Abstract
The innate immune system must coordinate elaborate signaling pathways to turn on expression of hundreds of genes to provide protection against pathogens and resolve acute inflammation. Multiple genes within distinct functional categories are coordinately and temporally regulated by transcriptional on and off switches in response to distinct external stimuli. Three classes of transcription factors act together with transcriptional coregulators and chromatin-modifying complexes to control these programs. In addition, newer studies implicate long noncoding RNA (lncRNA) as additional regulators of these responses. LncRNAs promote, fine-tune, and restrain the inflammatory program. In this study, we provide an overview of gene regulation and the emerging importance of lncRNAs in the immune system.
Introduction
The acute inflammatory response is induced as a first line of defense against microbial infection. This response must be appropriately scaled and regulated to avoid devastating consequences for the host and diseases such as atherosclerosis, arthritis, and cancer (Aringer and others 2013; Moore and others 2013). The inflammatory program is coordinated through germ-line encoded receptors, including the Toll-like receptors (TLRs), Nod-like receptors, Aim2-like receptors, Rig-I-like receptors, and C-type lectins (Kumar and others 2011; Moore and others 2013). These sensors are present in distinct locations within cells of the innate immune system and are activated by microbial products or endogenous danger signals released from damaged or dying cells. Once activated they trigger complex signaling cascades resulting in changes in expression of hundreds of genes involved in antimicrobial defense, phagocytosis, cell migration, metabolic reprogramming, tissue repair, and regulation of adaptive immunity. At the level of transcription many of these genes are tightly controlled by on and off switches accounting for the specificity of gene expression in response to distinct external stimuli.
Enormous progress has been made in understanding how pathogen or danger signals are detected and in elucidating both the signaling pathways and transcription factors (TFs) that underlie the development and activation of immune cells. Defined signaling pathways lead to the deployment of TFs that couple target gene selection to the recruitment of the transcription apparatus. Multiple layers of regulation control these pathogen or danger-induced cell-lineage and signal-specific gene expression programs. Chromatin interacting proteins enable or prevent access to DNA sequences by TFs and are also essential for the direct recruitment of the transcription apparatus for transcriptional initiation and elongation. The TFs themselves require posttranslational modifications as well as interaction with coregulators to regulate the expression of their target genes. Collectively, the combination of chromatin state, histone or DNA modifications, use of enhancers, and recruitment of TFs, all lead to differential gene expression in a kinetically defined and cell-type specific manner.
In this review, we cover the molecular mechanisms that coordinate and fine-tune the transcriptional regulation of inflammatory gene expression. Additionally, we highlight the emerging role of long noncoding RNAs (lncRNAs) as a new layer of regulation in these processes. A better understanding of this circuitry could facilitate the development of selective therapeutics to prevent damaging inflammation while maintaining antimicrobial defenses.
Transcriptional Regulation in Macrophages
Lineage specificity
Gene regulation within innate immune cells such as macrophages is controlled by TFs that are lineage specific, those basally expressed and activated in a signal-dependent manner or those that are themselves transcriptionally regulated to amplify and/or dampen inflammatory cascades. The key TFs associated with macrophage specification include PU.1 (also known as SPI1), C/EBPα, and runt-related TF (RUNX1), which are induced during macrophage development (Valledor and others 1998; Heath and others 2004; Cai and others 2008; Kumar and others 2011; Thompson and others 2011). These regulators are considered Pioneer factors in that they are the first to engage target sites in chromatin and generate regions of open chromatin that enable the subsequent recruitment of TFs activated in response to external cues in differentiated cells (Ostuni and others 2013). At the same time, they promote the silencing of genes associated with alternative cell fates (Ghisletti and others 2010; Heinz and Glass 2012; Pham and others 2012; Ostuni and others 2013). Although these TFs are expressed in other cell types it is their combinatorial expression together with their unique interactions that defines the monocyte lineage. The second class of TFs important in inflammation are those constitutively expressed, but activated in a stimulus-specific manner in myeloid cells. These factors bind subsets of genes and coordinate expression of genes with shared biological functions (Smale 2012). The best-characterized include NFκB, interferon (IFN)-regulatory factor (IRF), AP1, and cAMP-responsive-element-binding protein 1 (CREB1) families (Medzhitov and Horng 2009). The signaling pathways that control the activation of these TFs have been worked out in significant detail and reviewed elsewhere (Sasai and Yamamoto 2013). The genes that are induced most rapidly (the so called primary response genes, PRGs) are regulated directly by these TFs acting alone or in combination. Many of these PRGs are induced within minutes, followed by the induction of secondary response genes (SRGs) and the initiation of autocrine and paracrine feedback loops (Amit and others 2009).
Primary response genes
PRGs are induced in the absence of new protein synthesis and mostly contain CpG islands within their promoters (Yamamoto and Alberts 1976; Smale 2010; Fowler and others 2011). Such genes are associated with poised RNA polymerase II even in unstimulated cells and have histone tail modifications commonly found at the promoters of actively transcribed genes (Sims and others 2004; Suzuki and Bird 2008; Hargreaves and others 2009; Ramirez-Carrozzi and others 2009). They have open chromatin and do not require chromatin remodeling by the SWI–SNF (SWItch/Sucrose NonFermentable) remodeling complex (Sims and others 2004; Ramirez-Carrozzi and others 2009; Fowler and others 2011). Chromatin Immunoprecipitation was recently used in a high-throughput manner to define the dynamics of DNA binding by 25 TFs and 4 chromatin marks in dendritic cells (DCs). These studies revealed that TFs vary substantially in their temporal binding characteristics and revealed the hierarchically organized nature of these factors into cell differentiation factors, factors that bind targets before cellular activation (so called Pioneer TFs), and factors that regulate specific gene programs (Kumar and others 2011; Thompson and others 2011; Garber and others 2012). PRGs are permissive for very rapid induction (Ghisletti and others 2010; Fowler and others 2011; Heinz and Glass 2012).
