Abstract
Quantitative analysis of computed tomography (CT) is essential to the study of acute lung injury. However, quantitative CT is made difficult by poor lung aeration, which complicates the critical step of image segmentation. To overcome this obstacle, this study sought to develop and validate a semiautomated, multilandmark, registration-based scheme for lung segmentation that is effective in conditions of poor aeration. Expiratory and inspiratory CT images were obtained in rats (n = 8) with surfactant depletion of incremental severity to mimic worsening aeration. Trained operators manually delineated the images to provide a comparative landmark. Semiautomatic segmentation originated from a single, previously segmented reference image obtained at healthy baseline. Deformable registration of the target images (after surfactant depletion) was performed using the symmetric diffeomorphic transformation model with B-spline regularization. Registration used multiple landmarks (i.e., rib cage, spine, and lung parenchyma) to minimize the effect of poor aeration. Then target images were automatically segmented by applying the calculated transformation function to the reference image contour. Semiautomatically and manually segmented contours proved to be highly similar in all aeration conditions, including those characterized by more severe surfactant depletion and expiration. The Dice similarity coefficient was over 0.9 in most conditions, confirming high agreement, irrespective of poor aeration. Furthermore, CT density-based measurements of gas volume, tissue mass, and lung aeration distribution were minimally affected by the method of segmentation. Moving forward, multilandmark registration has the potential to streamline quantitative CT analysis by enabling semiautomatic image segmentation of lungs with a broad range of injury severity.
Keywords: semiautomatic segmentation, image registration, ARDS, computed tomography, image analysis
quantitative analysis of computed tomography images (qCT) is a key method for the examination of acute respiratory distress syndrome (ARDS) (15). qCT can quantify several important aspects of ARDS pathophysiology, including lung injury severity, as indicated by lung mass (i.e., edema) (22) or gas content (11), degree of atelectasis and lung strain (deformation) (9, 13), and the regional heterogeneity of lung mechanics (10, 14). These capabilities are highly valuable in both research and clinical settings, where qCT can be used to trace the progression of lung injury, detect regional responses of lung mechanics to respiratory maneuvers (18) [i.e., recruitment, positive end-expiratory pressure (PEEP), and tidal volume], and independently predict ARDS mortality (10, 14).
The impressive findings of recent qCT studies have increased the interest in clinically practical methods of image processing. A prerequisite for any form of qCT lung analysis is the separation of pulmonary from nonpulmonary tissue. Lung masks must be precisely segmented so that they contain only lung parenchyma. However, this task is particularly difficult in ARDS. Automatic threshold-based segmentation (16), using the high contrast between lung gas and surrounding tissue, performs poorly in ARDS due to its exclusion of high-density (injured) tissue from the image; only gas contents can be measured using this method. Advanced automatic and semiautomatic segmentation techniques are available (31) that use intrathoracic and extrathoracic landmark structures to detect the lung boundary (23, 28). However, lung hyperdensities often mask these landmarks when injury is severe, making corrections by skilled human operators necessary.
As a consequence of the shortcomings of automatic techniques, manual segmentation (i.e., the manual delineation of lung borders from surrounding structures) remains integral to most clinical and laboratory work using qCT in ARDS. This method is time consuming and cumbersome, limiting the use of qCT to laboratories and research centers. Interpolation techniques have been developed to streamline manual segmentation (21, 24). In this approach, a reduced sample of slices is segmented manually and followed by interpolation of missing information in the interposed slices. Unfortunately, this modified technique still requires heavy human involvement and is intrinsically prone to missing detail when injury is heterogeneous.
Registration-based segmentation provides a potential means of making qCT processing more rapid while retaining image accuracy. In registration, all studied images are matched with a reference image (IR), which can be either a previous acquisition from the same subject, or a probabilistic atlas generated from a broad population of subjects (19, 20, 32). The registration seeks to generate a transform function that maximizes the similarity between the target images (IT) and IR. If the IR is segmented, the lung boundaries can be tracked in all other images by utilizing the transform function acquired from the registration. This methodology streamlines the processing of large sets of images and can be particularly useful when serial imaging is used to track disease progression or when multiple respiratory maneuvers are tested.
Here, we describe a semiautomated scheme for lung segmentation originating from healthy baseline images. All images were obtained in a surfactant depletion rat model of incremental severity to mimic worsening ARDS; while this model may not reproduce the inflammatory characteristics of ARDS in the short term, it mimics its radiological features by producing atelectasis and edema. To assess the usefulness of our approach, we tested two hypotheses: 1) that there is a high similarity between semiautomatically and manually segmented images; and 2) that the values of standard qCT measurements of lung injury are not altered by the proposed segmentation method. We have validated our registration-based method using manually segmented images and report that both hypotheses have been confirmed.
