Abstract
Background
A human betaretrovirus (HBRV) has been linked with primary biliary cirrhosis (PBC) following the detection of viral particles in biliary epithelium by electron microscopy and cloning of the betaretrovirus genome from biliary epithelium and peri-hepatic lymph nodes. Evidence for viral infection was found in the majority of PBC patients' peri-hepatic lymph node samples. However, less than a third of the liver samples had detectable HBRV, whereas others were unable to detect betaretrovirus infection or noted the presence of virus in the liver of patients with other diagnoses.
Aims
To address the hypothesis that the betaretrovirus may be below the limits of detection in the liver, biliary epithelial cells (BEC) were investigated for the evidence of infection.
Methods
Ligation-mediated PCR and next generation sequencing were used to detect proviral integrations in liver, lymph nodes and BEC isolated from liver transplant recipients. Hybridisation-based assays were used to detect betaretroviral RNA in BEC.
Results
Unique HBRV integrations and betaretrovirus RNA were detected in the majority of biliary epithelia derived from patients with PBC, autoimmune hepatitis and cryptogenic liver disease but rarely in other liver transplant recipients with primary sclerosing cholangitis and other hepatic disorders. HBRV integrations were commonly found in PBC patients' lymph nodes but rarely in whole liver samples.
Conclusions
Human betaretrovirus infection is frequently observed at the site of disease in patients with primary biliary cirrhosis and also in biliary epithelium of patients with autoimmune hepatitis and cryptogenic liver disease.
Introduction
It is thought that primary biliary cirrhosis (PBC) occurs as a result of a combination of genetic and environmental factors.1, 2 Bacteria, viruses and xenobiotics have all been proposed as potential aetiological agents but none has been causally associated with PBC.1 We have focused attention on studying a viral association with PBC following the discovery of serum reactivity to retroviral proteins, detection of virus-like particles in biliary epithelial cells (BEC) and identification of viral sequences from patients' samples.3–5 We cloned a proviral genome sharing 91–99% nucleotide identity with the mouse mammary tumour virus (MMTV) and the ‘human mammary tumour virus’ cloned from human breast cancer samples.4–6 As the latter are betaretroviruses, we referred to the agent found in PBC patients as the human betaretrovirus (HBRV).
Of interest, HBRV has been connected with a disease-specific phenotype of PBC associated with the production of anti-mitochondrial antibodies (AMA). It is well described that AMA recognise pyruvate dehydrogenase complex-E2 (PDC-E2) and related oxo-acid dehydrogenase complexes. In healthy cells, PDC-E2 is mainly located within the mitochondrial inner membrane, whereas antigens resembling PDC-E2 are markedly increased in PBC patients' biliary epithelium and peri-hepatic lymph nodes.5, 6 In our preliminary studies, HBRV proteins were found in the same PBC patients' cells that also displayed increased expression of the PDC-E2-like proteins.5 In vitro, BEC co-cultivated with HBRV or pure isolates of MMTV developed increased expression of the PDC-E2-like protein, whereas other viruses had no such effect.5, 7
Clinical studies from our laboratory and other groups have suggested a very low level of betaretrovirus in the liver of patients with PBC.5, 8 This is somewhat similar to observations in mice, where the virus is trafficked in lymphocytes and not found as a free particle in blood.9 Prior studies from our laboratory determined that only one in four patients had detectable HBRV in serum representing a copy number that rarely exceeded 102/mL, which likely represents viral nucleic acid liberated from lysed cells rather than a true viremia.8 In comparison, the majority of patients with untreated HIV usually have a two-log fold higher or greater viral load of 104–107/mL10 and a viral load of 104–105/mL may be found in peripheral blood mononuclear cells from patients with HTLV.11
Our original tissue studies in liver disease patients using reverse transcription (RT)-PCR and immunochemistry reported the presence of HBRV RNA and HBRV proteins in the majority of PBC peri-hepatic lymph nodes, whereas less than a third of frozen liver samples from PBC patients had detectable HBRV RNA and none had demonstrable protein.4, 5 Two further studies assessing hepatic DNA had conflicting results; one laboratory was unable to detect HBRV DNA using a single round of PCR,12 whereas another laboratory detected HBRV using nested PCR.13 However, the latter group did not confirm a specific association of HBRV with PBC, as patients with alcohol-related cirrhosis had a similar prevalence of infection.13 In accord with both studies, we could not detect HBRV DNA with a single round of PCR and only detected virus in less than 20% of PBC liver samples using nested PCR. Accordingly, the reports from all laboratories were in agreement that HBRV was difficult to detect in the liver and the main differences with reports concerned the interpretation of the data.4–6, 12, 13 The main procedural differences reported from the different laboratories were the methods and samples employed. Specifically, no other group investigated the presence of HBRV RNA, which is more abundant than the proviral DNA; neither have other laboratories sought evidence of HBRV in BEC at the site of the disease, nor within peri-hepatic lymph nodes, a major reservoir of viral infection.4–6
Taken together, the data suggest that either HBRV is either not present in the liver of PBC patients or is below the limits of detection. Given the conflicting data, several commentaries have suggested that the gold standard for viral detection should be applied by demonstrating the junction of the retrovirus long terminal repeat (LTR) integrating into the human genome in a large number of PBC patients.12, 14, 15 We therefore used ligation-mediated (LM) PCR16 with a HBRV LTR primer and an adaptor primer to the adjacent human DNA sequence (Figure S1) and then deep sequenced the amplification products with more than 2 million reads per sample to identify as many of the virus/human junction regions as possible.11 Herein, we address the hypothesis that HBRV may reside in the biliary epithelium and report the frequent detection of HBRV RNA and proviral integrations in BEC extracted from patients with autoimmune and cryptogenic liver disease.
Materials and Methods
Study design
The study was designed as a descriptive prevalence study to determine the distribution HBRV in subjects with liver disease. Liver and lymph node samples were collected from December 2004 to January 2013 from 90 patients undergoing liver transplantation and four patients undergoing hepatic resection at the University of Alberta, Edmonton, Canada. Biliary epithelial cell extractions were performed within 24 h of collection using previously established protocols employing immune-selection with anti-HEA125, as described.5, 17 The liver samples prioritised for biliary epithelial preparations included those from patients with PBC, primary sclerosing cholangitis (PSC), autoimmune hepatitis (AIH), and cryptogenic liver disease as well as patients with nonviral liver disease. Accordingly, the available BEC samples influenced the comparison group of patients used for contrasting the prevalence HBRV in patients with PBC. As patients with AIH had previously been shown to harbour HBRV,8 they were assessed as a separate group of patients as were patients with cryptogenic liver disease with unknown disease aetiology. Accordingly, the liver disease ‘control’ group mainly consisted of patients with PSC, the nonviral liver diseases listed below as well as the four patients with nondiseased BEC from patients with focal nodular hyperplasia (FNH) or hemangioma resection.