A group of intermediately expressed genes known as the late primary response genes (LPRGs) (e.g., IL1α and Ccl5) are induced later in the absence of new protein synthesis. Unlike classical PRGs they do require some degree of chromatin remodeling (Saccani and others 2001; Ramirez-Carrozzi and others 2006, 2009). Sixteen out of 55 of these genes require key components of the SWI–SNF complex (Brg, BRM) for their activation whereas the remaining genes act as true PRGs, independent of the SWI–SNF complex. Many LPRGs require the TF IRF3 for their activation (Saccani and others 2001; Foster and Medzhitov 2009; Ramirez-Carrozzi and others 2006, 2009).
Secondary response genes
Following induction of PRGs and LPRGs, a second wave of gene expression, the so-called SRGs, require de novo protein synthesis and comprehensive chromatin remodeling and activity of enhancers for the activation of transcription to occur. PRGs include cytokines and chemokines that in turn amplify gene regulation in autocrine and paracrine manners (Saccani and others 2001; Ramirez-Carrozzi and others 2006, 2009; Foster and Medzhitov 2009; Smale 2012).
Litvak and colleagues have provided significant insight into the control of early (PRG), intermediate (LPRG), and late (SRG) genes using systems approaches to predict and map regulatory networks. They identified a critical regulatory circuit consisting of NFκB, CEBPδ, and ATF3. NFκB regulates the induction of PRGs and also regulates expression of CEBPδ, which they show acts as an amplifier of NFκB signaling and an essential mediator of SRGs such as IL6 and acute phase genes. CEBPδ also discriminates between transient and persistent TLR4 signals, ensuring that only dangerous insults initiate persistent responses (Litvak and others 2009). In addition to defining these amplifiers they also identified ATF3 as a critical negative regulator of the TLR4 pathway, where induction of ATF3 functions as a postinduction feedback regulator of the NFκB/Cebpδ pathway (Gilchrist and others 2006). Miz1 has also been identified as a negative regulator of C/EBPδ-driven inflammation (Do-Umehara and others 2013). It is known that macrophages express more than 500 TFs, 100 of which are regulated by lipopolysaccharide (LPS). Some of these TFs likely function as master regulators to positively or negatively control distinct functional modules. A schematic detailing the role of Pioneer factors and the mechanisms controlling PRG, LPRG, and SRG is shown in Fig. 1. Characteristics associated with each class of genes are outlined in Table 1.
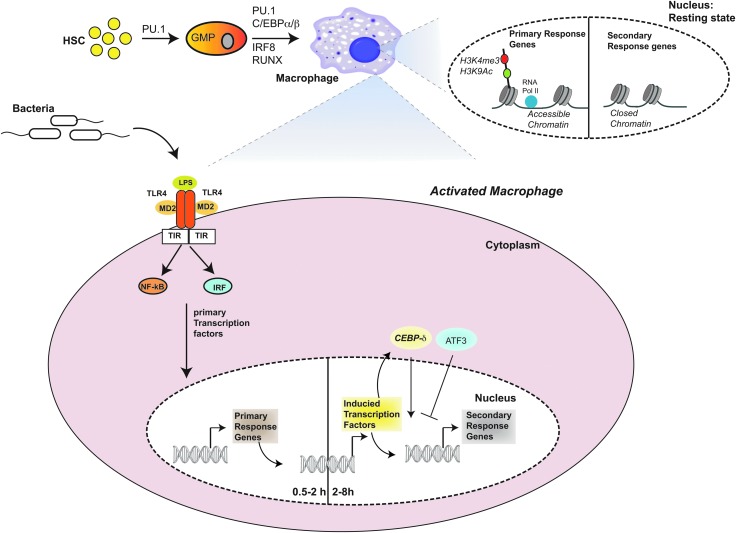
FIG. 1.
Transcription factors (TFs) involved in the development and activation of macrophages. The Pioneer TFs involved in macrophage development include PU.1, C/EBPα and β, RUNX1, and IRF8. These TFs engage target sites in chromatin generating regions of open chromatin that enable rapid recruitment of TFs activated in response to external cues in differentiated cells. Microbial activation of macrophages initiates the inflammatory signaling pathway first resulting in translocation of TFs such as NFκB and IRFs from the cytosol to the nucleus rapidly activating the primary response genes (PRGs). PRGs are associated with poised RNA polymerase and possess histone tail modifications commonly found at the promoters of actively transcribed genes (H3K4me3 and H3K9ac). The final class of TFs include C/EBPδ and ATF3 that get induced after the PRGs and are critical for the induction and control of the secondary response genes (SRGs). The SRGs require chromatin remodeling mediated by the SWI–SNF complex. CEBPδ acts as an amplifier of NFκB signaling and is critical for SRG induction whereas ATF3 plays a role in negatively regulating the pathway enabling efficient termination of the inflammatory response.
