (Invitrogen) for RNA extraction. Complementary (c)DNA was synthesized by using a qScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD). The sequences of the primer sets for WNV envelope (WNVE), toll-like receptor (TLR)s and retinoic acid-inducible gene 1(RIG-I) gene cDNA and Q-PCR reaction conditions were described previously [20-23]. The primer sequences for innate cytokines and A20 and IκBα genes were listed in Table 1. The assay was performed in a CFX96 real-time PCR system (Bio-Rad, Hercules, CA). Gene expression was calculated based on Ct values as described before [24].
Table 1.
Q-PCR primer sequence
| Gene |
Forward primer |
Reverse primer |
|---|---|---|
| IFNα | 5’CCTGATGAAGGAGGACTCCATT3’ | 5’AAAAAGGTGAGCTGGCATACG3’ |
| IFNβ | 5’GTCTCCTCCAAATTGCTCTC3’ | 5’ACAGGAGCTTCTGACACTGA3’ |
| IL-1β | 5’ACGAATCTCCGACCACCACT3’ | 5’CCATGGCCACAACAACTGAC3’ |
| IL-6 | 5’GCAACACCAGGAGCAGCC3’ | 5’AACTCCTTCTCCACAAGCGC3’ |
| IL-12 | 5’TGGAGTGCCAGGAGGACAGT3’ | 5’TCTTGGGTGGGTCAGGTTTG3’ |
| TNFα | 5’GCCCAGGCAGTCAGATCATCT3’ | 5’TTGAGGGTTTGCTACAACATGG3’ |
| A20 | 5’TCCTCAGGCTTTGTATTTGAGC3’ | 5’TGTGTATCGGTGCATGGTTTTA3’ |
| IκBα | 5’CTCCGAGACTTTCGAGGAAATAC3’ | 5’GCCATTGTAGTTGGTAGCCTTCA3’ |
| GAPDH | 5’CTCAAGATCATCAGCAATGCCT3’ | 5’AAGTTGTCATGGATGACCTTGG3’ |
2.4. Cytokine assays
Culture supernatant was collected for analysis of cytokine production using a Bio-Plex Pro Mouse Cytokine Assay (Bio-Rad).
2.5. PCR Arrays
A 32-gene PrimePCR assay (Biorad) was performed following the manufacturer's protocol. Briefly, RNA was purified from non-infected and WNV-infected cells using an RNeasy extraction kit (Qiagen). cDNA was synthesized using the iScript family reverse transcription kits and then loaded onto 96-well PCR array plates for amplification on the CFX96 real-time PCR system (Biorad). A list of 32 genes is provided in Supplementary Table 1. Data analysis was performed by using CFX manager software. Four genes, including GAPDH, were selected as reference genes. Data were presented as clustergram and scatterplot.
2.6. siRNA knockdown for retinoic acid-inducible gene (RIG)-I, Toll-like receptor (TLR)4 and TLR7
Cells were transfected with 75 nM TLR4 specific siRNA, or 50 nM TLR7 specific siRNA, or 75 nM RIG-I specific siRNA (Sigma) using Superfect (Qiagen) per the manufacturer's instructions. Scrambled siRNAs (Sigma) were used as a negative control. Transfected cells were grown in RPMI medium containing 10% FBS. Q-PCR analysis of the TLR4, TLR7 and RIG-I mRNA was used to confirm the effects of the siRNA knockdown. At 48 h post-treatment, cells were infected with WNV (multiplicity of infection (MOI) of 0.5).
2.7. Statistical analysis
Data were analyzed by using Prism software (Graph-Pad) statistical analysis. Values for viral burden and cytokine production experiments were presented as means ± SEM. The P values of these experiments were calculated with a non-paired Student's t test. Statistical significance was accepted at P < 0.05.
3. Results
3.1. WNV NS4B-P38G mutant produced higher levels of viral RNA, but not infectious virus in both THP-1 cells and THP-1 macrophages
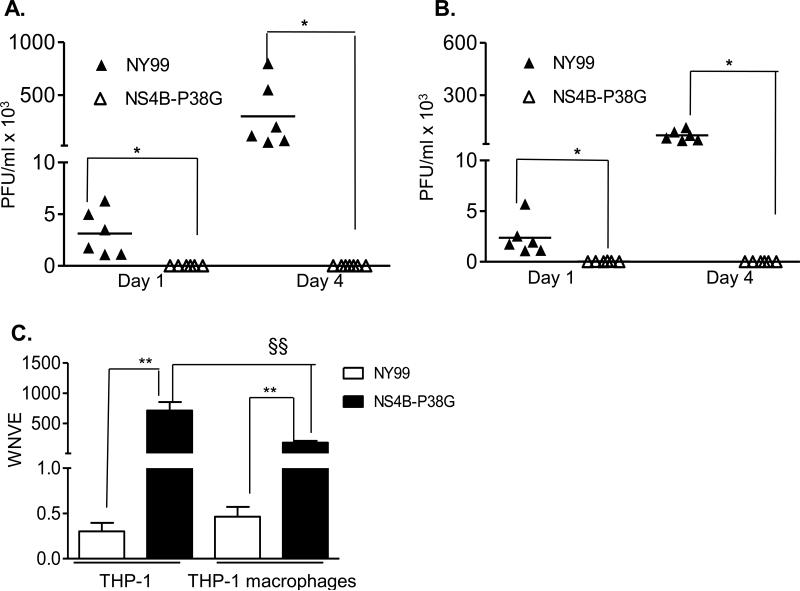
THP-1 is a well-characterized monocytic cell line and is known to be permissive to flavivirus infection [25-26]. Initial studies involved infection of THP-1 cells with the NS4BP38G mutant and its parent WNV NY99 strains at a MOI of 5. Multiplication kinetics was measured on days 1 and 4 post-infection by a plaque assay. As shown in Figure 1A, viral titers in supernatants of WNV NY99–infected THP-1 cells increased more than 100-fold from day 1 to day 4; whereas there was no evidence of detectable infectivity in the supernatant of THP-1 cells- infected with WNV NS4B-P38G mutant on days 1 and 4. The differentiation of THP-1 monocytes into macrophages can be achieved with the treatment of PMA [19]. Therefore, THP-1 cells were next stimulated with PMA and the experiment was repeated. WNV NY99 had a similar replication rate in THP-1 macrophages as THP-1 cells, and again no viral infectivity was detected by plaque assay on either day 1 or 4 post infection, following infection with the WNV NS4B-P38G mutant (Figure 1B). Significantly, although it failed to produce detectable infectious virus, NS4B-P38G mutant-infected THP-1 cells and THP-1 macrophages had 2373 and 395 fold higher viral RNA levels, respectively, than WNV NY99-infected cells at 24 h (Figure 1C). Overall, these results indicate that the NS4B-P38G mutant multiplies poorly, if at all, in THP-1 monocyte and macrophage cells.