METHODS
Animal Preparation
The experiments were performed on male Sprague-Dawley rats (n = 8, 320 ± 30 g) following the protocol approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Anesthesia was induced with 50 mg/kg intraperitoneal pentobarbital and maintained with one-third of the initial dosage injected every 60 min. Animals were transorally intubated with a 14-gauge, 2-in. catheter with sealant placed at the level of the vocal cords (DAP Products, Baltimore, MD) to prevent gas leakage. The rats were then connected to a custom-built small-animal ventilator, set with tidal volume of 10 ml/kg, respiratory rate of 60 breaths/min, and inspiratory time fraction of 0.3. PEEP of 5 cmH2O and inspired fraction of O2 of 1.0 were maintained throughout the experiment to maintain acceptable oxygenation. The intubated, ventilated animals were placed supine in the CT scanner for the duration of the experiment and removed only to undergo pulmonary saline lavage (SL) procedures. Airway pressure was measured with a fiber-optic sensor (Samba Sensors, Vastra Frolunda, Sweden). Animals were paralyzed to provide immobility during imaging by injecting 1 mg/kg intravenous pancuronium bromide through a 24-gauge tail vein catheter. Heart rate and peripheral oxygen saturation levels were monitored by a veterinary pulse oximeter (Nonin Medical, Plymouth, MN). Body temperature was maintained at 37°C by a circulating warm water pad placed underneath the animal. After the last set of image acquisitions, animals were euthanized by lethal intravenous pentobarbital injection.
Surfactant Depletion
Baseline CT imaging was performed after the animal underwent a recruitment maneuver to eliminate atelectasis, which was performed by temporarily increasing PEEP to 9 cmH2O (7). Then rats underwent three SL procedures to induce surfactant depletion of increasing severity, each followed by image acquisition. For each SL, 10 ml/kg warmed (to body temperature) normal saline solution were instilled in the rat's lungs through the endotracheal tube and recovered by gentle suction using a 15-ml syringe. The animals were reconnected to the ventilator and repositioned in the scanner. Imaging was performed after a recruitment maneuver and a 3-min period of stabilization. SL, recruitment, and imaging were repeated in this sequence two additional times. The ranges of observed peripheral oxygen saturation were 90–95% after one SL, 83–88% after two SL, and 72–78% after three SL instillations. Heart rate ranged between 300 and 380 beats/min throughout the experiment. The weight of saline solution recovered after each SL was measured and used to estimate residual fluid left in the lung by subtracting it from the administered amount (1.20 ± 0.31, 0.16 ± 0.23, and 0.20 ± 0.37 g following the first, second, and third SL procedures, respectively).
CT Imaging
High-resolution CT scans were acquired using an eXplore CT120 scanner (Gamma Medica, Northridge, CA). To avoid blurring due to respiratory motion, respiratory-gated images were acquired at end-inspiration (EI) and end-expiration (EE) during 500-ms breath holds. Two hundred and twenty views over 180° were acquired for each EI and EE image. Only a single view per breath hold was acquired, and the total imaging time required to acquire the combination of EI and EE images was ∼11 min. The image resolution adopted for this study was 50 μm with 100 kV, 50 mA. All of the images were reconstructed to a 200-μm isotropic resolution to decrease the number of slices and the time required for manual segmentation of the lungs.
Three-Dimensional Lung Segmentation
Manual segmentation.
Manual segmentation of whole lung three-dimensional masks was performed by trained operators using Microview version 2.3 software. All available 200-μm-thick coronal slices were sequentially analyzed (from ventral to dorsal); 100–150 slices were thus segmented in each animal, which required about 1.5 h per three-dimensional image. In each slice, operators manually outlined the lung borders, excluding the heart, the central airways, and the major blood vessels, but including all lung tissue, irrespective of its density. Specifically, the operators set image contrast to a fixed intensity window [2,000 Hounsfield units (HU)] and level (−1,000). After crude manual outlining, the software shrank this boundary to the closest density gap. The operators then refined the new boundary to correct for errors due to poor aeration. All of the operators received verbal and written instructions and practiced on a set of images obtained during previous experiments in healthy and surfactant-depleted animals.
Semiautomatic segmentation.
Using a straightforward segmentation-by-registration approach as discussed in Ref. 25, the CT scan of the baseline healthy subject served as the IR, and the IT was scanned after each subsequent SL procedure. In addition, a binary lung mask was obtained from the IR (MR); this can be done either by manual segmentation, as was used in this study, or using existing segmentation algorithms (16). The image registration approach computes an image transformation Φ to match IR with IT. This transformation Φ is then applied to the MR to obtain the segmentation of the lungs in the IT: MT = MR (Φ), where MT is binary lung mask obtained from the IT. However, lung injury causes variable degrees of regional hyperdensity and of lung deformation that render such direct registration unreliable. For this reason, we strengthened our approach by creating a multilandmark registration method that integrates a deformable registration technique with the use of bone masks as cues to predict the location of the lungs. The entire pipeline of image analysis is shown in Fig. 1.
Fig. 1.
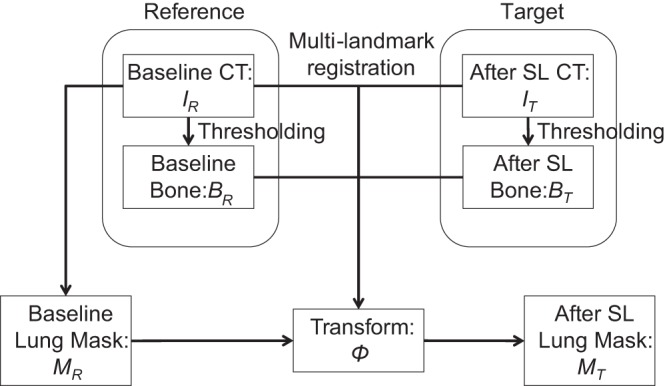
The pipeline of automated segmentation. The process of lung segmentation is shown, starting with thresholding the computed tomography (CT) images to obtain the bone mask. A multilandmark registration was then computed using both the pair of CT images and the pair of bone masks (after distance transform for the deformable registration). The resulting lung mask after saline lavage (SL) was computed by applying the transform to the baseline lung mask. IR, reference image; IT, target image; BR, bone mask for the reference IR; BT, bone mask for the target SL image IT; MR, binary lung mask obtained from the IR; MT, binary lung mask obtained from the IT.