Table 1.
Characteristics Associated with Gene Classes
| Gene class | Gene examples | Activating transcription factors | Promoter characteristics | Nucleosome remodeling | Basal RNA Pol II association |
|---|---|---|---|---|---|
| Primary response genes | TNF, PTGS2, CXCL1/2 | NFκB | CpG island promoters | None, low nucleosome occupancy | Yes |
| IL1α/β, IL123a, Ccl3 | NFκB | Non CpG island promoters | None, low nucleosome occupancy | No | |
| Late primary response genes | Vcam1, Ccl2, IL10 | NFκB | Non CpG island promoters | SWI/SNF-dependent | No |
| IFIT 1/2/3, Ccl5/12 | NFκB | Non CpG island promoters, IRF3 dependent | SWI/SNF-dependent | No | |
| Secondary response genes | IRF7, IL6, IL12b, Nos2 | NFκB, Cebpδ | Non CpG island promoters | SWI/SNF-dependent | No |
Epigenetic Control of Gene Transcription
The histone code
Chromatin remodeling involves the dynamic modification of chromatin architecture to facilitate access of condensed genomic DNA to TFs and the transcriptional machinery to control gene expression. Such remodeling is carried out by covalent histone modifications and ATP-dependent chromatin remodeling complexes such as the SWI–SNF complex, which move, eject, or restructure nucleosomes.
Histone-containing nucleosomes limit the accessibility of TFs to promoter/enhancer regions of genes (Yamamoto and Alberts 1976; Foster and others 2007; Smale 2010; Fowler and others 2011). Histone-modifying enzymes (so-called writers, erasers, and readers) modify histones and in so doing create access to binding sites for TFs. In addition, histone reader enzymes that dock to modified histones through defined protein domains are essential for the recruitment of additional components of the transcriptional machinery (Sims and others 2004; Suzuki and Bird 2008; Hargreaves and others 2009; Ramirez-Carrozzi and others 2009). Covalent modifications of the NH2-terminal tails of the 4 core histones (H2A, H2B, H3, and H4) facilitate the activation as well as the silencing of gene expression (Martin and Zhang 2005; Li and others 2013). These dynamic modifications include acetylation, methylation, phosphorylation, sumoylation, citrullination, and ubiquitination and occur in characteristic temporal and spatial patterns that are associated with different transcriptional activities (Struhl 1999). The wide array of potential epigenetic marks is quite astounding. Table 2 outlines the known histone modifications within the mammalian system.
Table 2.
Histone Modifications Including Examples Within the Toll-Like Receptor Signaling Pathway
| Modifications | Position | Enzyme | Target promoters | Functions on transcription |
|---|---|---|---|---|
| Methylation | H3 K4 | SET, MLL | actively transcribed genes | Activation |
| H3 K9 | Suv39h,G9, ESET, EuHMTase | IL12b, Ccl22 | Repression | |
| H3 K27 | EZH2 | Genes involved in: | Repression | |
| Linage commitment and development | Activation | |||
| H3 K36 | Set2, NSD1, Symd2 | Actively transcribed genes | Activation | |
| Active genes | ||||
| H3 K79 | Dot | Activation | ||
| H3 R2 | CARM1 | Activation | ||
| H3 R17 | CARM1 | Activation | ||
| H3 R26 | CARM1 | Activation | ||
| H4 R3 | PRMT1, p300 | Activation | ||
| H4 K20 | SUV420H2, SET | Repression | ||
| Demethylation | H3 K4 | LSD1 JHDM, JMJD | All active genes | Repression |
| H3 K9 | JMJD | IL12b, Ccl22 | Activation | |
| H3 K27 | JHDM, JMJD3 | HoxA, BMP2 | Activation | |
| H3 K36 | All genes | Repression | ||
| Acetylation | H2A K5 | CBP/p300 | Activation | |
| H2B K12 | CBP/p300 | Activation | ||
| H2B K15 | CBP/p300 | Activation | ||
| H3 K9 | PCAF/GCN | Activation | ||
| H3 K14 | CBP/p300, PCAF/GCN5, TIP60 | Activation | ||
| H3 K18 | CBP/p300, PCAF/GCN5 HAT1 | TNF, IL6, IL12b | Activation | |
| H3 K27 | CBP/p300 | Activation (IFNγ + additional signal) | ||
| H4 K5 | CBP/P300 HAT1, TIP60, HB01 | Activation | ||
| H4 K8 | CBP/P300, TIP60, HB01 | Activation | ||
| H4 K12 | HAT1, TIP60 | Activation | ||
| H4 K16 | TIP60 | Activation | ||
| Deacetylation | H4 | HDAC1 | IL6, IL12b | Repression |
| Phosphorylation | H3 S10 | p38, MSK1 | IL6, IL12p40, Ccl2 | Activation |
| Ubiquitination | H2AK119 | Ccl5, CXCL2, CXCL10 | Repression | |
| Deubiquitination | H2AK119 | 2A-HUB | Ccl5, CXCL2, CXCL10 | Activation |
To date only a fraction of these modifications have been catalogued within the inflammatory signaling pathway. It will be interesting to further characterize the exact subsets of genes controlled by each of these modifications. Histone modifications with the exception of methylation result in a change in the net charge of nucleosomes, loosening interactions between histones and DNA. These modifications can directly affect chromatin structure allowing remodeling to occur and greater access of TFs to promoters (Foster and others 2007; Smale 2010; Li and others 2013). Chromatin remodeling complexes use ATP to slide nucleosomes relative to DNA or to alter nucleosome–DNA contacts, thereby modulating the accessibility of chromatin-associated DNA to transcriptional regulators.