Fig. 1.
WNV infection in THP-1 cells and THP-1 macrophages. THP-1 cells (A) and THP-1 macrophages (B) were infected with WNV NY99 and WNV NS4B-P38G mutant at a MOI of 5. A- B. The viral titers at indicated time points post-infection were measured by using a plaque assay. C. Viral titer at 24h post-infection was measured by a Q-PCR assay. Data are presented as means ± SEM, n = 4 -6. *P < 0.05 or **P < 0.01 for NY99 vs. NS4B-P38G. §§ P < 0.01 for THP-1 vs. THP-1 macrophages. Results presented are one representative of two similar experiments.
3.2. The WNV NS4B-P38G mutant induced higher innate cytokine responses than WNV NY99 in THP-1 cells and THP-1 macrophages
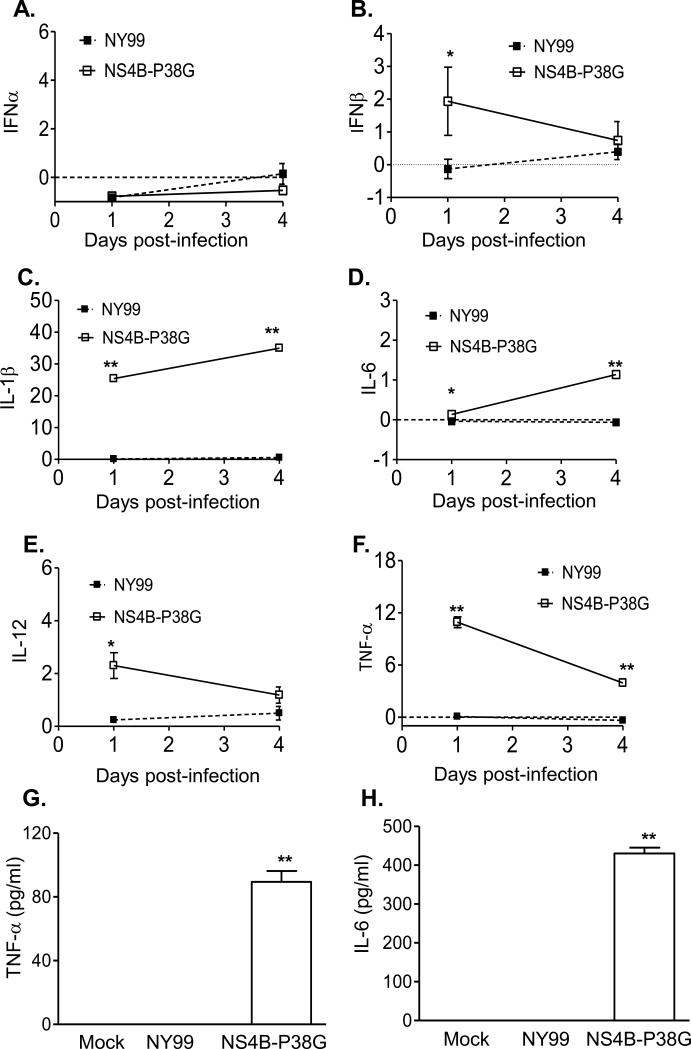
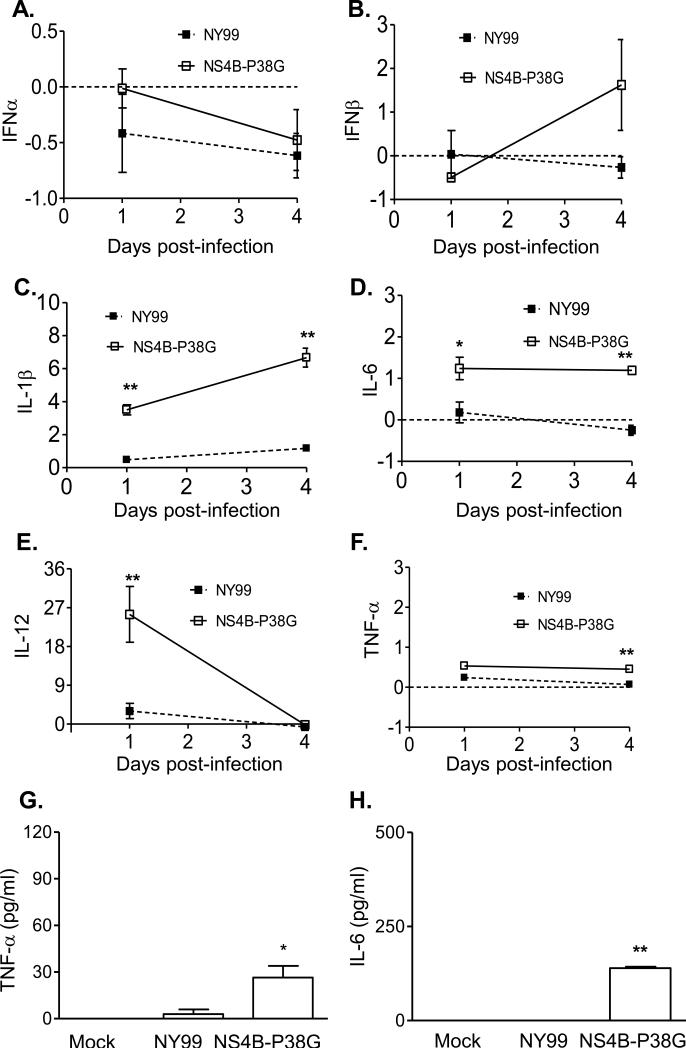
We have previously shown that the NS4B-P38G mutant induces stronger innate and adaptive immune responses in mice than did wild-type NY99 strain [17]. Here, we examined anti-viral innate cytokine RNA and protein production following WNV NY99 and NS4B-P38G mutant infection in THP-1 cells. Neither virus induced IFN-α expression (Figure 2A). Although WNV NY99 did not induce IFN-β expression, a 2-fold increase in IFN-β gene expression was observed on day 1 post- NS4B-P38G mutant infection (Figure 2B). Furthermore, there were significant increases in IL-1β, IL-6, IL-12 and TNF-αgenes (Figure 2C-2F) and TNF-α and IL-6 production (Figure 2G-2H) in THP-1 cells following infection with the WNV NS4B-P38G mutant; whereas WNV NY99 induced very low, if any, of these cytokines. In THP-1 macrophages, neither WNV NY99 nor NS4B-P38G mutant could enhance IFN-α and IFN-β gene expression (Figure 3A and 3B). Nevertheless, the NS4B-P38G mutant triggered the upregulation of IL-1β, IL-6, IL-12 and TNF-α gene expression and TNF-α and IL-6 production in THP-1 macrophages (Figure 3C-3H). Significantly, similar to results of the infection in THP-1 cells, WNV NY99 barely induced any gene expression, such as that described above. In comparison to THP-1 cells, induction of IL-1β and TNF-α gene levels in THP-1 macrophages following NS4B-P38G mutant infection was significantly reduced.
Fig. 2.