The ribs and spine were automatically segmented by thresholding the CT images for gray-scale values above +100 HU. The bone mask for the reference IR is denoted as BR, and for the target SL image IT, it is denoted as BT. We computed the distance transform of BR and BT to derive D(BR) and D(BT).
To account for the effects of lung deformation, we executed two steps. In the first step, a rigid transform was computed by registering the binary bone masks, BR and BT, using mutual information as the similarity function. The second step computed a refined deformable transformation that added the distance transform of BR and BT, D(BR) and D(BT), which were integrated with BR and BT. We performed this deformable registration step using the symmetric diffeomorphic transformation model (2, 27) with third-order B-spline regularization (29). Mean squared difference was used as the similarity metric between D(BR) and D(BT). Normalized cross correlation was used as the similarity metric between IR and IT. Implementation details of parameters can be found in the Supplemental Data. (The online version of this article contains supplemental data.) The deformation was initialized as the rigid transform from the first step. The final result transform Φ matched both the pair of IR and IT and the pair of D(BR) and D(BT). The multilandmark registration was implemented using the open-source ANTS package (version 1.9.x) (3) with the newly refactored Insight Toolkit (version 4) (4).
All images obtained after SL were registered to their corresponding IR. The semiautomatic segmentation, MT, of the post-SL image IT was subsequently obtained by applying the transform Φ to the lung segmentation mask MR of the baseline image as MT = MR (Φ).
Quantitative Image Analysis
Quantitative CT density analysis was performed using standard methodology on each image obtained through manual and semiautomatic segmentation. Key parameters of gas volume, aeration distribution, and tissue content were calculated following standard methodology, using custom Matlab code (8). Lung gas volume in each region of interest (ROI) (Gas volumeROI) was estimated at EI and EE using the following equation (6):
The total weight of the lungs was then obtained using the equation:
where the term Total volumeROI included both gas and tissue volumes, and Mean CTROI denoted the mean of the lung density in HU within each ROI. The lung tissue density was considered as 1 g/ml in the equation.
Whole lung ROIs were partitioned into tissue compartments with increasing aeration, as previously described. These were defined based on density ranges: 1) nonaerated (−100 to +100 HU); 2) poorly aerated (−500 to −101 HU); 3) normally aerated (−900 to −501 HU); and 4) hyperinflated (−1,000 to −901 HU). The size of each compartment was measured as the percentage of lung volume it occupies.
Statistical Analysis
All data are shown as means ± SD. To compare the results of semiautomatic and manual segmentation, two approaches were chosen.
1) Overlap between manually and semiautomatically segmented images was quantified using the Dice similarity coefficient (DSC) (12), which was calculated according to:
where X and Y represent the two segmentation masks. A DSC with value “1” corresponds to the exact overlap of two masks.
2) Agreement between the results of quantitative density analysis obtained from manually and semiautomatically segmented images was tested using Bland-Altman plots (5) and by calculating the 95% limits of agreement (LOA).
The effects of repeated SL and inspiration on lung volumes and weights were tested using ANOVA for repeated measures.
RESULTS
Similarity Between Segmentation Techniques
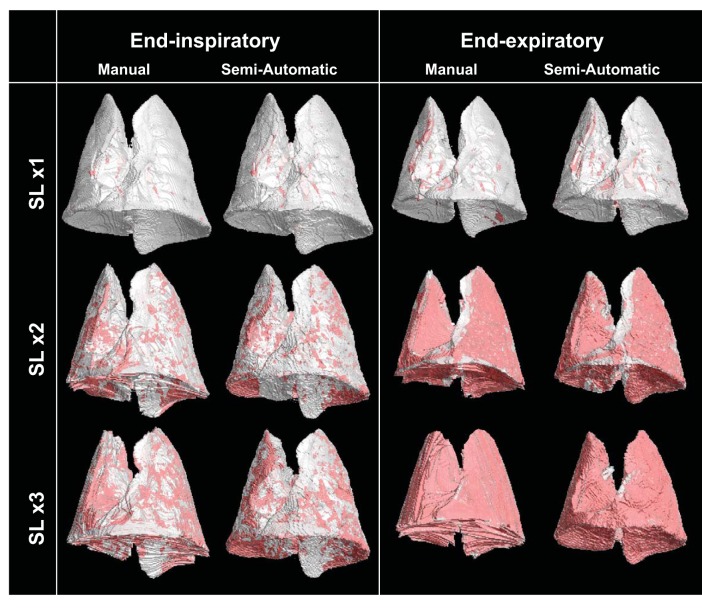
Three-dimensional lung masks obtained by manual and semiautomatic segmentation are displayed for the same representative rat after three SLs at EI and at EE (Fig. 2). These three-dimensional rendering images show the outer lining of the lungs and, being partly transparent, allow viewing of high-density tissue (in red) inside the lung. The images showed adequate overlap of lung borders and similar masks of the lungs outlined by the two methodologies. The DSC confirmed high agreement between semiautomatic and manual segmentation (Table 1). Similarity was more than adequate in all conditions and was marginally affected by the increasing severity of surfactant depletion (i.e., the number of SLs) and by imaging at EI vs. EE: we observed 7.6% difference between the best (after one SL at EI) and the worst (after three SLs at EE) imaging conditions. To test interobserver and intraobserver agreement for manual segmentation (the ground truth), all images from a randomly chosen animal were segmented once by two separate operators and twice by the same operator. DSC was higher than 0.965 for the same operator and higher than 0.931 for the two separate operators.