Active epigenetic marks
The histone code or epigenetic landscape surrounding genes determines the transcriptional output that occurs following a given signal (Jenuwein and Allis 2001). Chromatin modifications act as a rate-limiting step in the activation of gene expression during infection. The fate of gene expression through chromatin structure is largely established within cells during development. Enhancers for inducible genes are known to associate with Pioneer TFs involved in lineage commitment. The histone mark histone 3 lysine 4 monomethylation (H3K4me1) corresponding to active enhancers is found at basal levels and following stimulation in macrophages, suggesting this modification occurs during lineage commitment. PU.1 has been shown to promote H3K4me1 and in doing so prepares some genes for transcription to occur following stimulation of macrophages (Ghisletti and others 2010; Heinz and Glass 2012). These exciting findings indicate that cell type specificity is determined early in development.
Pathogen-induced histone modifications also occur, some of which appear to be maintained to affect subsequent responses to the same or a different pathogen (Ghisletti and others 2010; Heinz and Glass 2012). TLRs and other sensors modify histones in a manner associated with the activation of transcription. Two of the main histone modifications associated with active transcripts are histone H3 Lysine 4 trimethylation (H3K4me3) denoting an active promoter and histone H3 Lysine 36 trimethylation (H3K36me3) indicating active transcription of an open reading frame (Mikkelsen and others 2007). Studies in DCs stimulated with LPS revealed that H3K4me3 is very stable during the first 2 h following stimulation (Garber and others 2012). LPS-stimulated macrophages also show increased histone H4 acetylation (H4Ac), an indicator of open chromatin at numerous sites across the genome. Intensive investigation of the IL6 and IL12 promoters reveals these modifications within 1 h following stimulation, decreasing after 2 h (Gilchrist and others 2006). In DCs, H4K27 acetylation varied over an LPS time course correlating with Pol II binding (Garber and others 2012). Acetylation of histones leads to more relaxed chromatin and, therefore, greater access for TFs whereas deacetylation limits access to chromatin. Some modifications in macrophages, particularly H3K4me1 at distal enhancers, persist long after the stimulation has ceased, providing some epigenetic memory of exposure to pathogens, at least in the short term (Ostuni and others 2013). Further, BCG vaccination or exposure to Candida albicans initiates trained immunity in monocytes for up to 3 months through epigenetic reprogramming (Saccani and Natoli 2002; Arbibe and others 2007; De Santa and others 2007). How long these induced histone modifications can be maintained in mature cells or potentially in stem cells for bona fide epigenetic memory that could determine subsequent responses of individuals to the same or different pathogen is an intriguing area that needs further investigation.
The transcription of primary and SRGs have a differential requirement for chromatin remodeling. Promoters of PRGs with CpG islands have H3K4me3 marks and preassociated RNA polymerase II before cell activation (Smale 2012). In contrast SRGs with low CpG content promoters require the SWI–SNF complex for chromatin remodeling and display low H3K4me3 marks and limited RNA polymerase II occupancy. PRGs produce low levels of unspliced transcripts, but their induction can be controlled through signal-dependent recruitment of the elongation factor positive elongation factor (P-TEFb). Histone H4 acetylation at positions 5,8, and 12 (H4K5/8/12Ac) are detected by the bromodomain-containing protein Brd4, which in turn recruits P-TEFb and removes the pausing complex negative elongation factor (NELF) and DRB sensitivity-inducing factor to allow induction of PRGs (Hargreaves and others 2009; Patel and others 2013). Knockdown of BRD2, 3, and 4 suggest that they are involved not only in the control of PRGs, but also for the regulation of all inducible genes (Nicodeme and others 2010). However, chemical inhibition of BRD reduced transcription of only a subset of genes, mainly SRGs that possess lower levels of histone acetylation in a naive cell. This suggests that histone acetyl-mimics of BRD proteins cannot outcompete the already high levels of histone acetylation on poised PRGs (Nicodeme and others 2010; Smale 2012). Epigenetic control of gene expression is outlined in Fig. 2.
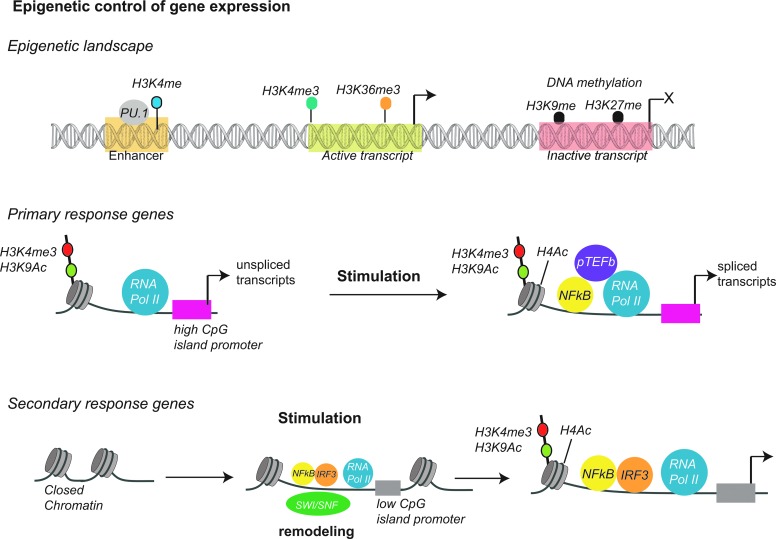
FIG. 2.