Patients and samples
For this study, 136 samples were collected from 94 patients that included 38 peri-hepatic lymph nodes, 36 liver samples and 61 BEC isolates. Where possible, an attempt was made to match samples from the same patient. In total, one patient had all three samples available for study, 17 patients provided both liver and lymph node, 11 supplied liver and BEC, four patients provided lymph node and BEC and the remainder supplied a solitary sample. The samples were derived from patients with the following diagnoses: PBC (n = 39), AIH (n = 8), cryptogenic liver disease (n = 5), PSC (n = 19), and patients without biliary or autoimmune disease (n = 23). The latter included the following diagnoses, nonalcoholic steatohepatitis (NASH, n = 4), FNH (n = 3), alcoholic cirrhosis (n = 3), glycogen storage disease (n = 2) and one patient each with acetaminophen induced fulminant hepatic failure, biliary atresia, Budd Chiari syndrome, erythropoietic protoporphyria, factor 2 deficiency, fibrolamellar hepatocellular carcinoma, giant cell hepatitis, hyperoxaluria, hemangioma, sarcoidosis and short gut syndrome. All patients with PSC and cryptogenic liver disease that were tested for AMA were found to be AMA negative, whereas one of the four patients with AIH that were tested for AMA was positive. All procedures were conducted in accordance with the institutional ethics review board at the University of Alberta.
Detection of betaretrovirus RNA in biliary epithelium
Hybridisation-based QuantiGene (Panomics/Affymetrix, Inc., Freemont, CA, USA) technologies were used to detect HBRV RNA in BEC, either from the cell lysates (QG 2.0 assay) or by in situ hybridisation (ViewRNA assay). The probes were designed and synthesised by Panomics, using highly conserved regions in the gag-pro-pol genes from RefSeq NC_001503.1. For the QG 2.0 assay, approximately 1 million BEC from a low passage (<4) were processed with lysis buffer, loaded in duplicates into a 96-well assay plate and hybridised with probe. The resulting luminescence was quantified with a VICTOR3 plate reader (PerkinElmer, Waltham, MA, USA) and reported as the intensity above a blank solution background. Samples with a positive value were reported as having HBRV RNA, whereas samples with a negative value were assigned a value of zero. For the ViewRNA in situ hybridisation assay, 103–104 cells from BEC were loaded in duplicates into a 96-well assay plate, fixed with 4% formaldehyde, digested with Proteinase K and hybridised in solution containing the probe set for 3 h and stained with DAPI. HBRV RNA was assessed in available BEC using the QG 2.0 assay (n = 54) and by ViewRNA (n = 26). The MMTV producing cell line, Mm5MT, was used as a positive control for the ViewRNA assay.
Ligation mediated-PCR
Genomic DNA was extracted from Mm5MT, BEC, lymph nodes and liver for processing by LM PCR to identify retroviral integration sites, as described.16 Genomic DNA was digested with MseI, and double stranded DNA adaptors, Linker-1 (GTAATACGACTCA CTATAGGGCTCCGCTTAAGGGAC) and Linker-2 (PO4-TAGTCCCTTAAGCGGAG-NH2) were ligated onto the DNA. The ligation products were then amplified using the HBRV LTR Outer Primer (CGTCTCCGCTCGTCACTTAT) and the Linker Outer Primer (GTAATACGACTCACTATAGGGC). Then nested PCR was performed with the HBRV-LTR Inner Primer (GCAGACCCCGGTGACCCTCAG) and the Linker Inner Primer (AGGGCTCCGCTTAAGGGAC). As the integrated provirus contains two LTRs, the LM-PCR process has the capacity to amplify an internal proviral fragment as well as the region where the LTR integrated into the human genome (Figure S1a). The LM-PCR products were cloned into a pGEM-T Easy vector for capillary sequencing with the ABI 3730 DNA Analyzer that generated long sequences encompassing the entire length of the LM-PCR product (Figure S1b). Subsequently, the LM-PCR products were cloned into Illumina libraries using the Genomic DNA Sample Preparation Kit for paired end sequencing with the HiSeq 2000 or the MiSeq platforms. The next generation sequencing generated a shorter read length that demonstrated the important features of the LM-PCR product in either a single end read (Figure S1c) or in the reads derived from each of the paired ends (Figure S1d and e).
Informatics pipeline for detection of proviral integrations
By definition, each integration site contained the HBRV 3′ LTR fragment, and within three bases of the LTR, a human sequence with at least 95% identity to the human genome (hg19 assembly) that was then followed by the linker sequence.16 Whereas complete integration sites were identified in the longer capillary reads, this was observed to a lesser degree in the Illumina sequences given their 75–250 bp read lengths (Figure S1). Most of the integrations in the Illumina libraries contained the HBRV LTR in one paired-end and the linker sequence in the other.