Innate cytokine gene expression in THP-1 cells following infection with WNV NS4B-P38G and WNV NY99. Cytokine levels at days 1 and 4 post-infection were determined by using Q-PCR (A- F) and Bioplex assays (G- H). The fold increase compared to that in the mock-infected group is shown. Data are presented as means ± SEM, n = 3. * P < 0.05 or **P < 0.01 for NY99 vs. NS4B-P38G. Results presented are one representative of three similar experiments.
Fig. 3.
Innate cytokine gene expression in THP-1 macrophages following infection with WNV NS4B-P38G and WNV NY99. Cytokine levels at days 1 and 4 post-infection were determined by using Q-PCR (A- F) and Bioplex assays (G- H). The fold increase compared to that in mock-infected group is shown. Data are presented as means ± SEM, n = 3. * P < 0.05 or **P < 0.01 for NY99 vs. NS4B-P38G. Results presented are one representative of three similar experiments.
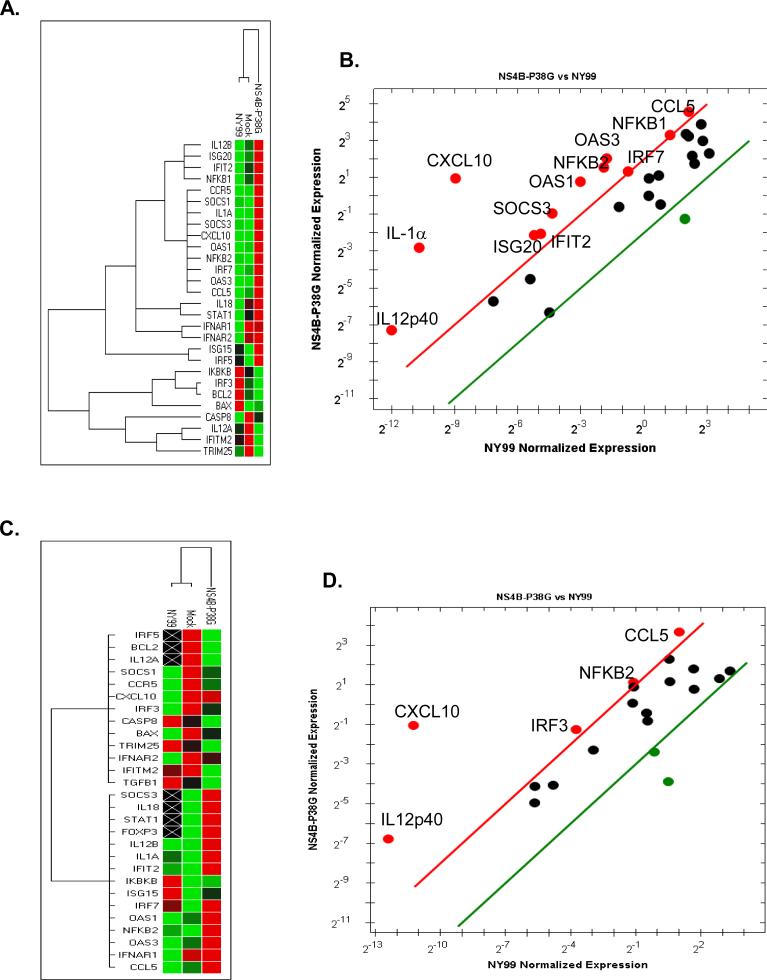
A 32-gene Q-PCR array was used to further identify the anti-viral gene profiles in these two cell types. Twenty genes were found to be upregulated following infection of THP-1 cells with the NS4B- P38G mutant (Figure 4A, marked in red color); whereas fewer genes were induced in both mock and WNV NY99-infected THP-1 cells (Figure 4A). Comparison of WNV NY99- to NS4B-P38G mutant-infected cells showed that 12 genes were upregulated more than 4-fold higher in WNV NS4B-P38G mutant-infected cells, which included genes encoding interferon-stimulated genes (ISGs), proinflammatory cytokines, and chemokines (Figure 4B). In THP-1 macrophages, there were 14 genes that had significant expression following WNV NS4B-P38G mutant infection, and 5 genes induced by wild-type WNV NY99 infection (Figure 4C, marked in red color). Among them, only 5 genes were found to have more than a 4-fold increase in WNV NS4B-P38G mutant-infected THP-1 macrophages compared to those induced by WNV NY99, including IL-12, CXCL10, CCL5, NFKB and IRF3 genes (Figure 4D). Taken together, these results suggest that WNV NS4B-P38G mutant infection induced a more diverse and robust innate anti-viral responses in THP-1 cells than in THP-1 macrophages, and was superior to WNV NY99 in both cell types.