Fig. 2.
Representative three-dimensional rendering of lung images obtained with manual and automatic segmentations after each SL repetition at end inspiration (EI; left) and end expiration (EE; right). The outer boundaries of the lungs are shown in gray; parenchyma with high density [nonaerated, Hounsfield units (HU) more than −100] is shown in red.
Table 1.
Dice's coefficients of similarity between manually and automatically segmented lung images
| Rat 1 | Rat 2 | Rat 3 | Rat 4 | Rat 5 | Rat 6 | Rat 7 | Rat 8 | Means ± SD | |
|---|---|---|---|---|---|---|---|---|---|
| Saline lavage 1 | |||||||||
| End of inspiration | 0.983 | 0.982 | 0.983 | 0.983 | 0.983 | 0.982 | 0.980 | 0.978 | 0.982 ± 0.002 |
| End of expiration | 0.977 | 0.978 | 0.978 | 0.979 | 0.982 | 0.979 | 0.979 | 0.973 | 0.978 ± 0.002 |
| Saline lavage 2 | |||||||||
| End of inspiration | 0.945 | 0.953 | 0.982 | 0.980 | 0.981 | 0.975 | 0.955 | 0.955 | 0.966 ± 0.015 |
| End of expiration | 0.914 | 0.928 | 0.976 | 0.973 | 0.981 | 0.966 | 0.966 | 0.949 | 0.956 ± 0.024 |
| Saline lavage 3 | |||||||||
| End of inspiration | 0.930 | 0.935 | 0.950 | 0.968 | 0.951 | 0.951 | 0.931 | 0.932 | 0.944 ± 0.014 |
| End of expiration | 0.930 | 0.856 | 0.925 | 0.890 | 0.926 | 0.929 | 0.891 | 0.911 | 0.907 ± 0.027 |
Coefficients were calculated for each set of images obtained after the first saline lavage, at end inspiration and end expiration.
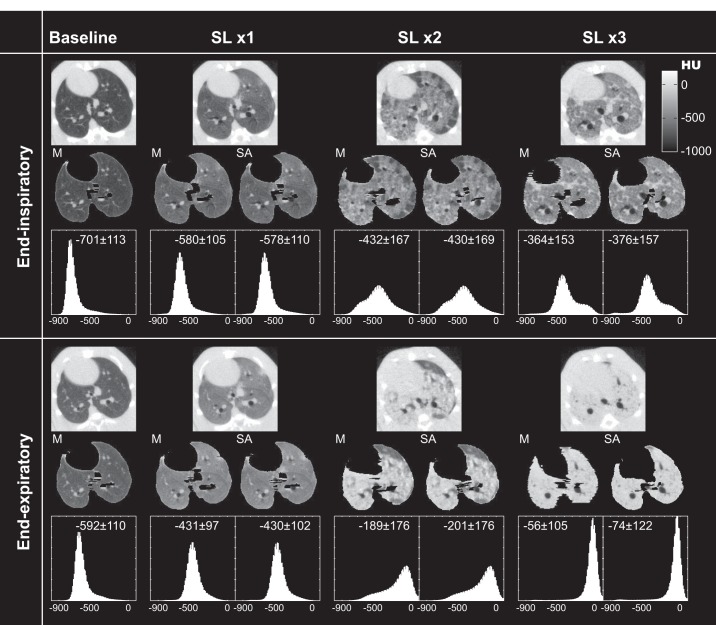
Figure 3 shows axial EI (top) and EE (bottom) images obtained in a representative animal at baseline and after each incremental SL procedure. The unprocessed images are shown in the top row of each panel; in the middle row, manually and semiautomatically segmented images are shown for the same axial slices (only manual segmentations are available at baseline). In the bottom row, the frequency distributions of CT densities (in HU) of each segmented image are displayed (mean ± SD density is also reported). CT intensity increased with the number of SLs. The density shift between EI and EE images documents the occurrence of tidal recruitment. The frequency distribution and dispersion of CT densities were affected by the number of SL procedures and by EI vs EE status, but they were always comparable between manual and semiautomatic images.
Fig. 3.
Axial images (midlung level) in a representative rat obtained at EI (top) and EE (bottom) after each repetition of SL. The top row of each panel shows raw images (including lung and nonpulmonary tissue); the middle row shows segmented images obtained manually (M) and semiautomatically (SA); the bottom row displays frequency distributions (with mean ± SD) of entire lung tissue densities in HU. Only M segmented images are available at baseline.
Agreement of Quantitative CT Analysis Results
Total lung weight and gas volume.