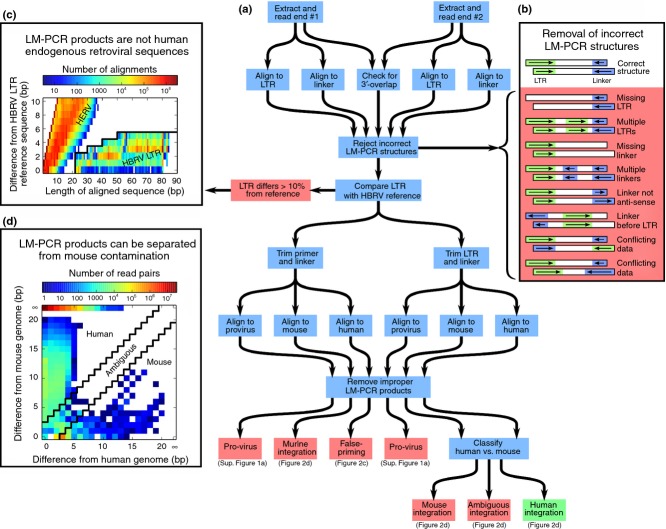
Therefore, the informatics pipeline was constructed to read and combine both ends of the LM-PCR product, while also excluding proviral internal fragments primed from the LTR, false-primed human genomic sequences and also any sequence that may have arisen as a result of contamination from mouse DNA (Figure 1). The pipeline began by removing the primer/adapter sequences as well as bases with a low quality score (Phred score <13) from the 3′ end of each Illumina read. Bowtie2 was used to align the paired end reads against the reference linker sequence and a viral reference (NCBI RefSeq accession: NC_001503.1). Incorrect LM-PCR structures were discarded (Figure 1b) and LM-PCR products were removed if the linker sequence did not match or if the LTR sequence had less than 90% identity with the HBRV LTR reference sequence. All false-primed human genomic sequences including human endogenous retrovirus sequences were removed (Figure 1c). As murine genomic DNA is commonly found in laboratory reagents,18 the pipeline searched for MMTV-mouse genomic integrations. These could be readily differentiated from human integrations because syntenic regions for the two genomes are only 69% identical on average (for example, see table 16 of ref.19). However, for the shortest of the sequences that we considered (21 bp) much higher degrees of identity were sometimes seen. Therefore, the pipeline required that the human region within the LM-PCR sequence had to share a 95% identity to the human genome and also had to include three more matching base-pairs than any alignment found within the mouse genome (mm10). This requirement of a high quality human alignment with no comparable murine sequence ensured that mouse genomic sequences that varied from their reference were not confused with human data-even at the cost of rejecting some legitimate human cases (Figure 2d). Libraries with more than 200 potential MMTV-murine genome integrations were discarded.
Figure 1.
Pipeline used to detect HBRV integrations from next generation sequencing data. (a) The paired ends were analyzed for the correct structure containing the HBRV 3′LTR fragment, a human sequence within three bases demonstrating at least 95% homology to the human genome and the linker sequence. (b) High-quality Illumina sequences were aligned against a viral reference and the reference linker sequence using bowtie2 and incorrect structures were discarded. (c) All sequences were removed if the LTR sequence demonstrated less than 90% homology to the reference HBRV LTR. To evaluate the possibility of false priming to endogenous retrovirus and other human sequences, the distribution of observed HBRV LTR sequences (under the back line) was plotted against a simulation of all human genomic sequences similar to the HBRV LTR. None of the human sequences coincided with HBRV LTR and only 18 of the 50 million human sub-sequences were found adjacent to the HBRV region. (d) Sequences were removed if they resembled MMTV mouse DNA integrations. In the plot of aligning LM-PCR sequences to human and murine genomes, the majority of sequences only aligned to the human genome (98.1% of all reads observed on the top edge, reported as infinitely different from mouse). Most of the other sequences were observed on the left edge (1.4%), demonstrating only a few base differences from the human genome and these sequences were retained; whereas the ambiguous (0.1%) or more mouse-like sequences (0.4%) on the bottom edge were rejected. Once the improper reads with mouse-like sequences, false primed human products and internal provirus LM-PCR products were removed, the reads were determined to be a human integration and these were plotted onto the human genome (Figure 2).
Figure 2.
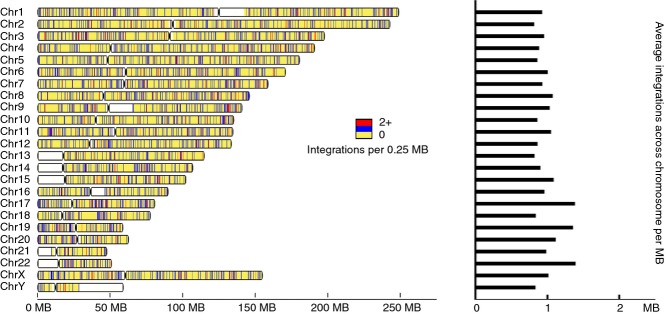
Human betaretrovirus integration sites. (a) HBRV integrations were mapped to human genome 19 and shown in blue as single integrations and in red for >two per 250 kb on a yellow background for known human chromosomal regions, where white represents unsequenced regions. The density of integration sites per Mb averaged across the chromosome is displayed in the histogram.
Following the removal of false-primed human genomic products lacking HBRV LTR, mouse like sequences and the internal HBRV proviral product (Figure S1a), duplicate sequences were merged and mapped to the human genome for reference (Figure 2). Integrations found in more than one library were identified as another potential source of contamination from sample to sample and removed. The final ‘unique’ integration data set was restricted to HBRV human genome integrations found in only one sample.
Sequence variability in HBRV LTR
Variations within the recovered HBRV LTR fragments were assessed to check for the sequence variability that is expected from a virus with an error-prone polymerase. LTR variants with a frequency less than 1% of the reads from any sample were rejected as likely sequencing errors (Figure S2). The human-derived LTR fragments were compared with their mouse counterparts. A consensus phylogenetic tree including all HBRV LTR sequences was generated by the MEGA4 software using a neighbour-joining method with 1000 replicates.
Statistics
Categorical differences between groups were evaluated by Fisher's exact test or χ2 and continuous variables by paired t-test using prism 5.0 software. P values <0.05 were considered significant and two sided P values were used in circumstances where no prior association had been established.
Results
Confirmation of HBRV integration
As more than 8% of the human genome contains human endogenous retrovirus sequences, a rule was inserted into the pipeline that HBRV LTR sequence found in the LM-PCR product had to demonstrate 90% or greater similarity to the reference betaretrovirus LTR. This rule helped to ensure that inadvertent amplification of human endogenous retroviruses did not impact on detecting true HBRV integrations. This cut-off was established because a search of the human genome showed the closest nucleotide similarity of any human endogenous retrovirus sequence was 84% with the HBRV LTR. We then conducted a survey to find sequences resembling the HBRV LTR in the human genome and identified more than 50 million sequences. An alignment of these human sequences with the HBRV LTR sequences detected by LM-PCR showed that the pipeline rejected all the human sequences similar to HBRV LTR (Figure 1c). However, 18 short human sequences were closely related to the HBRV LTR and even accounting for the potential of sequencing error, these data suggest that the vast majority of any human endogenous retroviral sequences amplified by the LM-PCR process would be discarded by the pipeline (calculated as less than 1.4 × 10−7).
As part of the established pipeline, we searched for MMTV-mouse genomic integrations as murine genomic DNA is commonly found in laboratory reagents.18 During this process, we identified libraries contaminated by LM-PCR products from the MMTV producing MM5MT mouse breast cancer cells inadvertently sequenced with clinical samples (Figure S3). As a result, we discarded three libraries with more than 200 mouse integrations. All other LM-PCR sequences were removed if they resembled murine-like integrations or ambiguous sequences, with either 95% homology to both human and murine genomes or within 3 bp of the two genomes. In the process, 0.5% of the reads were removed representing approximately 2 (median) mouse-like sequences in 61% of the libraries. While true HBRV human integrations were probably removed by the procedure, it minimised the concern of contaminating mouse DNA impacting on the final data set.
