Abstract
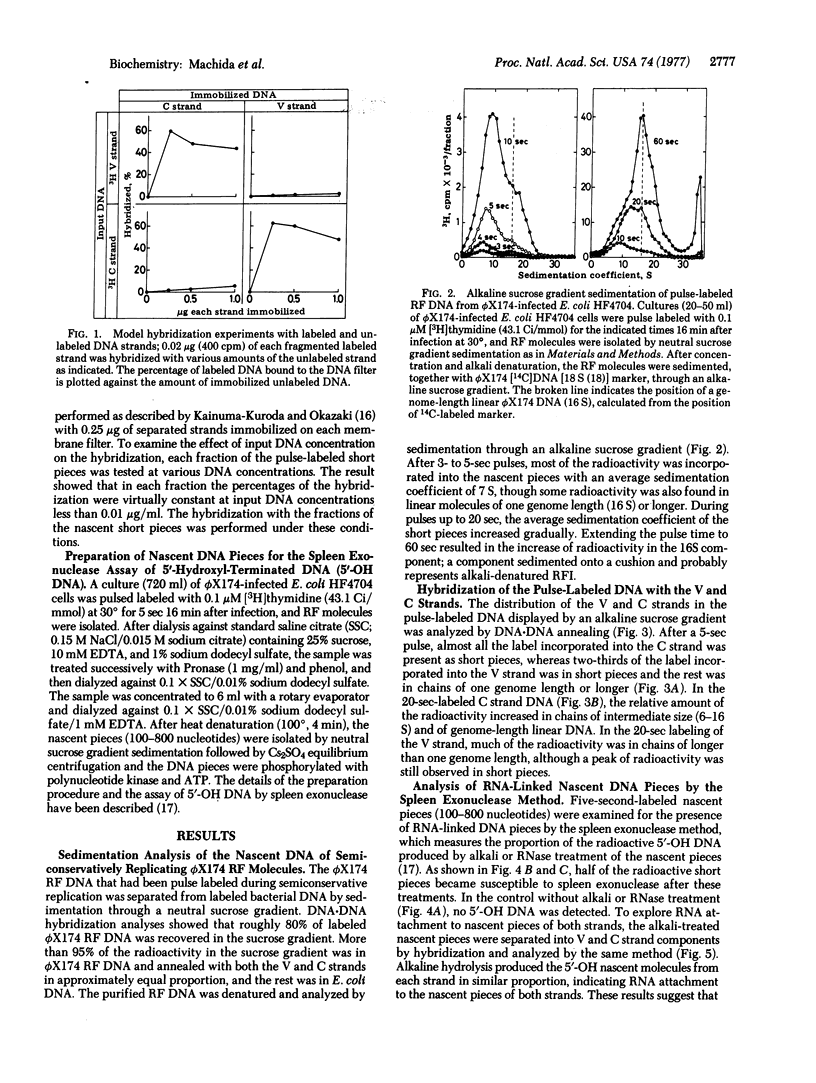
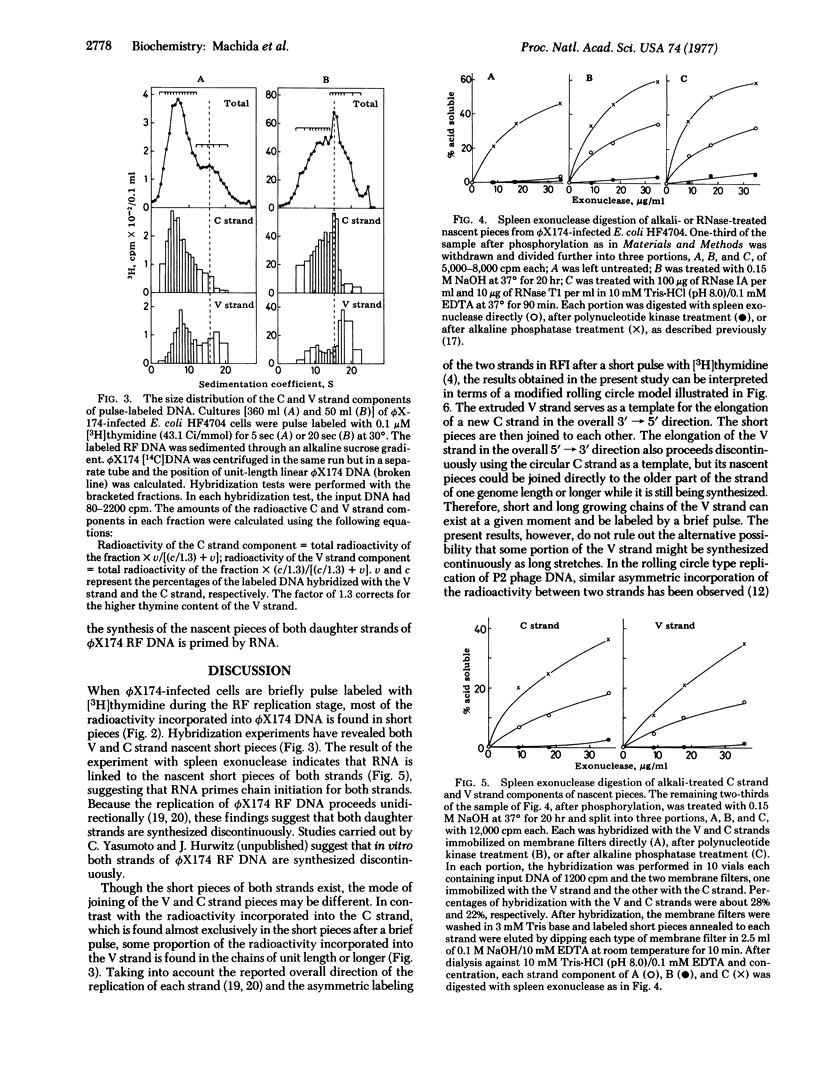
Bacteriophage phiX174 DNA has been labeled with short pulses of [3H]thymidine during synthesis of replicative form molecules in infected Escherichia coli HF4704 cells. The replicating phiX174 DNA was isolated and analyzed by sedimentation in an alkaline sucrose gradient. During a brief pulse (5 sec at 30 degrees), the radioactivity incorporated into the complementary strand was found in chains much shorter than one genome length. Of the radioactivity incorporated into the viral strand, two-thirds was in the short pieces and the rest was in chains of one genome length or longer. RNA attachment to the 5' end of both strand components of the nascent short pieces was shown by the appearance of spleen exonuclease-digestable nascent molecules after alkali treatment. These observations suggest that the viral as well as the complementary strand is synthesized by the discontinuous mechanism with RNA primers during replication of duplex phiX174 DNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Denhardt D. T. Evidence for preferential breakage of the minus strand of phi-X174 replicative form DNA by a T4-induced endonuclease. Biochim Biophys Acta. 1970 Nov 12;224(1):21–28. doi: 10.1016/0005-2787(70)90616-7. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. PhiX174 replicative form DNA replication, origin and direction. J Mol Biol. 1972 Feb 14;63(3):569–576. doi: 10.1016/0022-2836(72)90448-2. [DOI] [PubMed] [Google Scholar]
- Calendar R., Lindqvist B., Sironi G., Clark A. J. Characterization of REP- mutants and their interaction with P2 phage. Virology. 1970 Jan;40(1):72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. Formation of ribosylthymine in Escherichia coli. Studies on pulse labeling with thymine and thymidine. J Biol Chem. 1969 May 25;244(10):2710–2715. [PubMed] [Google Scholar]
- Dressler D. The rolling circle for phiX DNA replication. II. Synthesis of single-stranded circles. Proc Natl Acad Sci U S A. 1970 Dec;67(4):1934–1942. doi: 10.1073/pnas.67.4.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler D., Wolfson J. The rolling circle for phi X DNA replication. 3. Synthesis of supercoiled duplex rings. Proc Natl Acad Sci U S A. 1970 Sep;67(1):456–463. doi: 10.1073/pnas.67.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A., Sinsheimer R. L. Process of infection with bacteriophage phi X 174 XXXVIII. Replication of phi chi 174 replicative form in vivo. J Virol. 1976 Mar;17(3):776–787. doi: 10.1128/jvi.17.3.776-787.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Origin and direction phiX174 double- and single-stranded DNA synthesis. J Mol Biol. 1974 Nov 25;90(1):127–141. doi: 10.1016/0022-2836(74)90261-7. [DOI] [PubMed] [Google Scholar]