Epigenetic regulation in innate immunity. The TF PU1 promotes H3K4me1 histone mark, which is strongly associated with enhancer regions. H3K4me3 is associated with active promoters whereas H3K36me3 marks active transcripts. DNA methylation is associated with silencing of genes and the histone marks H3K9 and H3K27 are strongly associated with transcriptional silencing. PRGs already possess histone marks associated with active transcription, including H3K4me3 and H3K9ac before microbial activation. PRGs possesses poised RNA polymerase II and can produce unspliced transcripts before stimulation, whereas SRGs require chromatin remodeling by the SWI–SNF complex following stimulation to become transcriptionally active.
As mentioned earlier NFκB/Cebpδ and ATF3 coordinate the inflammatory program. NFκB alone is a weak inducer of IL6 and requires induction and activity of Cebpδ to amplify responses. Cebpδ recruits the histone acetyl transferase CBP to target promoters leading to histone acetylation, open chromatin, and increased transcription (Kovacs and others 2003; Litvak and others 2009). ATF3 in turn recruits HDAC1 to attenuate transcription of the same gene program (Gilchrist and others 2006; Litvak and others 2009). STAT1 has also been linked to chromatin remodeling in IFN-γ treated macrophages (Qiao and others 2013). Activation of macrophages with IFN-γ followed by a second signal such as TLR ligand results in sustained activation of TNF, IL6, and IL12b. IFN-γ priming marks these genes with STAT1 and H3K27 acetylation, enabling rapid transcription once a second signal is initiated. These results reveal the ability of IFN-γ to alter the epigenetic landscape such that it primes promoters and enhancers to reprogram subsequent responses to environmental cues. Phosphorylation of Histone H3 on Serine 10 (H3S10), demethylation of Lys9, and acetylation of Lys9 and 14 (H3K9/K14Ac) are also associated with transcriptional regulation and activation (Saccani and Natoli 2002; Ramirez-Carrozzi and others 2009). H3S10 phosphorylation through p38MAPK is specifically associated with activation of SRGs such as IL6 and IL12p40 (Saccani and Natoli 2002). Interestingly, pathogens such as Shigella Flexniri can alter H3S10 phosphorylation to block the activation of a subset of NFκB-responsive genes for their own benefit (Arbibe and others 2007).
Repressive epigenetic marks
Repressive histone modifications are equally important in the regulation of innate immunity. The histone methyltransferase G9a directs methylation of histone H3 on Lysine 9 (H3K9me) influencing DNA methylation and heterochromatin formation resulting in gene silencing (Ghisletti and others 2009; Stender and others 2012). H4K20me3 is an important repressive histone modification that modulates the expression of inflammatory genes (Stender and others 2012). DNA methylation occurs at cytosine residues within CpG dinucleotides. There is a strong relationship between DNA methylation and histone methylation in particular with H3K9me and H3K27me. These modifications are strongly associated with gene repression. DNA methylation mediated by DNMT3a and b results in strong repression of target genes and it is not as easily reversed (Cedar and Bergman 2009). H3K9 methylation is found at a subset of promoters of inducible genes such as IL12b and Ccl22, but this repressive mark is removed rapidly following LPS stimulation (Saccani and Natoli 2002). H3K27 trimethylation mediated by Ezh2 and the polycomb complex is essential for the maintenance of gene repression mainly for genes controlling cell fate and differentiation (De Santa and others 2007). Ubiquitination of histone 2A at lysine 9 (H2AK119) is another repressive mark inhibiting the basal expression of LPS inducible genes including Ccl5, CXCL2, and CXCL10 in macrophages. This inhibitory mark is reversed following signal-dependent induction of the deubiquitinating enzyme 2A-HUB (Zhou and others 2008).
Transcriptional Corepressors
Gene expression can also be controlled through transcriptional corepressors. Corepressors regulate transcription but do not by themselves bind DNA. Nuclear receptor corepressor (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) associate with a broad array of inflammatory gene promoters at basal levels in macrophages and their repressor functions are mediated through recruitment of histone deacetylases (Ghisletti and others 2009). Subsets of inflammatory genes are regulated by NCoR or SMRT and a select group of genes can be regulated by both corepressors. NCoR is directed to promoters in part through c-jun whereas SMRT is directed through translocation-ETS-leukemia. The dismissal of NCoR and SMRT are essential to allow signal-induced transcription of inflammatory genes (Ghisletti and others 2009).