At completion of the analysis, a total of 2223 HBRV-human proviral integration sites were identified in patient samples. These were mapped onto the human genome (Figure 2) and the distribution was random without apparent clustering in specific regions, as predicted from prior studies.16 To identify unique integrations, all HBRV-human integrations found in more than one library were removed to create the final data set of 1518 human integrations used for determining the frequency of infection within patients' samples. This stringent process also eliminated true HBRV human integrations but reduced concern that data may be attributable to PCR carry-over.
Sequence variability in HBRV LTR
A further analysis was performed to investigate the sequence variability in HBRV LTR in all samples. Retrovirus polymerase genes are error prone, so one would expect to see variance in the genomic sequence of individual viral particles, whereas little divergence would occur with PCR artefact. Therefore, single nucleotide polymorphisms within the recovered HBRV LTR fragments were evaluated from all the LM-PCR products. To determine the impact of sequencing errors leading to single nucleotide polymorphisms, we performed an assessment of all the HBRV LTR reads found in the LM-PCR products. As a result, sequences found with single nucleotide polymorphisms at a frequency of less than 1% of the reads from individual samples were excluded as possible sequencing artefacts (Figure S2). Multiple variants were observed in the phylogenetic analysis of HBRV LTR sequences, where 16 of 19 LTRs were found to be novel in comparison to known MMTV LTR sequences (Figure 3). The variability in LTR sequences in different samples provided reassurance against a single source of viral contamination in cell culture from a virus producing cell line, as suggested in a recent report concerning gammaretroviral sequences detected in human samples.18
Figure 3.
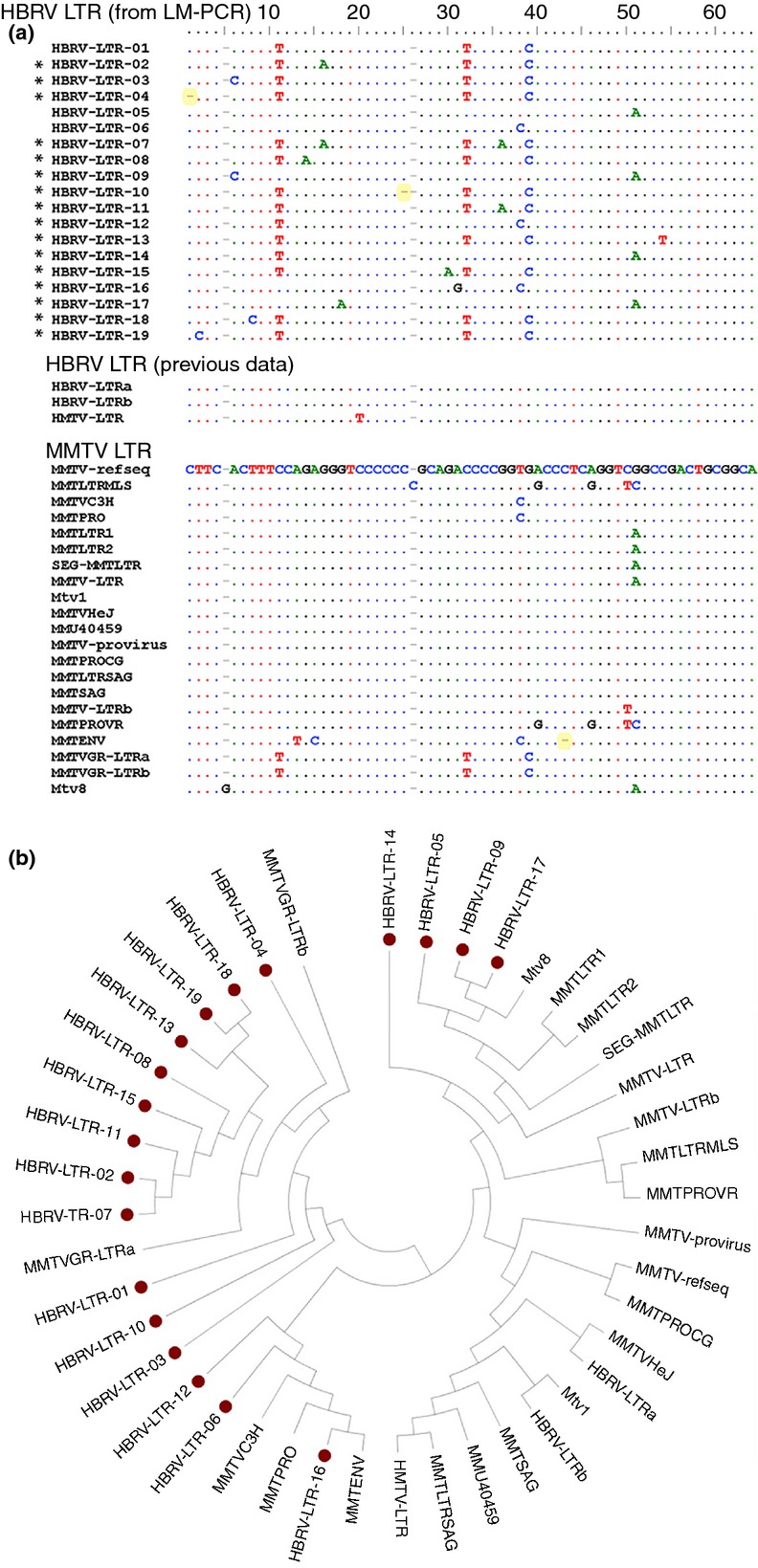
Variable HBRV LTR sequences detected by LM-PCR. (a) Alignment of HBRV LTR sequences from the LM-PCR studies with MMTV LTR derived from NCBI, demonstrating that 84% of the human LTR sequences differ from known mouse LTRs based on SNP and InDel combinations [InDels labelled yellow and novel HBRV LTR unrelated to MMTV labelled with *]. (b) Phylogenetic analysis of the same sequences showing a degree of clustering of human-derived LTR sequences (red circle denotes HBRV LTR derived by LM-PCR). The consensus phylogenetic tree was generated with the MEGA4 software using a neighbour-joining method with replication of 1000.