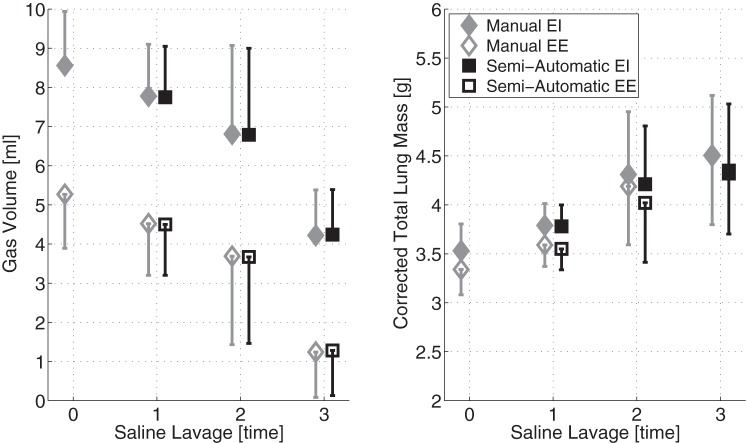
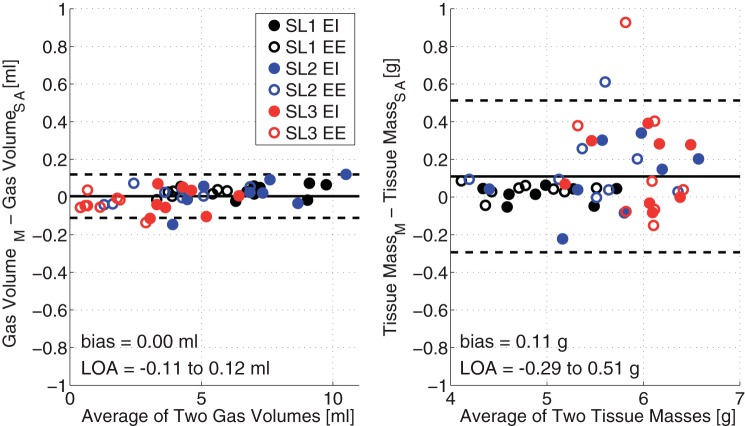
Both lung weight and gas volumes were consistently comparable between manual and semiautomatic segmentation (Fig. 4). Changes in lung volume (P < 0.01) reflected worsening injury severity due to SL repetitions and to the effects of inspiration (P < 0.01). Total lung mass, which was corrected for the weight of unrecovered saline, increased (P < 0.01) with each subsequent lavage, but was stable between EI and EE. We observed excellent agreement, as shown by a low LOA (2.80% of the median), and negligible bias (<0.10% of the median) between the two segmentation methods in the measurement of lung volumes (Fig. 5, left). In the measurement of lung weights (Fig. 5, right), bias was minimal (<2.00% of the median), but the LOA was relatively larger (9.25% of the median), although acceptable, with limited outlier data beyond LOA. The differences between manual and semiautomatic segmentation were larger when weight was bigger, i.e., after three lavages, particularly at EE. The presence of a small proportional bias in the weight measurement was tested by regression analysis, and a positive correlation was found (slope = 0.07, offset = −0.25).
Fig. 4.
Mean ± SD (cross animals) values of total lung volume (left) and of lung mass (right) in all experimental conditions, measured after each SL repetition at EI and EE. Lung volumes and weights (corrected for retained SL solution) were obtained from manually and automatically segmented images.
Fig. 5.
Bland-Altman plots of the agreements between manual and automatic segmentation in the measurement of total lung gas volume (left) and total lung mass (right). Images were obtained after each SL repetition at EI and EE. Limits of agreement (LOA) are shown.
Distribution of lung aeration.
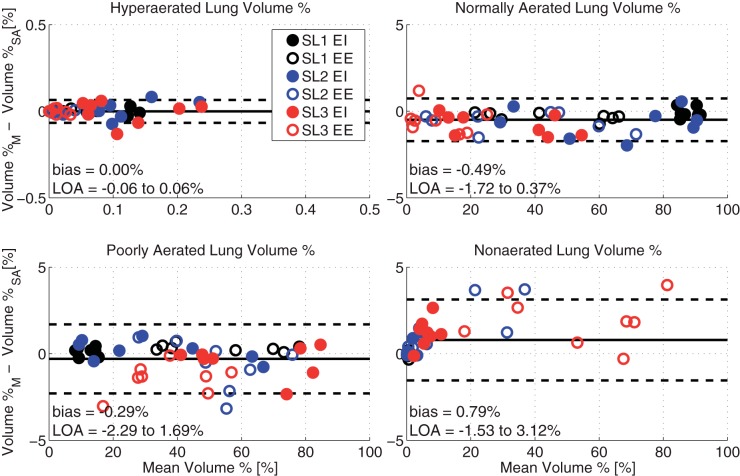
We also found excellent agreement (bias < 0.79% and LOA < 3.12% of the median values) between manual and semiautomatic segmentation in the measurement (obtained in each segmented image) of the amounts of lung tissue in each of the hyperaerated, normally aerated, poorly aerated, and nonaerated tissue compartments (Fig. 6). As expected, the quota of poorly aerated and nonaerated tissue increased with the number of SL and at EE, at the expense of normally aerated lung. However, the LOA between semiautomatic and manual techniques remained very small in all conditions, and no relevant bias was detectable. The amount of hyperinflated tissue was minimal and nonrelevant in all conditions.