Control of Basal Gene Expression
Although much attention is focused on the molecular basis of inducible gene expression, basal control of gene expression is equally important. Given the enormous proinflammatory potential of cytokines and other immune mediators, their expression must be kept in check to maintain cellular and tissue homeostasis. Any disruption in the basal control of inflammatory genes can have devastating effects on the host. For example, dysregulated TNF, IL1, or type I IFNs contribute to arthritis and autoimmunity. Elegant recent work from Aderem and colleagues have provided important insights into the regulatory networks controlling baseline expression of immune genes in macrophages (Litvak and others 2012). The TF FOXO3 was identified as a basal regulator of IRF7. FOXO3 deficient macrophages have more than 5 times the levels of IRF7 than their wild-type counterparts. This increase in IRF7 is associated with an increase in histone acetylation at the IRF7 locus. It appears that FOXO3 forms a complex with nuclear corepressor 2 (NCOR2) and histone deacetylase 3 (HDAC3) to control basal levels IRF7. Not surprisingly, FOXO3-deficient mice are more resistant to virus infection, however, this dysregulation of IRF7 comes at a cost as these animals have increased hemorrhaging and tissue damage (Litvak and others 2012). These animals may also be prone to autoimmunity as they age. Type I IFN expression is repressed by additional mechanisms, including H3K9me2. Levels of H3K9me2 at IFN and ISGs correlated inversely with the scope and amplitude of IFN and ISG expression in fibroblasts and DCs. Genetic ablation of G9a results in robust expression of type I IFN in cells that normally do not produce large amounts of this antiviral cytokine (Fang and others 2012). These studies once again highlight the importance of maintaining careful balance within all inflammatory pathways. Moderation of all signals is the key to maintaining cellular and tissue homeostasis.
Transcriptional Control of Inflammatory Gene Expression by LncRNA
Recent advances in deep sequencing technologies have provided new insights into the organization and regulation of the genome. Current estimates indicate that only 1%–2% of the genome has protein coding potential whereas 85% of the genome is transcribed (Hangauer and others 2013). One of the largest groups of RNA transcribed from the genome are lncRNAs. LncRNA are larger than 200 nucleotides in length and do not encode proteins. A number of lncRNA were discovered and characterized before 2005 (Pachnis and others 1988; Brannan and others 1990; Penny and others 1996), however, thousands of these transcripts have since been discovered in diverse cell types (Mortazavi and others 2008; Guttman and others 2009, 2010; Guttman and Rinn 2012; Rinn and Chang 2012). LncRNAs are emerging as major regulators of chromatin remodeling, transcription, and posttranscriptional regulation of gene expression in diverse biological contexts (Rinn and Chang 2012). LncRNA are now emerging as important regulators of gene expression within the immune system (examples are shown in Fig. 3).
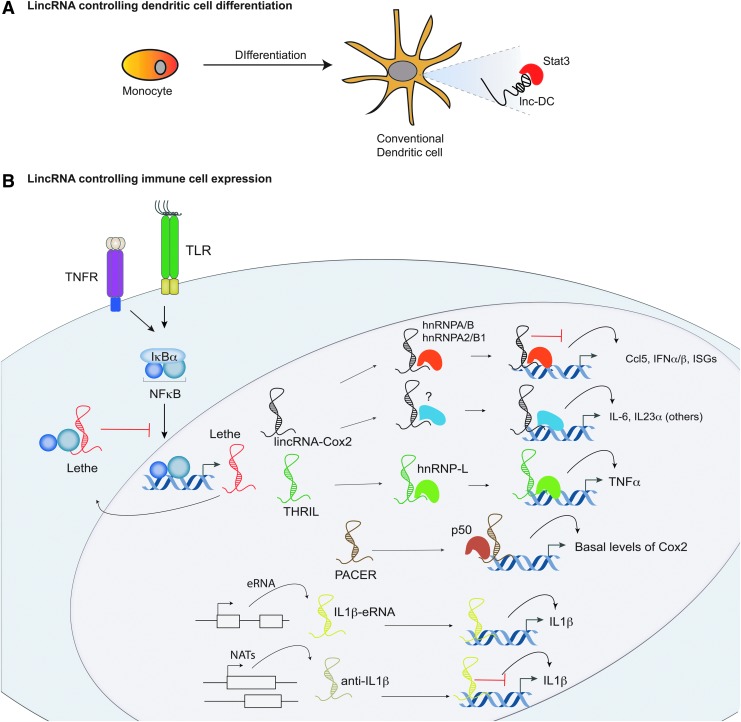
FIG. 3.
Long noncoding RNA (lncRNA) regulation within the Toll-like receptor (TLR) signaling pathway. A, Lnc-dendritic cell (DC) is the first lncRNA identified to function in immune cell differentiation. Its role is in monocyte to conventional DC differentiation. B, Recent studies have identified lncRNAs that play critical roles in the TLR signaling pathway. LincRNA-Cox2 is required to maintain basal levels of interferon-stimulated genes (ISGs) through interactions with hnRNP-A/B and A2/B1. LincRNA-Cox2 also possesses activating functions and is critical for the induction of proinflammatory genes following microbial challenge. A long noncoding pseudogene RNA named Lethe can act as a negative regulator of TLR signaling. Lethe can bind to RelA, a subunit of NFκB heterodimeric complex preventing NFκB from binding to promoter regions of target genes. A lincRNA termed THRIL acts to regulate TNF-α expression in human monocytes through its interactions with hnRNP-L. LincRNA PACER controls basal levels of Cox2 (PTGS2) through binding to the p50 subunit of NFκB. An enhancer RNA termed IL1β-eRNA functions to positively regulate IL1β production in human monocytes. Finally overexpression of a natural antisense transcript anti-IL1β inhibits IL1β production through alteration of the chromatin structure surrounding the IL1β promoter.