Frequency of detecting HBRV in tissue samples
The patient samples were assessed as four separate groups that included PBC, AIH and cryptogenic liver disease (that may potentially harbour HBRV) as well as the remainder of ‘other liver’ group of PSC and predominantly nonviral liver disease patients and BEC from four healthy subjects with either FNH or hemangioma. In the biliary epithelium analyses, we found that 58% of BEC from patients with PBC had detectable HBRV integrations as compared to 7% of the liver controls (P = 0.0002, Figure 4). A similar proportion of PBC patients had detectable HBRV RNA in BEC lysates using the QG 2.0 assay (59% vs. 15% of liver control BEC, P < 0.001, Figure 5) or by in situ hybridisation using the ViewRNA assay (75% vs. 13% liver controls, P < 0.02, Figure 5). Of note, 50% of the BEC from the patients with AIH (including the one AMA positive patient) also had an increased frequency of HBRV integrations as well as evidence of HBRV RNA in BEC, consistent with previous reports of detecting HBRV in blood samples from patients with AIH.8 HBRV proviral integrations and HBRV RNA were also observed in a proportion of BEC derived from patients with cryptogenic liver disease (Figures 4 and 5). BEC with HBRV integrations from the liver disease group were derived from two patients with PSC undergoing their first and second liver transplant, respectively. In the HBRV RNA studies, the four positive liver disease patients included the latter PSC patients, as well as two other patients with NASH and FNH, respectively.
Figure 4.
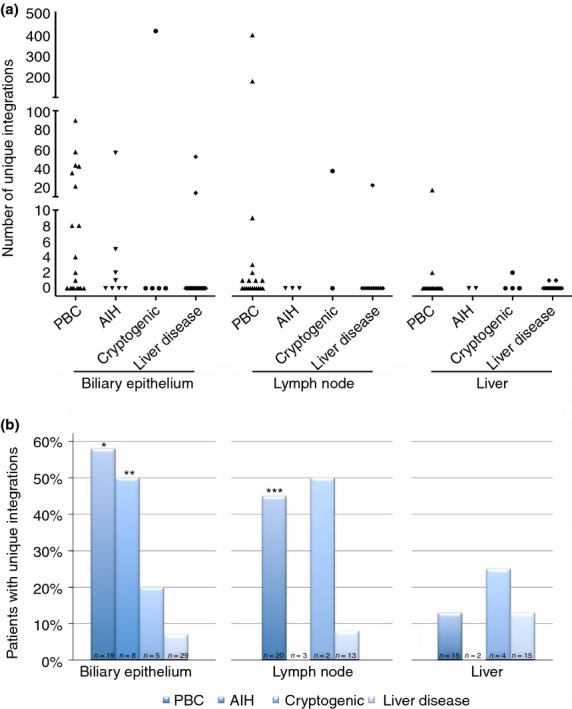
Frequency and proportion of HBRV integration sites among liver disease patients. (a) The highest frequency of proviral integrations was observed in BEC and lymph nodes from patients with PBC, AIH or cryptogenic liver disease. (b) Proviral HBRV integrations were found in the majority of BEC from PBC and AIH patients, less so lymph nodes and rarely in the liver (*P = 0.0002, **P = 0.01 and ***P < 0.05 vs. liver disease control subjects).
Figure 5.
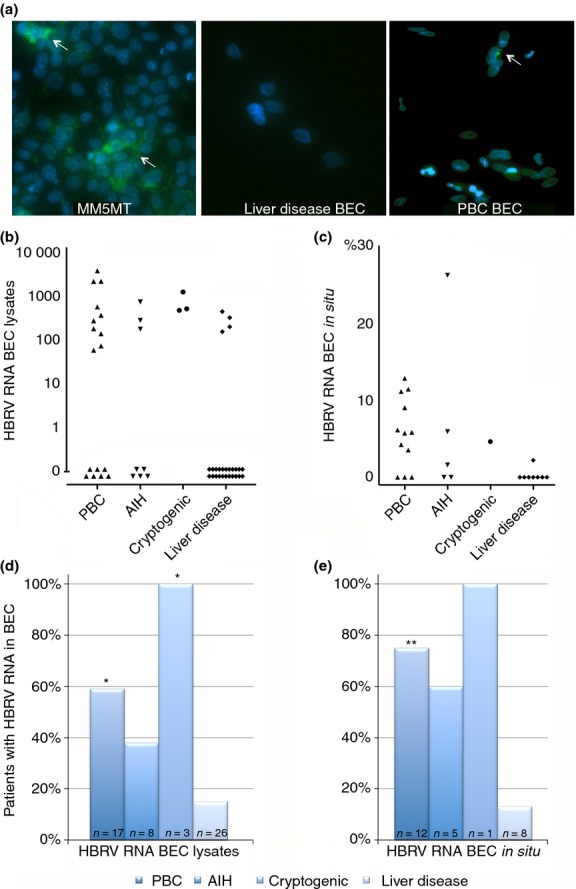
Detection of HBRV RNA in BEC. (a) Green signal (white arrows) indicative of HBRV RNA was observed in the cytoplasm of MMTV producing MM5MT cells, biliary epithelium from a patient with PBC, but not in the uninfected BEC (DAPI blue stain for nuclei; magnification ×200). (b) HBRV RNA was detected in BEC lysates using QG 2.0 assay and recorded as optical density units increased over baseline. (c) The percentage of BEC with positive in situ hybridisation signal was assessed using the ViewRNA assay. HBRV RNA was found in a similar proportion of BEC from PBC patients as those with proviral integrations by either (d) the QG 2.0 assay or (e) the View RNA assay (*P < 0.01 and **P < 0.02 vs. liver disease control subjects).
The frequency of detecting HBRV integrations in BEC was consistently higher than those observed within the liver. Indeed, proviral HBRV integrations were seldom detected in liver samples from PBC patients or controls and found at a frequency previously reported in prior studies.5, 12, 13 We observed a higher proportion of PBC lymph nodes contained HBRV integrations as compared to liver disease control lymph nodes samples. However, the detection rate was at a lower level than previously reported using RT-PCR and immunohistochemistry (45% with HBRV integrations vs. 75% using RT-PCR and immunohistochemistry5); the differences in frequency of detection were possibly due to the stringent pipeline procedure of eliminating integrations and sensitivities of the different assays. The HBRV integrations found in the liver control samples were a liver and lymph node sample from a patient undergoing liver transplantation for erythropoietic protoporphyria and a liver sample from a patient undergoing liver transplantation for biliary atresia.