Fig.4.
PCR array following WNV infection. THP-1 cells (A- B) and THP-1 macrophages (C-D) were infected with WNV NY99 and WNV NS4B-P38G mutant at a MOI of 5, harvested at 24h, and analyzed by using a PrimePCR array. A & C. The clustergram image depicts relative expression of a sample as follows: upregulation (higher expression) by a red square, downregulation (lower expression) by a green square, and no regulation by a black square. The lighter the shade of color the greater the relative expression difference. On the outer edges of the data plot is a dendrogram, which indicates the clustering hierarchy. B & D. The scatter plot shows the normalized expression of targets for NY99-infected versus NS4B-P38G infected samples. The plot image shows the following changes in target expression based on the threshold set of 4: upregulation as red circles, downregulation as green circles; and no change as black circles.
3.3. Pathogen recognition receptor (PRR) signaling pathways involved in differential innate immune responses in THP-1 cells and THP-1 macrophages
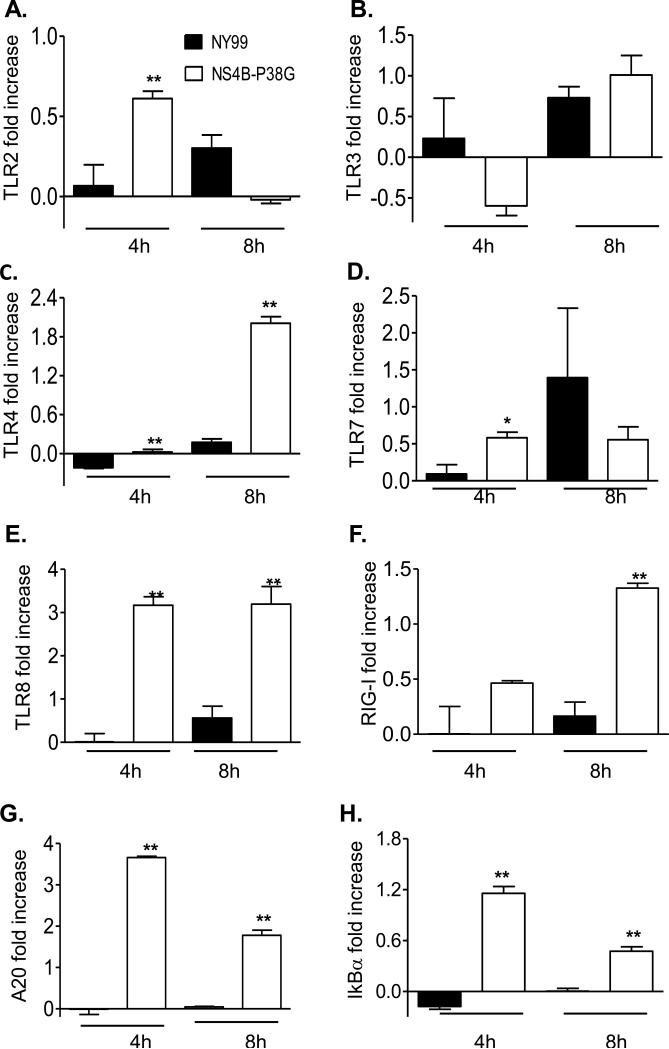
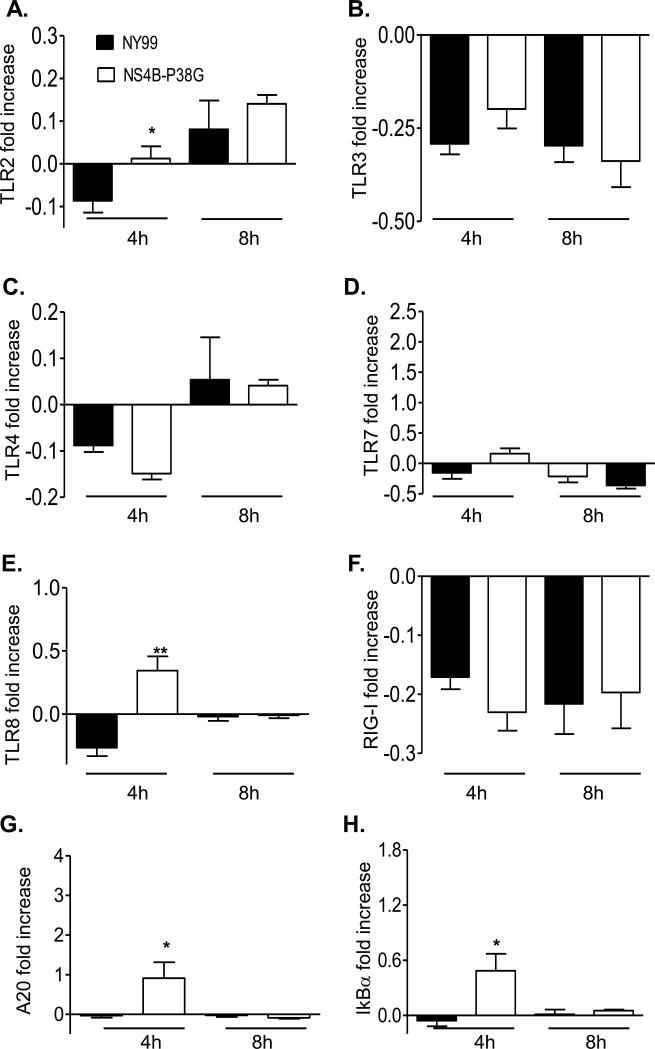
THP-1 cells are known to express various types of pathogen recognition receptors (PRRs), including TLRs and RIG-I-like receptors (RLRs) [27-29]. To understand the underlying mechanisms by which the NS4B-P38G mutant induced higher innate cytokine responses in these cells, we next examined PRR gene expression at 4 and 8 h post-infection by a Q-PCR assay. Although both viruses upregulated PRR expression in THP-1 cells, gene expression of 5 PRRs (TLR2, TLR4, TLR7, TLR8 and RIG-I) were much more augmented by the NS4B-P38G mutant than by WNV NY99 strain (Figure 5A, 5C-5F). TLR3 induction was similar in both virus-infected cells (Figure 5B). The induction of transcription factor NF-κB is important for expression of a variety of viral-induced IFNs and proinflammatory cytokine genes. There were higher levels of A20 and IκBα two regulators involved in NFkB-signaling pathways, in NS4BP38G infected cells (Figure 5G& 5H). In THP-1 macrophages, upregulation of TLR2 and TLR8 genes was significantly higher following NS4B-P38G mutant infection when compared with that from WNV NY99 strain (Figure 6A & 6E), though the magnitude was much lower compared to that of THP-1 cells. Neither TLR3 nor