- Kainuma-Kuroda R., Okazaki R. Mechanism of DNA chain growth. XII. Asymmetry of replication of P2 phage DNA. J Mol Biol. 1975 May 15;94(2):213–228. doi: 10.1016/0022-2836(75)90079-0. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y., Ogawa T., Hirose S., Okazaki T., Okazaki R. Mechanism of DNA chain growth. XV. RNA-linked nascent DNA pieces in Escherichia coli strains assayed with spleen exonuclease. J Mol Biol. 1975 Aug 25;96(4):653–664. doi: 10.1016/0022-2836(75)90144-8. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y., Okazaki R. Mechanism of DNA chain growth. XIII. Evidence for discontinuous replication of both strands of P2 phage DNA. J Mol Biol. 1975 May 15;94(2):229–241. doi: 10.1016/0022-2836(75)90080-7. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Sinsheimer R. L. Process of infection with bacteriophage phi-X174. XIV. Studies on macromolecular synthesis during infection with a lysis-defective mutant. J Mol Biol. 1967 Aug 28;28(1):87–94. doi: 10.1016/s0022-2836(67)80079-2. [DOI] [PubMed] [Google Scholar]
- McFadden G., Denhardt D. T. The mechanism of replication of phiX174 DNA. XIII. Discontinuous synthesis of the complementary strand in an Escherichia coli host with a temperature-sensitive polynucleotide ligase. J Mol Biol. 1975 Nov 25;99(1):125–142. doi: 10.1016/s0022-2836(75)80163-x. [DOI] [PubMed] [Google Scholar]
- Newbold J. E., Sinsheimer R. L. The process of infection with bacteriophage phiX174. XXXI. Abortive infection at low temperatures. J Mol Biol. 1970 Apr 14;49(1):23–47. doi: 10.1016/0022-2836(70)90374-8. [DOI] [PubMed] [Google Scholar]
- Ray D. S., Schekman R. W. Replication of bacteriophage M13. I. Sedimentation analysis of crude lysates of M13-infected bacteria. Biochim Biophys Acta. 1969 Apr 22;179(2):398–407. [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Sinsheimer R. L., Knippers R., Komano T. Stages in the replication of bacteriophage phi X174 DNA in vivo. Cold Spring Harb Symp Quant Biol. 1968;33:443–447. doi: 10.1101/sqb.1968.033.01.051. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., Weiss B. Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):154–157. doi: 10.1073/pnas.74.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]