A 75% agreement was observed in the detection of HBRV proviral integrations and HBRV RNA in BEC (Table 1). Samples from four patients were HBRV integration positive and HBRV RNA negative, suggesting that these patients may not have had transcriptionally active HBRV in the isolated BEC. In contrast, nine BEC samples were found to be HBRV RNA positive but negative for HBRV integrations. While some of these BEC had a low percentage of HBRV positive cells by in situ hybridisation, the HBRV RNA levels were clearly above the cut-off, suggesting that a false positive HBRV RNA analysis was unlikely. The alternative explanation of a false negative LM-PCR result may be attributable to the strict process of eliminating HBRV integrations for derivation of the unique integration data set.
Table 1.
Comparison of detection of HBRV RNA and HBRV integrations in BEC samples
| HBRV integration positive | HBRV integration negative | |
|---|---|---|
| HBRV RNA positive | 12 | 9 |
| HBRV RNA negative | 4 | 28 |
Discussion
This descriptive study demonstrating HBRV RNA and proviral integrations in biliary epithelium provides credible evidence of betaretrovirus infection at the site of disease in patients with PBC. To confirm these data, however, case–control studies involving coded samples will be required with corroboration of other laboratories. Moreover, methodological improvements are needed to increase the specificity and to decrease the risk of contamination, by indexing the libraries to uniquely identify the source of individual integrations.11 We observed more patients with PBC had detectable HBRV integrations within the biliary epithelium as compared with their liver samples, which may be related to increased replication of HBRV within biliary epithelium as compared to the other hepatic parenchymal cells, as observed in the NOD.c3c4 mouse model of autoimmune biliary disease.20, 21 Also, the use of cultured BEC may permit HBRV replication due to the lack of immune surveillance in vitro. Notably, the detection of HBRV integrations within 13% of PBC liver samples is consistent with prior studies that identified HBRV in 12–17% of liver DNA using nested PCR5, 13 and those that were unable to detect HBRV using a single round of PCR.12
Herein, we observed HBRV in 50% of BEC from patients with AIH consistent with previous reports of detecting HBRV and reverse transcriptase activity in serum of AIH patients.8 While both PBC and AIH are considered separate entities, up to 19% of PBC patients show definite or probably clinical and histological features of AIH22 and the EASL guidelines recommend immunosuppression for PBC patients with clinical criteria for AIH.23 Indeed, the phenotype of increased AMA reactivity in BEC has also been linked with AIH, even though patients with AIH rarely develop severe cholangitis or ductopenia.1, 24, 25
Observations from the NOD.c3c4 mouse model of autoimmune biliary disease help to shed light on the possible role of betaretrovirus infection in the development of hepatitis.21 In this model, MMTV was observed in the intrahepatic lymphocytes, possibly serving a dual purpose of delivery of the virus to the bile ducts as well as propagating interface hepatitis. With regard to the latter, a pathophysiological comparison can be made with infectious mononucleosis, where the Epstein–Barr virus is mainly restricted to the infiltrating lymphocytes to propagate the hepatitis as the virus is rarely detected in hepatocytes.26, 27 In this scenario, it has been suggested that the hepatitis is triggered by trapping of virally infected CD8+ cells in the hepatic sinusoids followed by their removal by apoptosis.28, 29 As betaretrovirus usually replicated and spread in lymphocytes prior to reaching their target organ,9 it is conceivable that removal of these infected lymphocytes may directly impact on disease. Indeed, we noted the serial reduction and then disappearance of HBRV serum levels in a newly diagnosed AIH patient with concomitant improvement in hepatic biochemistry following immunosuppressive therapy, supporting the hypothesis that infected HBRV lymphocytes was propagating the hepatitis.8 These data suggest that betaretrovirus infection in the liver may promote hepatitis and cholangitis depending on genetic background and other modulating factors.
In this study, HBRV was found in BEC from patients with cryptogenic liver disease and control subjects with PSC, NASH and FNH. In previous reports, HBRV was detected in patients with cryptogenic cirrhosis and alcohol-related liver disease.13 At this juncture, one could draw a parallel with early observations in the detection of hepatitis C virus infection. Following discovery, evidence for HCV infection was observed in patients with other hepatic disorders such as autoimmune hepatitis, hemochromatosis, alcoholic cirrhosis, alpha-1 anti-trypsin deficiency and other hepatic disorders;30 this observation was to be expected because the non-A non-B hepatitis virus had already been established as a hepatitis virus. In contrast, the role of the HBRV in liver disease remains to be resolved. Moreover, in this study one patient without parenchymal liver disease (FNH) had detectable HBRV, suggesting that the virus may not be associated with inflammatory disease in specific circumstances. These data suggest that HBRV infection is common and insufficient to trigger PBC. As the prevalence of infection likely exceeds the prevalence of PBC, further investigations will be necessary to address a more specific hypothesis, that the combined effects of HBRV infection on the background of genetic predisposition are required to trigger autoimmune liver disease.1, 2 Indeed, it is interesting to note that the genetic predisposition may be in part related to a relative immunodeficiency in both patients and mouse models of PBC that in turn leads to diminished control of infectious disease.2, 21
We took stringent measures to exclude data that may be attributable to contamination because of the recent reports of a gammaretrovirus infection in clinical samples was subsequently attributed to PCR carry-over with mouse DNA18 or spread from in vitro infection from a mouse gammaretrovirus derived from a human prostate cancer cell line previously passaged through mice.31 To our knowledge, no such human cell line exists for MMTV-like betaretroviruses. We do not believe that mouse integrations had any serious impact on our results as it is easy to differentiate human integrations from any murine integrations – the sequences of the two genomes are only 69% identical.19 Furthermore, all ambiguous cases that could not confidently identify as nonmouse were removed from the analysis. It was also reassuring that the phylogenetic evaluation of the HBRV LTRs demonstrated multiple variants observed above the frequency expected for sequencing artefacts, consistent with novel HBRV sequences unrelated to known MMTV LTRs.