RIG-I was induced by the two viruses (Figure 6B & 6F) and there were no differences in TLR4 and TLR7 gene expression between the two virus–infected groups (Figure 6C & 6D). Furthermore, A20 and IκBα levels were enhanced in NS4B-P38G mutant –infected cells compared to that of the infected wild-type NY99 strain, but the magnitude was reduced, when compared with that in the THP-1 cells (Figure 6G & 6H). Thus, TLR7, RIG-I and TLR4 -mediated signaling pathways may contribute to a differential anti-viral response observed in THP-1 cells and THP-1 macrophages by the attenuated NS4B-P38G mutant.
Fig. 5.
PRR and NF-κB -related gene expression in WNV-infected THP-1 cells. THP-1 cells were infected with WNV NY99 and WNV NS4B-P38G mutant at a MOI of 0.5. TLRs 2, 3, 4, 7 and 8 (A- E), RIG-I (F), A20 (G), and IκBα (H) gene expression at 4 and 8 hrs post infection were determined by using a Q-PCR assay. The fold increase compared to that in the mock-infected group is shown. Data are presented as means ± SEM, n = 4. * P < 0.05 or **P < 0.01 for NY99 vs. NS4B-P38G. Results presented are one representative of two similar experiments.
Fig. 6.
PRR and NFkB-related gene expression in WNV-infected THP-1 macrophages. THP-1 macrophages were infected with WNV NY99 and WNV NS4B-P38G mutant at a MOI of 0.5. TLRs 2, 3, 4, 7 and 8 (A- E), RIG-I (F), A20 (G), and IκBα (H) gene expression at 4 and 8 hrs post-infection were determined by using a Q-PCR assay. The fold increase compared to the mock-infected group is shown. Data are presented as means ± SEM, n = 4. * P < 0.05 or **P < 0.01 for NY99 vs. NS4B-P38G. Results presented are one representative of two similar experiments.
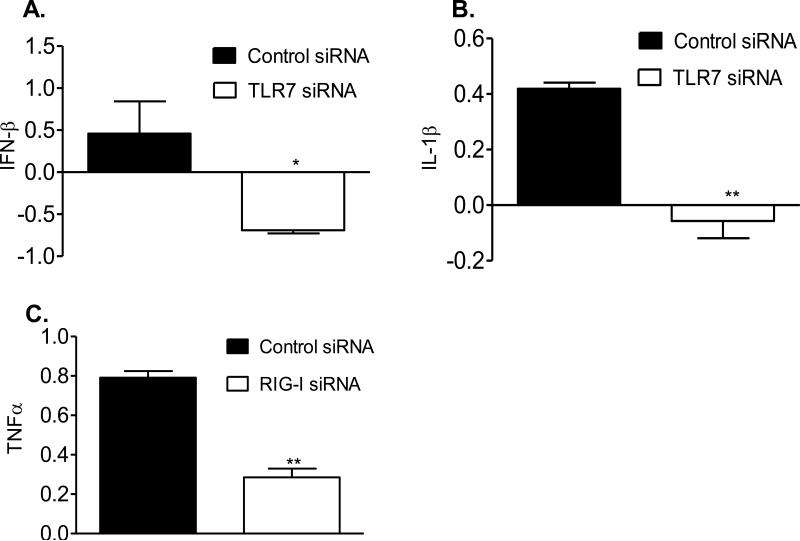
siRNA knockdown was used to investigate the TLR7, RIG-I and TLR4 -mediated signaling pathways in more detail. Knockdown of TLR7 signaling abolished expression of IFN-β and IL-1β in WNV NS4B-P38G-infected THP-1 cells (Figure 7A & 7B) and knockdown of RIG-I signaling also reduced expression of TNF-α in NS4B-P38G-infected THP-1 cells (Figure 7C). However, knockdown of TLR4 signaling did not affect the expression of the above genes (data not shown). Thus, the NS4B-P38G mutant triggered more robust expression of IFN and proinflammatory cytokine genes than WNV NY99 via TLR7 and RIG-I- mediated innate signaling pathways.