Fig. 6.
Bland-Altman plots of the agreements between measurements of lung aeration obtained by M and SA segmentation of whole lung images. Each image was partitioned in compartments with increasing CT density: hyperinflated, normally aerated, poorly aerated, nonaerated. The size of each compartment is expressed as the percent quota of total lung volume it occupies.
DISCUSSION
In this study, we designed a pipeline for the semiautomatic segmentation of the lung parenchyma in CT images of a rat model of ARDS. This approach utilized multilandmark registration to mitigate the clinically problematic labor intensity of the manual techniques and circumvent the limitations of existing automatic methods when used in ARDS. Our semiautomatic segmentations of three-dimensional images were superimposable to those obtained by trained operators using standard manual techniques. The high similarity between semiautomatic and manual images was maintained at all levels of radiological injury severity. Furthermore, our semiautomatic segmentation method had a minimal effect on the values of common qCT measurements of lung pathology.
Contour Similarity Between Methods of Segmentation
In our studies, similarity between semiautomatic and manual images was more than adequate (>0.856) in all conditions: the Dice's coefficient was always high (values > 0.85 are typically considered acceptable). Similarity was only slightly decreased when injury severity was highest, but only three images had values < 0.9, which were obtained in low-aeration conditions that would challenge any method of segmentation, including a manual approach.
Visual analysis of segmented three-dimensional images (Fig. 2) shows that semiautomatic segmentation was able to obtain high contour similarity with the reference method. However, manually segmented masks had more surface irregularities than the semiautomatic ones, the latter appearing more realistic. This was particularly evident after the second and third SLs at EE. Existing smoothing algorithms (i.e., time/frequency domain filters) could potentially improve the continuity of the contour. However, in conditions of poor aeration, high-density tissue in the lung periphery was more often mislabeled and lung boundaries more often overstepped by the operators. Therefore, when agreement between methodologies was lower, this may have been due to uncertainty in the reference manual measure rather than imprecision in the tested methodology.
Agreement Between qCT Measurements
Measurements of lung gas content, weight, and aeration partition had satisfactory agreement between the two methodologies in most conditions. This finding confirms that our segmentation technique enables for the measurement of physiologically meaningful variables, and that such measurements are minimally affected by the chosen technique.
A relatively higher (compared with the mean values) LOA was observed for lung weight, with increased dispersion when its mean value was elevated. Furthermore, a small positive proportional bias was observed for lung weight; using semiautomatic segmentation for lung weight resulted in slightly lower values than manual when injury was more severe. However, in these conditions (at EE after three SL), CT densities were so extreme that nonaerated tissue reached 60–80% of total lung volume (a clinically uncommon condition). Otherwise, the agreement between methods was more than acceptable.
Comparison With Other Techniques
As shown in Table 2, our proposed technique reduces operational time dramatically compared with a wholly manual segmentation approach; it streamlines data processing, allowing analysis of a large number of images, and it is minimally operator dependent. With current computing technology, semiautomatic segmentation does require significant time (4–6 h per image) for offline computations. However, rapid improvement in computing power and algorithms will likely decrease offline computation time considerably in the near future.
Table 2.
Processing times required by manual and automatic segmentation techniques
| Method | Offline per Image (Semiautomatic) | Online per Image (Manual) | Offline for 5 Images (Semiautomatic) | Online for 5 Images (Manual) |
|---|---|---|---|---|
| Manual segmentation | 1.5 h | 7.5 h | ||
| Automatic segmentation | ||||
| Baseline segmentation | 1.5 h | 1.5 h | ||
| Preregistration | 2 min | 10 min | ||
| Registration | 4–6 h | 20–30 h | ||
| Postregistration | 2 min | 10 min |
Time spent by a human operator while delineating images (online) and by the computer (2.4 GHz Intel Xeon CPU, 2 GB RAM) during automatic processing (offline) is shown per image and for a series of five images.
The use of registration-based methodologies to segment the lungs has been reported previously (25, 30). In these studies, a refined deformable registration (i.e., B-spline) was used after a global coarse (i.e., rigid) registration to optimize the similarity between target and reference. These image registration approaches in general assume that the intensities of the IR and the IT have similar patterns. However, in severe injury, the pathological regions may exhibit very different patterns of intensity and texture compared with the baseline image. This makes a direct registration between the baseline image IR and the target image IR\T unreliable.
Conversely, the bone structure in the CT images, i.e., the ribs and spine, is stable before and after injury, which creates a good cue to predict the location of the lungs. Prior studies have used the rib cage and spine to classify potential lung voxels (17) or as the boundary location constraints (23, 26). Our method, by adding the invariant features of the rib cage and spine positions into the registration framework, largely improves the registration accuracy, despite distinct intensity differences between IT and IR. These features are particularly helpful when no pattern or texture can be reliably identified (e.g., in nonaerated regions of the ARDS lung). They are also helpful when the pulmonary shape is deformed by atelectasis and variable lung inflation.