As over half the PBC patients tested have evidence of HBRV infection in BEC, the collective data suggest that HBRV is below the limit of PCR detection in the liver of most patients. This brings up a question of whether this low level viral burden could play any role in the disease process or whether the presence of virus is an epiphenomenon? In patients with HTLV infection, for example, proviral integrations may be found in up to 500 copies per cell.11 However, the vial burden of MMTV is low in mice and rarely detected in serum.9 Previously, we have shown that MMTV and the low level of HBRV in lymph node homogenates from PBC patients can trigger the disease specific phenotype in biliary epithelium by increasing expression of proteins reactive to AMA.5, 7 Accordingly, the viral infection is linked with a disease specific phenotype. While the mechanism for increased PDC-E2-like protein expression has yet to be resolved, the link with HBRV infection is important as it demonstrates that MMTV like viruses can infect human biliary epithelium. In fact, it was previously thought that MMTV could only infect murine cells expressing the MMTV entry receptor, characterised as the mouse transferrin receptor.32 However, human cells such as Hs578T, HeLa and HEK-293 cells have recently been shown to be permissive for MMTV in vitro.33, 34
We emphasise that this study does not provide evidence to causally link HBRV infection with autoimmune liver disease but supporting data suggest that presence of betaretrovirus infection is pathogenic rather than an epiphenomenon. In mouse models, we have shown that the expression of MMTV proteins in the same distribution with PDC-E2-like antigens and that the development anti-betaretrovirus antibodies corresponds with the development of AMA suggesting that viral infection contributes to breaking tolerance.6, 20, 21 Indeed, we have used the NOD.c3c4 mouse model to demonstrate that specific HIV reverse transcriptase and protease inhibitors decrease betaretrovirus load in the liver and ameliorate cholangitis, suggesting a more direct role for viral infection in the autoimmune biliary disease.20, 35 While randomised controlled trials in PBC patients with zidovudine/lamivudine 300/150 mg lead to a significant but not substantial reduction in alkaline phosphatase levels, it is notable that the interim analysis of combination anti-retroviral therapy using lopinavir/ritonavir 800/200 mg and tenofovir/emtricitabine 300/200 mg resulted in a significant and substantial reduction in hepatic biochemistry36, 37 equivalent to improvements observed with the new Farnesoid-X receptor agonist, Obeticholic acid.38 Further studies demonstrating immune responses to HBRV are required to demonstrate that the detection of HBRV integrations is more than an epiphenomenon. Also, randomised controlled trials with potent anti-retroviral therapies capable of linking reduction in viral load with biochemical and histological improvement will help determine whether HBRV infection has a role in the development of liver disease. In summary, further investigation of HBRV is warranted in patients with liver disease.
Acknowledgments
Declaration of personal interests: Dr. Mason has served on advisory boards for Novartis and Intercept and has received research support from AbbVie and Gilead Sciences. Weiwei Wang, Stanislav Indik, Shawn T. Wasilenko, Alexander Faschinger, Eric Carpenter, Zhijian Tian, Yong Zhang and Gane Ka-Shu Wong have no conflicts of interest to declare.
Declaration of funding interests: This study was supported by a grant from the Alberta Cancer Foundation, the Canadian Institutes for Health Research, the Canadian Liver Foundation and the Center of Excellence for Gastrointestinal Inflammation and Immunity Research, supported by the Canadian Foundation for Innovation. The study sponsors had no role in the design and conduct of the study, collection, management, analysis, interpretation of the data, drafting of the manuscript or decision to submit the manuscript for publication. Drs. Mason and Wang were supported by awards from Alberta Innovates Health Solutions and Dr. Wong by an award from Alberta Innovates Technology Futures.
Authorship
Guarantor of article: Dr. Andrew Mason
Author contributions: ALM contributed to study design, data collection, data analysis and manuscript drafting and editing. WW, SI, STW and GKSW contributed to data collection, data analysis, manuscript drafting and editing. AF, EJC, ZT and YZ contributed to data production, data analysis and manuscript drafting. All authors have approved the final version of the manuscript, including the authorship list.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Nucleic acid sequencing of the LM-PCR products by either capillary or Illumina next generation sequencing methodologies.
Figure S2. Assessment of LTR variation and likely sequencing errors.
Figure S3. Matrix of cross-contaminations by MMTV mouse-like integrations.
References
- 1.Poupon R. Primary biliary cirrhosis: a 2010 update. J Hepatol. 2010;52:745–58. doi: 10.1016/j.jhep.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Hirschfield GM, Chapman RW, Karlsen TH, Lammert F, Lazaridis KN, Mason AL. The genetics of complex cholestatic disorders. Gastroenterology. 2013;144:1357–74. doi: 10.1053/j.gastro.2013.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mason A, Xu L, Guo L, et al. Detection of retroviral antibodies in primary biliary cirrhosis and other idiopathic biliary disorders. Lancet. 1998;351:1620–4. doi: 10.1016/S0140-6736(97)10290-2. [DOI] [PubMed] [Google Scholar]
- 4.Mason A, Xu L, Shen Z, et al. Patients with primary biliary cirrhosis make anti-viral and anti-mitochondrial antibodies to mouse mammary tumor virus. Gastroenterology. 2004;127:1863–4. doi: 10.1053/j.gastro.2004.10.024. author reply 4–5. [DOI] [PubMed] [Google Scholar]
- 5.Xu L, Shen Z, Guo L, et al. Does a betaretrovirus infection trigger primary biliary cirrhosis? Proc Natl Acad Sci U S A. 2003;100:8454–9. doi: 10.1073/pnas.1433063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason AL. The evidence supports a viral aetiology for primary biliary cirrhosis. J Hepatol. 2011;54:1312–4. doi: 10.1016/j.jhep.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Sadamoto T, Joplin R, Keogh A, Mason A, Carman W, Neuberger J. Expression of pyruvate-dehydrogenase complex PDC-E2 on biliary epithelial cells induced by lymph nodes from primary biliary cirrhosis. Lancet. 1998;352:1595–6. doi: 10.1016/S0140-6736(05)61042-2. [DOI] [PubMed] [Google Scholar]
- 8.McDermid J, Chen M, Li Y, et al. Reverse transcriptase activity in patients with primary biliary cirrhosis and other autoimmune liver disorders. Aliment Pharmacol Ther. 2007;26:587–95. doi: 10.1111/j.1365-2036.2007.03402.x. [DOI] [PubMed] [Google Scholar]