Fig. 7.
TLR7 and RIG-I are responsible for higher IFN and proinflammatory cytokine production in WNV NS4B-P38G –infected THP-1 cells. Cells were transfected with siRNAs to TLR7 (A-B, siTLR7), or siRNA to RIG-I (C, siRIG-I) as described in Materials & Methods. Cells treated with the same concentration of scrambled siRNA were used as controls. Cells were harvested from mock and WNV NS4B-P38G-infected cells at day 1 post infection. The fold increase compared to that of the mock-infected group was shown. Data are presented as means ± SEM, n = 4. * P < 0.05 or **P < 0.01 compared to control vector-treated cells.
4. Discussion
Development of live attenuated vaccine candidates is based on a combination of attenuation and immunogenicity. The WNV NS4B-P38G mutant has an excellent balance of attenuation and immunogenicity. Consequently, understanding the mechanisms of these phenotypes will aid live attenuated virus vaccine development as a whole. Studies to date with the NS4B-P38G have focused on the mouse system [17-18]. In this study, we have characterized the infection and innate cytokine responses of the attenuated WNV mutant in two human cell lines. There were three main conclusions from the work. First, the NS4B-P38G mutant produced viral RNA, but not infectious virus in both cell types; second, the attenuated mutant triggered a higher innate cytokine expression than the virulent WNV NY99 in both cells; and lastly, the mutant-induced anti-viral response was more diverse and robust in THP-1 cells than in THP-1 macrophages.
Although THP-1 cells are permissive to wild-type WNV infection as described previously [25, 30], we noted that the WNV NS4B-P38G mutant failed to produce detectable infectious virus in either THP-1cells or THP-1 macrophages. Previous studies have shown that the WNV NS4B is a major contributor to interferon antagonism [31-33]; while the studies here suggest that NS4B has a larger role in control of the host innate immune response. However, we cannot exclude that the NS3 helicase is contributing to the phenotype since the NS4B-P38G mutant was not viable as a single mutation. Directed mutagenesis of NS4B-P38G in a WT NY99 infectious clone resulted in two other compensatory mutations (NS4B-T116I and NS3-N480H), which either alone or in combination did not result in an attenuated phenotype [15], implying that NS4B-P38G is the critical residue.
The flavivirus nonstructural proteins, together with uncharacterized cell proteins constitute the replication complex [34]. There is only rudimentary understanding of this complex. Recent studies indicate the NS4B protein of flaviviruses, contribute to viral replication either directly or indirectly via interactions with other NS proteins [35-37]. For example, the 2k signal peptide of WNV NS4B protein was reported to induce endoplasmic reticulum-derived, replication-competent membrane structures [37-40]. NS4B protein could interact physically with other NS proteins, such as NS1, which conveys signals to the cytoplasm and regulates early viral RNA replication [41]. Furthermore, a P101L mutation in DENV4 NS4B confers a smaller plaque size in mosquito cells and a larger plaque size in mammalian cells, and this is associated with NS4B interaction with the NS3 protease [36, 42]. In the present studies, there was accumulating viral RNA without release of infectious virions from the NS4B-P38G mutant-infected THP-1cells and THP-1 macrophages indicating that mutation in NS4B results in a defective viral life cycle presumably due to alterations in the interactions of the viral proteins in the complex. Future studies will be focused on the role of NS4B-P38G alone or together with NS4B-T116I and NS3-N480 in late stage of viral life cycle.
The mechanisms by which NS4B-P38G mutant induced higher innate cytokine responses than WNV NY99 in THP-1 cells and THP-1 macrophages are still under investigation. Both THP-1 cells and THP-1 macrophages infected by the NS4B-P38G mutant accumulated higher levels of viral RNA than did cells infected by WNV NY99, which could serve as pathogen-associated molecular patterns (PAMP)s to activate potent TLR or RLR –mediated anti- viral responses. THP-1 cells express many types of PRRs [27-29]. Compared to WNV NY99 infection, NS4B-P38G mutant induced a higher expression of TLRs 2, 4, 7, 8 and RIG-I on THP-1 cells and TLR2, and TLR8 on THP-1 macrophages, respectively. Thus, excessive viral products due to a defective life cycle may lead to stronger innate immune responses in these cells. The NS4B protein of flaviviruses is also associated with evasion of host immune responses [14, 31-33]. The N-terminal domain of NS4B protein (amino acids 35- 60) is highly conserved among flaviviruses and is centered around a tyrosine residue. This domain bears a resemblance to an immunomodulatory tyrosine inhibitory motif found in various components of mammalian cell-signaling cascades [43], which is thought to contribute to its antagonist activities for IFN and inflammatory cytokine signaling [14, 32-33]. Thus, it is also likely that the P38G mutation of NS4B protein abolishes or partially reduces its ability to antagonize innate cytokine responses during WNV infection.