In three randomly chosen animals, we compared our method with a simpler affine registration algorithm. The DSC between the affine method and manually segmented images was smaller than with the proposed method after one SL (EI: 0.950 ± 0.009 vs. 0.982 ± 0.001) and two SL (EI: 0.865 ± 0.063 vs. 0.960 ± 0.019); it further decreased after the third SL (EI: 0.758 ± 0.111 vs. 0.938 ± 0.010), particularly at EE (0.717 ± 0.129 vs. 0.904 ± 0.041). This finding could be due to nonisotropic lung deformation after injury and during breathing, for which a simpler registration algorithm may not account.
Limitations
The most obvious limitation of our method is the need for a baseline mask of the healthy lung to allow the segmentation of registered IT; this necessity reduces the clinical applicability of this approach. Baseline images can easily be obtained via automatic intensity thresholding, if lung parenchyma is mostly healthy, as most lung tissue has markedly lower density than surrounding structures. While this is not typically a problem in the experimental setting when a baseline is available, it is a serious limitation for this method's applicability in humans. Images may be available of many patients wherein the lung is totally or mostly healthy, e.g., preadmission, posttrauma screening, but this is not true for the majority of subjects.
The use of a probabilistic atlas could productively address this limitation (25). Atlas-based segmentation has been widely used in segmenting various organs, particularly in brain mapping (1). Atlases are built from a large pool of sample images and matched with IT for segmentation. This process typically requires a number of parenchymal landmarks, which are often obscured by dense opacities in ARDS. Introducing rib and spine landmarks in the atlas, as described in this paper, decreases the reliance on intrapulmonary landmarks and will make segmentation from atlases possible, despite sample limitations. However, the agreement with the ground truth may be lower than in the present study, where our method was likely favored by using a IR of the same subject.
Another important limitation of our approach is that local ventilation and strain may not be accurately measurable when local pathology causes irregular distortion of lung tissue within its outer boundaries. Furthermore, the accuracy of our method is unclear when large positional changes (i.e., between supine and prone position) modify the relationship between lung surface and surrounding thoracic structures. These problems could be addressed in the future by adding more internal landmarks (i.e., conductive airways, large blood vessels) to better constrain the registration.
Conclusion
This preliminary study in surfactant-depleted rats tested the feasibility of a registration-based method of semiautomatic lung segmentation for quantitative CT analysis. The results show sufficient similarity between the images generated by our technique and those produced through manual segmentation by expert observers. The registration-based technique described here significantly reduces operational time compared with the manual approach. These findings confirm that semiautomatically segmented images can be used to measure physiologically meaningful data on disease severity in an animal model of mild-to-severe injury. This method is thus ready for use in ventilated rodent models of injury where healthy IR are available. Future studies are needed to test this methodology in larger animals and humans. The development of a probabilistic atlas could facilitate the expansion of this method for human research.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute (Bethesda, MD) Grants R01-HL064741 and R01-HL077241. M. Cereda is supported by grants from the Foundation for Anesthesia Education and Research (Rochester, MN), the Society of Critical Care Anesthesiologists (Park Ridge, IL), and from the Transdisciplinary Awards Program in Translational Medicine and Therapeutics (TAPITMAT)-Translational Biomedical Imaging Center (TBIC).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.X., G.S., M.C., J.C.G., and R.R.R. conception and design of research; Y.X., M.C., and H.P. performed experiments; Y.X., G.S., M.C., H.H., Y.J., N.M., J.W., and N.J.T. analyzed data; Y.X., G.S., M.C., S.K., H.H., Y.J., J.W., N.J.T., J.C.G., and R.R.R. interpreted results of experiments; Y.X., G.S., and M.C. prepared figures; Y.X., G.S., and M.C. drafted manuscript; Y.X., G.S., M.C., S.K., J.R., J.C., and R.R.R. edited and revised manuscript; Y.X., M.C., S.K., J.C., J.C.G., and R.R.R. approved final version of manuscript.
REFERENCES
- 1.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46: 726–738, 2009. [DOI] [PubMed] [Google Scholar]
- 2.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12: 26–41, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54: 2033–2044, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The Insight ToolKit image registration framework. Front Neuroinform 8: 44, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1: 307–310, 1986. [PubMed] [Google Scholar]
- 6.Caironi P, Carlesso E, Gattinoni L. Radiological imaging in acute lung injury and acute respiratory distress syndrome. Semin Respir Crit Care Med 27: 404–415, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Cereda M, Emami K, Kadlecek S, Xin Y, Mongkolwisetwara P, Profka H, Barulic A, Pickup S, Mansson S, Wollmer P, Ishii M, Deutschman CS, Rizi RR. Quantitative imaging of alveolar recruitment with hyperpolarized gas MRI during mechanical ventilation. J Appl Physiol 110: 499–511, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cereda M, Xin Y, Emami K, Huang J, Rajaei J, Profka H, Han B, Mongkolwisetwara P, Kadlecek S, Kuzma NN, Pickup S, Kavanagh BP, Deutschman CS, Rizi RR. Positive end-expiratory pressure increments during anesthesia in normal lung result in hysteresis and greater numbers of smaller aerated airspaces. Anesthesiology 119: 1402–1409, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiumello D, Carlesso E, Cadringher P, Caironi P, Valenza F, Polli F, Tallarini F, Cozzi P, Cressoni M, Colombo A, Marini JJ, Gattinoni L. Lung stress and strain during mechanical ventilation for acute respiratory distress syndrome. Am J Respir Crit Care Med 178: 346–355, 2008. [DOI] [PubMed] [Google Scholar]