Some of the initial studies in this area found that lncRNAs were differentially regulated in virus-infected cells (Peng and others 2010) and in DCs following LPS stimulation (Guttman and others 2009). A lncRNA called NeST was identified and shown to control susceptibility to Theiler's virus and Salmonella infection in mice through epigenetic regulation of the IFN-γ locus (Collier and others 2012; Gomez and others 2013). NeST RNA binds WDR5 altering H3K4me3 at the IFN-γ locus, thereby increasing expression of IFN-γ. Our group has identified a number of immune regulated lncRNA genes that are differentially regulated following an inflammatory stimulus (Carpenter and others 2013). One of these lncRNAs, lincRNA-Cox2, was highly inducible in both macrophages and DCs exposed to TLR ligands. A series of loss and gain of function approaches showed that lincRNA-Cox2 in turn controlled basal levels of ISGs and inducible expression of proinflammatory cytokines following microbial challenge (Carpenter and others 2013). LincRNA-Cox2 represses expression of ISGs through interactions with hnRNP-A/B and A2/B1. The exact mechanisms involved in lincRNA-Cox2-mediated control of proinflammatory gene expression remains to be determined. Another lincRNA Lethe has recently been described as a negative regulator of NFκB. Lethe can directly bind to RelA, a subunit of NFκB and prevent NFκB from binding to promoter regions of target genes (Rapicavoli and others 2013). An additional lincRNA called THRIL [TNFα and heterogeneous nuclear ribonucleoprotein L (hnRNPL) related immunoregulatory LincRNA] was linked to regulation of TNFα expression in human monocytes through its interactions with hnRNP-L (Li and others 2014). Finally a lncRNA named PACER (p50-associated Cox2 extragenic RNA) was recently shown to act as a regulator of constitutive levels of Cox2 (PTGS2) expression. PACER was shown to directly interact with p50, the inhibitory component of NFκB complex, and prevent it from binding to the promoter of the adjacent Ptgs2 gene (which encodes Cox2) (Krawczyk and Emerson 2014). Collectively, these studies highlight the importance of lncRNAs in the regulation of gene expression in macrophages (Fig. 3).
LncRNA are classified based on their transcription relative to protein-coding genes. There are lncRNA that are transcribed from intronic regions within a protein-coding transcript, or divergent to a protein-coding gene (within 5′ proximal regions). LncRNA can be intergenic located between 2 protein-coding genes. Many lncRNA overlap with protein-coding genes and are referred to as natural antisense transcripts (NATs). Most recently, noncoding RNAs have been found to be transcribed from enhancer regions, adding yet another layer of complexity to the transcriptome (De Santa and others 2010; Garmire and others 2011). Indeed these enhancer RNAs (eRNA) have been implicated in innate immunity. IIott and others identified 221 differentially regulated lncRNA in primary human monocytes. Interestingly, they show that 58% of these lncRNA are marked by a high H3K4me1/H3K4me3 ratio suggesting these lncRNA are in fact eRNAs (IIott and others 2014). They show evidence for an eRNA downstream of IL1β as well as an upstream region of bidirectional transcription. Knockdown of these transcripts referred to as IL1β-eRNA and IL1β-RBT46 in turn attenuated LPS-induced IL1-induction and release (IIott and others 2014). Another study by Lu and others identified an antisense transcript to IL1β in murine macrophages that functions to inhibit IL1β expression by altering chromatin structure surrounding the IL1β promoter (Lu and others 2013). These authors identified 27 additional antisense transcripts that are antisense to a broad collection of innate immune genes. Collectively these studies raise the potential for a broader level of regulation of immune genes by NATs.
A lncRNA called lnc-DC was recently identified during monocyte to DC differentiation in humans. Wang and others identified lnc-DC in human conventional DCs. LncRNA-DC promotes the activation of STAT3 in the cytoplasm enabling the STAT3 transcriptional program to occur (Wang and others 2014). This work suggests that lncRNA are capable of interacting with signaling molecules within the cytoplasm to impact cellular signaling and cell differentiation.
Our understanding of lncRNA function in immunity is at its infancy. There is a growing literature from other fields defining the importance of lncRNA in various aspects of transcriptional and posttranscriptional regulation of gene expression. Indeed many lncRNA display aberrant expression patterns in disease contexts such as cancer. A recent study identified lncRNA SChLAP1, which is highly overexpressed in prostate cancer (Prensner and others 2013). Interestingly SChLAP1 negatively regulates the function of the SWI–SNF chromatin-remodeling complex. It will be interesting to determine if this lncRNA could impact the TLR signaling pathway especially SRGs that require SWI–SNF complex for their induction. The study of lncRNA in innate immunity is a fast growing area of research and we anticipate an exciting time ahead as researchers unravel the roles that these genes play in controlling inflammatory processes and their potential to be dysregulated in disease.
Conclusions and Future perspectives
This review highlights the vast array of mechanisms that have evolved to control the inflammatory pathway. This is a highly complex process which is critical to protect the host during infection. However, there are many opportunities for these systems to be inappropriately activated or regulated resulting in disease. We believe a major future goal should be to target factors important in transcription of different inflammatory gene subsets. Many of the studies described in this study have focused mainly on the specific signal transduction pathways activated by certain stimuli and many kinases involved in the downstream pathways have been targeted for therapeutic manipulation. However, events occurring in the nucleus involving, TFs, lncRNA, and chromatin modifiers are also all viable areas for manipulation. Obtaining a better understanding of the subsets of genes controlled by each of these factors will facilitate the rationale design of more specialized and accurate anti-inflammatory agents.