- 9.Acha-Orbea H, Finke D, Attinger A, et al. Interplays between mouse mammary tumor virus and the cellular and humoral immune response. Immunol Rev. 1999;168:287–303. doi: 10.1111/j.1600-065x.1999.tb01299.x. [DOI] [PubMed] [Google Scholar]
- 10.Bukhari SZ, Qazi JI, Ashshi AM, Zia N. Human immunodeficiency virus type-1 (HIV-1) disease progression and viral activity: a seroepidemiological and molecular study. J Coll Physicians Surg Pak. 2012;22:565–9. [PubMed] [Google Scholar]
- 11.Gillet NA, Malani N, Melamed A, et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–22. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selmi C, Ross SR, Ansari AA, et al. Lack of immunological or molecular evidence for a role of mouse mammary tumor retrovirus in primary biliary cirrhosis. Gastroenterology. 2004;127:493–501. doi: 10.1053/j.gastro.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Johal H, Scott GM, Jones R, Camaris C, Riordan S, Rawlinson WD. Mouse mammary tumour virus-like virus (MMTV-LV) is present within the liver in a wide range of hepatic disorders and unrelated to nuclear p53 expression or hepatocarcinogenesis. J Hepatol. 2009;50:548–54. doi: 10.1016/j.jhep.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Voisset C, Weiss RA, Griffiths DJ. Human RNA “Rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol Mol Biol Rev. 2008;72:157–96. doi: 10.1128/MMBR.00033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason AL, Gilady SY, Mackey JR. Mouse mammary tumor virus in human breast cancer red herring or smoking gun? Am J Pathol. 2011;179:1588–90. doi: 10.1016/j.ajpath.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faschinger A, Rouault F, Sollner J, et al. Mouse mammary tumor virus integration site selection in human and mouse genomes. J Virol. 2008;82:1360–7. doi: 10.1128/JVI.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joplin R. Isolation and culture of biliary epithelial cells. Gut. 1994;35:875–8. doi: 10.1136/gut.35.7.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearney MF, Spindler J, Wiegand A, et al. Multiple sources of contamination in samples from patients reported to have XMRV infection. PLoS ONE. 2012;7:e30889. doi: 10.1371/journal.pone.0030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 20.Sharon D, Chen M, Zhang G, et al. Impact of combination antiviral therapy in the NOD.c3c4 mouse model of autoimmune biliary disease. Liver Int. 2014 doi: 10.1111/liv.12699. doi: 10.1111/liv.12699. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Chen M, Graham D, et al. Mouse mammary tumor virus in anti-mitochondrial antibody producing mouse models. J Hepatol. 2011;55:876–84. doi: 10.1016/j.jhep.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Manns MP, Czaja AJ, Gorham JD, et al. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–213. doi: 10.1002/hep.23584. [DOI] [PubMed] [Google Scholar]
- 23.European Association for the Study of the L. EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol. 2009;51:237–67. doi: 10.1016/j.jhep.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Perrillo R, Mason A, Jacob S, Gerber M. Hepatitis and cholestasis in a middle-aged woman. Hepatology. 1996;24:730–4. doi: 10.1002/hep.510240342. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien C, Joshi S, Feld JJ, Guindi M, Dienes HP, Heathcote EJ. Long-term follow-up of antimitochondrial antibody-positive autoimmune hepatitis. Hepatology. 2008;48:550–6. doi: 10.1002/hep.22380. [DOI] [PubMed] [Google Scholar]
- 26.Kimura H, Nagasaka T, Hoshino Y, et al. Severe hepatitis caused by Epstein-Barr virus without infection of hepatocytes. Hum Pathol. 2001;32:757–62. doi: 10.1053/hupa.2001.25597. [DOI] [PubMed] [Google Scholar]
- 27.Hara S, Hoshino Y, Naitou T, et al. Association of virus infected-T cell in severe hepatitis caused by primary Epstein-Barr virus infection. J Clin Virol. 2006;35:250–6. doi: 10.1016/j.jcv.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Park S, Murray D, John B, Crispe IN. Biology and significance of T-cell apoptosis in the liver. Immunol Cell Biol. 2002;80:74–83. doi: 10.1046/j.1440-1711.2002.01065.x. [DOI] [PubMed] [Google Scholar]
- 29.Crispe IN, Dao T, Klugewitz K, Mehal WZ, Metz DP. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunol Rev. 2000;174:47–62. doi: 10.1034/j.1600-0528.2002.017412.x. [DOI] [PubMed] [Google Scholar]
- 30.Gish RG, Mason A. Autoimmune liver disease. Current standards, future directions. Clin Liver Dis. 2001;5:287–314. doi: 10.1016/s1089-3261(05)70167-7. [DOI] [PubMed] [Google Scholar]
- 31.Delviks-Frankenberry K, Cingoz O, Coffin JM, Pathak VK. Recombinant origin, contamination, and de-discovery of XMRV. Curr Opin Virol. 2012;2:499–507. doi: 10.1016/j.coviro.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross SR, Schofield JJ, Farr CJ, Bucan M. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc Natl Acad Sci U S A. 2002;99:12386–90. doi: 10.1073/pnas.192360099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Indik S, Günzburg WH, Kulich P, Salmons B, Rouault F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology. 2007;4:73. doi: 10.1186/1742-4690-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Indik S, Gunzburg WH, Salmons B, Rouault F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005;65:6651–9. doi: 10.1158/0008-5472.CAN-04-2609. [DOI] [PubMed] [Google Scholar]
- 35.Montano-Loza AJ, Wasilenko S, Bintner J, Mason AL. Cyclosporine A inhibits in vitro replication of betaretrovirus associated with primary biliary cirrhosis. Liver Int. 2010;30:871–7. doi: 10.1111/j.1478-3231.2010.02257.x. [DOI] [PubMed] [Google Scholar]
- 36.Mason AL, Lindor KD, Bacon BR, Vincent C, Neuberger JM, Wasilenko ST. Clinical trial: randomized controlled study of zidovudine and lamivudine for patients with primary biliary cirrhosis stabilized on ursodiol. Aliment Pharmacol Ther. 2008;28:886–94. doi: 10.1111/j.1365-2036.2008.03799.x. [DOI] [PubMed] [Google Scholar]
- 37.Mason AL, Montano-Loza AJ, Saxinger L. Letter: biochemical response to combination anti-retroviral therapy in patients with primary biliary cirrhosis. Aliment Pharmacol Ther. 2014;39:236–7. doi: 10.1111/apt.12575. [DOI] [PubMed] [Google Scholar]
- 38.Mason A, Luketic V, Lindor K, et al. Farnesoid-X receptor agonists: a new class of drugs for the treatment of PBC? An international study evaluating the addition of (INT-747) to ursodeoxycholic acid. Hepatology. 2010;52(4 Suppl):357A. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleic acid sequencing of the LM-PCR products by either capillary or Illumina next generation sequencing methodologies.
Figure S2. Assessment of LTR variation and likely sequencing errors.
Figure S3. Matrix of cross-contaminations by MMTV mouse-like integrations.