- 10.Cressoni M, Cadringher P, Chiurazzi C, Amini M, Gallazzi E, Marino A, Brioni M, Carlesso E, Chiumello D, Quintel M, Bugedo G, Gattinoni L. Lung inhomogeneity in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 189: 149–158, 2014. [DOI] [PubMed] [Google Scholar]
- 11.Denison DM, Morgan MD, Millar AB. Estimation of regional gas and tissue volumes of the lung in supine man using computed tomography. Thorax 41: 620–628, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dice LR. Measures of the amount of ecologic association between species. Ecology 26: 297–302, 1945. [Google Scholar]
- 13.Fuld MK, Easley RB, Saba OI, Chon D, Reinhardt JM, Hoffman EA, Simon BA. CT-measured regional specific volume change reflects regional ventilation in supine sheep. J Appl Physiol 104: 1177–1184, 2008. [DOI] [PubMed] [Google Scholar]
- 14.Gattinoni L, Caironi P, Cressoni M, Chiumello D, Ranieri VM, Quintel M, Russo S, Patroniti N, Cornejo R, Bugedo G. Lung recruitment in patients with the acute respiratory distress syndrome. N Engl J Med 354: 1775–1786, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Gattinoni L, Caironi P, Pelosi P, Goodman LR. What has computed tomography taught us about the acute respiratory distress syndrome? Am J Respir Crit Care Med 164: 1701–1711, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Hu S, Hoffman EA, Reinhardt JM. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging 20: 490–498, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Hua P, Song Q, Sonka M, Hoffman EA, Reinhardt JM. Segmentation of pathological and diseased lung tissue in CT images using a graph-search algorithm. In: Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium. New York: IEEE, 2011, p. 2072–2075. [Google Scholar]
- 18.Kaczka DW, Cao K, Christensen GE, Bates JH, Simon BA. Analysis of regional mechanics in canine lung injury using forced oscillations and 3D image registration. Ann Biomed Eng 39: 1112–1124, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Christensen GE, Hoffman EA, McLennan G, Reinhardt JM. Establishing a normative atlas of the human lung: computing the average transformation and atlas construction. Acad Radiol 19: 1368–1381, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Christensen GE, Hoffman EA, McLennan G, Reinhardt JM. Establishing a normative atlas of the human lung: intersubject warping and registration of volumetric CT images. Acad Radiol 10: 255–265, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Lu Q, Malbouisson LM, Mourgeon E, Goldstein I, Coriat P, Rouby JJ. Assessment of PEEP-induced reopening of collapsed lung regions in acute lung injury: are one or three CT sections representative of the entire lung? Intensive Care Med 27: 1504–1510, 2001. [DOI] [PubMed] [Google Scholar]
- 22.Mull RT. Mass estimates by computed tomography: physical density from CT numbers. AJR Am J Roentgenol 143: 1101–1104, 1984. [DOI] [PubMed] [Google Scholar]
- 23.Prasad MN, Brown MS, Ahmad S, Abtin F, Allen J, da Costa I, Kim HJ, McNitt-Gray MF, Goldin JG. Automatic segmentation of lung parenchyma in the presence of diseases based on curvature of ribs. Acad Radiol 15: 1173–1180, 2008. [DOI] [PubMed] [Google Scholar]
- 24.Reske AW, Reske AP, Gast HA, Seiwerts M, Beda A, Gottschaldt U, Josten C, Schreiter D, Heller N, Wrigge H, Amato MB. Extrapolation from ten sections can make CT-based quantification of lung aeration more practicable. Intensive Care Med 36: 1836–1844, 2010. [DOI] [PubMed] [Google Scholar]
- 25.Sluimer I, Prokop M, van Ginneken B. Toward automated segmentation of the pathological lung in CT. IEEE Trans Med Imaging 24: 1025–1038, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Sofka M, Wetzl J, Birkbeck N, Zhang J, Kohlberger T, Kaftan J, Declerck J, Zhou SK. Multi-stage learning for robust lung segmentation in challenging CT volumes. Med Image Comput Comput Assist Interv 14: 667–674, 2011. [DOI] [PubMed] [Google Scholar]
- 27.Song G, Tustison NJ, Avants BB, Gee JC. Lung CT Image Registration Using Diffeomorphic Transformation Models. Medical Image Analysis for a Clinic: A Grand Challenge. 13th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Beijing, China, September 20–24, 2010. [Google Scholar]
- 28.Sun S, Bauer C, Beichel R. Automated 3-D segmentation of lungs with lung cancer in CT data using a novel robust active shape model approach. IEEE Trans Med Imaging 31: 449–460, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tustison NJ, Avants BB. Explicit B-spline regularization in diffeomorphic image registration. Front Neuroinform 7: 39, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rikxoort EM, de Hoop B, Viergever MA, Prokop M, van Ginneken B. Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection. Med Phys 36: 2934–2947, 2009. [DOI] [PubMed] [Google Scholar]
- 31.van Rikxoort EM, van Ginneken B. Automated segmentation of pulmonary structures in thoracic computed tomography scans: a review. Phys Med Biol 58: R187–R220, 2013. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Hoffman EA, Reinhardt JM. Atlas-driven lung lobe segmentation in volumetric X-ray CT images. IEEE Trans Med Imaging 25: 1–16, 2006. [DOI] [PubMed] [Google Scholar]