The NS4B-P38G mutant induced a more diverse and robust anti-viral response in THP-1 cells, when compared to the response in THP-1 macrophages. For example, many ISGs were upregulated in NS4B-P38G mutant-infected THP-1 cells but not in THP-1 macrophages. There were also stronger proinflammatory cytokine responses in THP-1 cells. One possible explanation is that viral RNA levels in NS4B-P38G mutant- infected THP-1 cells were about 3- fold higher than those of THP-1 macrophages (Figure 1), which could induce more robust PRR – mediated anti-viral responses. Accordingly, the diversity and magnitude of upregulation of PRRs and NFkB transcription factor genes were reduced in THP-1 macrophages compared to that of THP-1 cells. Furthermore, SiRNA knockdown studies suggested to us that in THP-1 cells, TLR7 is involved in higher IFN-β expression; whereas TLR7 and RIG-I both contribute to proinflammatory cytokine induction.
WNV-induced acute viral encephalitis and neurological sequelae have been significant public health concern in the US during the past decade. No human vaccines are yet approved for use against WNV infection. Therefore, the development of effective human WNV vaccine remains a high priority. The susceptibility of human monocytes-macrophages to productive infection in vitro is compatible with a potential role in initial WNV replication and propagation after transmission by transfusion [44]. Therefore, study of infection and immunity to a potential vaccine candidate in human monocytic and macrophage cells should provide critical insights for vaccine efficacy in humans.
Supplementary Material
Highlights.
▶ NS4B-P38G mutant produced viral RNA, but not infectious virus in both cell types.
▶ NS4B-P38G mutant triggered a higher innate cytokine response in both cell types.
▶ NS4B-P38G mutant triggered diverse and robust anti-viral responses in THP-1 cells.
Acknowledgements
This work was supported in part by NIH grant R01AI072060 (T.W.) and R01 AI099123 (T.W.). G Xie was supported by a fellowship from the Sealy Center for Vaccine Development at UTMB. We thank Ms. Mardelle Susman for assisting in manuscript preparation.
Abbreviations used in this paper
- CNS
central nervous system
- DMEM
Dulbecco's modified eagle's medium
- E
envelope
- FBS
fetal bovine serum
- IFN
interferon
- IL
interleukin
- MOI
multiplicity of infection
- NFκB
nuclear factor kappa B
- PMA
phorbol 12-myristate 13-acetate
- Q-PCR
quantitative PCR
- RIG-I
retinoic acid inducible gene 1
- TLR
toll-like receptor
- TNF-α
tumor necrosis factor α
- WNV
West Nile virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petersen LR, Brault AC, Nasci RS. West Nile virus: review of the literature. JAMA. 2013 Jul 17;310(3):308–15. doi: 10.1001/jama.2013.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis. 2007 Oct 15;45(8):1039–46. doi: 10.1086/521911. [DOI] [PubMed] [Google Scholar]
- 3.Kramer LD, Styer LM, Ebel GD. A global perspective on the epidemiology of West Nile virus. Annu Rev Entomol. 2008;53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- 4.Carson PJ, Konewko P, Wold KS, Mariani P, Goli S, Bergloff P, et al. Long-term clinical and neuropsychological outcomes of West Nile virus infection. Clin Infect Dis. 2006 Sep 15;43(6):723–30. doi: 10.1086/506939. [DOI] [PubMed] [Google Scholar]
- 5.Ravindra KV, Freifeld AG, Kalil AC, Mercer DF, Grant WJ, Botha JF, et al. West Nile virus-associated encephalitis in recipients of renal and pancreas transplants: case series and literature review. Clin Infect Dis. 2004 May 1;38(9):1257–60. doi: 10.1086/383325. [DOI] [PubMed] [Google Scholar]
- 6.Ou AC, Ratard RC. One-year sequelae in patients with West Nile Virus encephalitis and meningitis in Louisiana. J La State Med Soc. 2005 Jan-Feb;157(1):42–6. [PubMed] [Google Scholar]
- 7.Cook RL, Xu X, Yablonsky EJ, Sakata N, Tripp JH, Hess R, et al. Demographic and clinical factors associated with persistent symptoms after West Nile virus infection. Am J Trop Med Hyg. 2010 Nov;83(5):1133–6. doi: 10.4269/ajtmh.2010.09-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadek JR, Pergam SA, Harrington JA, Echevarria LA, Davis LE, Goade D, et al. Persistent neuropsychological impairment associated with West Nile virus infection. J Clin Exp Neuropsychol. 2010 Jan;32(1):81–7. doi: 10.1080/13803390902881918. [DOI] [PubMed] [Google Scholar]
- 9.Anderson JF, Andreadis TG, Vossbrinck CR, Tirrell S, Wakem EM, French RA, et al. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science. 1999;286(5448):2331–3. doi: 10.1126/science.286.5448.2331. [DOI] [PubMed] [Google Scholar]
- 10.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286(5448):2333–7. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 11.Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008 Sep;82(17):8262–71. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholle F, Mason PW. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology. 2005 Nov 10;342(1):77–87. doi: 10.1016/j.virol.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Diamond MS. Mechanisms of Evasion of the Type I Interferon Antiviral Response by Flaviviruses. J Interferon Cytokine Res. 2009 Aug 20; doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- 14.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wicker JA, Whiteman MC, Beasley DW, Davis CT, McGee CE, Lee JC, et al. Mutational analysis of the West Nile virus NS4B protein. Virology. 2012 Apr 25;426(1):22–33. doi: 10.1016/j.virol.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Slyke GA, Jia Y, Whiteman MC, Wicker JA, Barrett AD, Kramer LD. Vertebrate attenuated West Nile virus mutants have differing effects on vector competence in Culex tarsalis mosquitoes. J Gen Virol. 2013 May;94(Pt 5):1069–72. doi: 10.1099/vir.0.049833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welte T, Xie G, Wicker JA, Whiteman MC, Li L, Rachamallu A, et al. Immune responses to an attenuated West Nile virus NS4B-P38G mutant strain. Vaccine. 