Author Disclosure Statement
No competing financial interests exist.
References
- Amit I, Garber M, Chevrier N, et al. . 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326(5950):257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, et al. . 2007. An injected bacterial effector targets chromatin access for transcription factor NF-|[kappa]|B to alter transcription of host genes involved in immune responses. Nat Immunol 8(1):47–56 [DOI] [PubMed] [Google Scholar]
- Aringer M, Gunther C, Lee-Kirsch MA. 2013. Innate immune processes in lupus erythematosus. Clin Immunol 147(3):216–222 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. . 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447(7146):799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol 10(1):28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. 2008. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene 27(19):2772–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, et al. . 2013. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341(6147):789–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10(5):295–304 [DOI] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. 2012. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol 189(5):2084–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, et al. . 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8(5):e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130(6):1083–1094 [DOI] [PubMed] [Google Scholar]
- Do-Umehara HC, Chen C, Urich D, et al. . 2013. Suppression of inflammation and acute lung injury by Miz1 via repression of C/EBP-δ. Nat Immunol 14(5):461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang TC, Schaefer U, Mecklenbrauker I, et al. . 2012. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med 209(4):661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447(7147):972–978 [DOI] [PubMed] [Google Scholar]
- Foster SL, Medzhitov R. 2009. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol1 30(1):7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T, Sen R, Roy AL. 2011. Regulation of primary response genes. Mol Cell 44(3):348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, et al. . 2012. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell 47(5):810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmire LX, Garmire DG, Huang W, Yao J, Glass CK, Subramaniam S. 2011. A global clustering algorithm to identify long intergenic non-coding RNA - with applications in mouse macrophages. PLoS One 6(9):e24051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, et al. . 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32(3):317–328 [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, et al. . 2009. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 23(6):681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, et al. . 2006. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441(7090):173–178 [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, et al. . 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152(4):743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, et al. . 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, et al. . 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28(5):503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482(7385):339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9(6):e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell1 38(1):129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath V, Suh HC, Holman M, et al. . 2004. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood 104(6):1639–1647 [DOI] [PubMed] [Google Scholar]
- Heinz S, Glass CK. 2012. Roles of lineage-determining transcription factors in establishing open chromatin: lessons from high-throughput studies. Curr Top Microbiol Immunol 356:1–15 [DOI] [PubMed] [Google Scholar]
- IIott NE, Heward JA, Roux B, et al. . 2014. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 5: 3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293(5532):1074–1080 [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. 2003. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem 278(38):36959–36965 [DOI] [PubMed] [Google Scholar]
- Krawczyk M, Emerson BM. 2014. p50-Associated COX2 Extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife (Cambridge) 3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int Rev Immunol 30(1):16–34 [DOI] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, et al. . 2013. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39(6):1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chao T-C, Chang K-Y, et al. . 2014. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 111(3):1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, et al. . 2009. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 10(4):437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ratushny AV, Lampano AE, et al. . 2012. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490(7420):421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu X, Hong M, Tobias P, Han J. 2013. A potential suppressive effect of natural antisense IL-1β RNA on lipopolysaccharide-induced IL-1β expression. J Immunol 190(12):6570–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9(10):692–703 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, et al. . 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448(7153):553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, Fisher EA. 2013. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol 13(10):709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628 [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, et al. . 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468(7327):1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, et al. . 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152(1–2):157–171 [DOI] [PubMed] [Google Scholar]
- Pachnis V, Brannan CI, Tilghman SM. 1988. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 7(3):673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MC, Debrosse M, Smith M, et al. . 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 33(12):2497–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Armour CD, et al. . 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio1 1(5):e00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379(6561):131–137 [DOI] [PubMed] [Google Scholar]
- Pham TH, Benner C, Lichtinger M, et al. . 2012. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 119(24):e161–e171 [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, et al. . 2013. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45(11):1392–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Giannopoulou EG, Chan CH, et al. . 2013. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity 39(3):454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, et al. . 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138(1):114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, et al. . 2006. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev 20(3):282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. 2013. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Natoli G. 2002. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev 16(17):2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. 2001. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med 193(12):1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Yamamoto M. 2013. Pathogen recognition receptors: ligands and signaling pathways by Toll-like receptors. Int Rev Immunol 32(2):116–133 [DOI] [PubMed] [Google Scholar]
- Sims RJ3, Belotserkovskaya R, Reinberg D. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18(20):2437–2468 [DOI] [PubMed] [Google Scholar]
- Smale ST. 2010. Selective transcription in response to an inflammatory stimulus. Cell 140(6):833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. 2012. Transcriptional regulation in the innate immune system. Curr Opin Immunol 24(1):51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Pascual G, Liu W, et al. . 2012. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell 48(1):28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98(1):1–4 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9(6):465–476 [DOI] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3(6):920–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor AF, Borras FE, Cullell-Young M, Celada A. 1998. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol 63(4):405–417 [DOI] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, et al. . 2014. The STAT3-Binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science 344(6181):310–313 [DOI] [PubMed] [Google Scholar]
- Yamamoto KR, Alberts BM. 1976. Steroid receptors: elements for modulation of eukaryotic transcription. Annu Rev Biochem 45:721–746 [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, et al. . 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29(1):69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]