2011 Jun 24;29(29-30):4853–61. doi: 10.1016/j.vaccine.2011.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie G, Welte T, Wang J, Whiteman MC, Wicker JA, Saxena V, et al. A West Nile virus NS4B-P38G mutant strain induces adaptive immunity via TLR7-MyD88-dependent and independent signaling pathways. Vaccine. 2013 Aug 28;31(38):4143–51. doi: 10.1016/j.vaccine.2013.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007 Jan;56(1):45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 20.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, et al. Rapid detection of West Nile virus from human clinical specimens, field- collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol. 2000;38(11):4066–71. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colmont CS, Raby AC, Dioszeghy V, Lebouder E, Foster TL, Jones SA, et al. Human peritoneal mesothelial cells respond to bacterial ligands through a specific subset of Toll-like receptors. Nephrol Dial Transplant. 2011 Dec;26(12):4079–90. doi: 10.1093/ndt/gfr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen N, Fu Q, Deng Y, Qian X, Zhao J, Kaufman KM, et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15838–43. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang NN, Shen SH, Jiang LJ, Zhang W, Zhang HX, Sun YP, et al. RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proc Natl Acad Sci U S A. 2008 Jul 29;105(30):10553–8. doi: 10.1073/pnas.0804895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welte T, Aronson J, Gong B, Rachamallu A, Mendell N, Tesh R, et al. Vgamma4+ T cells regulate host immune response to West Nile virus infection. FEMS Immunol Med Microbiol. 2011 Nov;63(2):183–92. doi: 10.1111/j.1574-695X.2011.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mather T, Takeda T, Tassello J, Ohagen A, Serebryanik D, Kramer E, et al. West Nile virus in blood: stability, distribution, and susceptibility to PEN110 inactivation. Transfusion. 2003 Aug;43(8):1029–37. doi: 10.1046/j.1537-2995.2003.00464.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsai YT, Chang SY, Lee CN, Kao CL. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol. 2009 Apr;11(4):604–15. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 27.Indoh T, Yokota S, Okabayashi T, Yokosawa N, Fujii N. Suppression of NF-kappaB and AP-1 activation in monocytic cells persistently infected with measles virus. Virology. 2007 May 10;361(2):294–303. doi: 10.1016/j.virol.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Uehara A, Iwashiro A, Sato T, Yokota S, Takada H. Antibodies to proteinase 3 prime human monocytic cells via protease-activated receptor-2 and NF-kappaB for Toll-like receptor- and NOD-dependent activation. Mol Immunol. 2007 Jul;44(14):3552–62. doi: 10.1016/j.molimm.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Verma R, Jung JH, Kim JY. 1,25-Dihydroxyvitamin D3 up-regulates TLR10 while down-regulating TLR2, 4, and 5 in human monocyte THP-1. J Steroid Biochem Mol Biol. 2014 May;141:1–6. doi: 10.1016/j.jsbmb.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, et al. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003 Jul;4(7):723–8. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, et al. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005 Jul;79(13):8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005 Feb;79(3):1934–42. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans JD, Seeger C. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J Virol. 2007 Nov;81(21):11809–16. doi: 10.1128/JVI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi PY. Structural biology. Unraveling a flavivirus enigma. Science. 2014 Feb 21;343(6173):849–50. doi: 10.1126/science.1251249. [DOI] [PubMed] [Google Scholar]
- 35.Davis CT, Beasley DW, Guzman H, Siirin M, Parsons RE, Tesh RB, et al. Emergence of attenuated West Nile virus variants in Texas, 2003. Virology. 2004 Dec 5;330(1):342–50. doi: 10.1016/j.virol.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Hanley KA, Manlucu LR, Gilmore LE, Blaney JE, Jr., Hanson CT, Murphy BR, et al. A trade-off in replication in mosquito versus mammalian systems conferred by a point mutation in the NS4B protein of dengue virus type 4. Virology. 2003 Jul 20;312(1):222–32. doi: 10.1016/s0042-6822(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 37.Puig-Basagoiti F, Tilgner M, Bennett CJ, Zhou Y, Munoz-Jordan JL, Garcia-Sastre A, et al. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology. 2007 Apr 25;361(1):229–41. doi: 10.1016/j.virol.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou J, Xie X, Lee le T, Chandrasekaran R, Reynaud A, Yap L, et al. Dimerization of flavivirus NS4B protein. J Virol. 2014 Mar;88(6):3379–91. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant D, Tan GK, Qing M, Ng JK, Yip A, Zou G, et al. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J Virol. 2011 Aug;85(15):7775–87. doi: 10.1128/JVI.00665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufusi PH, Kelley JF, Yanagihara R, Nerurkar VR. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One. 2014;9(1):e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, et al. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol. 2012 Jul;86(13):7360–71. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006 Sep;87(Pt 9):2605–14. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 43.Isakov N. ITIMs and ITAMs. The Yin and Yang of antigen and Fc receptor-linked signaling machinery. Immunol Res. 1997 Feb;16(1):85–100. doi: 10.1007/BF02786325. [DOI] [PubMed] [Google Scholar]
- 44.Rios M, Zhang MJ, Grinev A, Srinivasan K, Daniel S, Wood O, et al. Monocytesmacrophages are a potential target in human infection with West Nile virus through blood transfusion. Transfusion. 2006 Apr;46(4):659–67. doi: 10.1111/j.1537-2995.2006.00769.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.