Abstract
During the last decades, many studies have investigated the transcriptional and epigenetic regulation of lineage decision in the hematopoietic system. These efforts led to a model in which extrinsic signals and intrinsic cues establish a permissive chromatin context upon which a regulatory network of transcription factors and epigenetic modifiers act to guide the differentiation of hematopoietic lineages. These networks include lineage-specific factors that further modify the epigenetic landscape and promote the generation of specific cell types. The process of B lymphopoiesis requires a set of transcription factors, including Ikaros, PU.1, E2A, and FoxO1 to ‘prime’ cis-regulatory regions for subsequent activation by the B-lineage-specific transcription factors EBF1 and Pax-5. The expression of EBF1 is initiated by the combined action of E2A and FoxO1, and it is further enhanced and maintained by several positive feedback loops that include Pax-5 and IL-7 signaling. EBF1 acts in concert with Ikaros, PU.1, Runx1, E2A, FoxO1, and Pax-5 to establish the B cell-specific transcription profile. EBF1 and Pax-5 also collaborate to repress alternative cell fates and lock cells into the B-lineage fate. In addition to the functions of EBF1 in establishing and maintaining B-cell identity, EBF1 is required to coordinate differentiation with cell proliferation and survival.
Keywords: EBF1, B-cell differentiation, regulatory network, lineage specification, B-cell commitment
This article is part of a series of reviews covering Transcriptional and Epigenetic Networks Orchestrating Immune Cell Development and Function appearing in Volume 261 of Immunological Reviews.
Introduction
Hematopoiesis is one of the best characterized developmental systems for studying cell fate decisions, differentiation, lineage-specific gene expression, as well as the stability and plasticity of cellular phenotypes. In particular, B lymphopoiesis is an excellent paradigm for the stepwise differentiation of a multipotent progenitor (MPP) into a terminally differentiated effector cell. The differentiation process can be monitored by the expression of surface antigens, the rearrangement status of the heavy and light chain of the immunoglobulin genes and by the expression of certain genes like Rag1 and Rag2 1. Differentiation of a hematopoietic stem cell (HSC) into highly specialized antibody-producing B cells involves the acquisition of cell type-specific gene expression signatures (specification) and the loss of the ability to differentiate into alternative cell lineages (commitment) 2–4. Specialized stromal cells guide the developmental progress by providing cell–cell interactions and secreting cytokines and chemokines 5. The microenvironment influences not only the survival and proliferation of cells but also affects their responsiveness to external signals, thereby shaping the transcriptional network of the developing cell. A set of transcription factors, including Ikaros, Runx1/Cbfβ, E2A (Tcf3), and FoxO1, directs cells into the B-cell lineage by providing an epigenetic landscape that is permissive for the action of B cell-specific factors, including early B-cell factor 1 (EBF1) and paired box transcription factor 5 (Pax-5). Together with other transcription factors, EBF1 and Pax-5 activate the transcriptional program that eventually leads to the generation of mature B cells, and they repress alternative lineage choices 6–9. During the last few years, it has become clear that the regulatory system underlying B-cell differentiation does not involve a simple linear hierarchy in which transcription factors are sequentially activated. Instead, B lymphopoiesis requires a complex regulatory network in which transcription factors are interconnected via feed-forward and feedback loops and cross-antagonism. Moreover, their expression or activity can be further modulated by signaling pathways and epigenetic regulation. In this review, we present the current view of the regulatory network governing B-cell differentiation with a focus on one key determinant, EBF1.
B-cell lymphopoiesis
B-cell differentiation starts in the fetal liver or adult bone marrow with an asymmetric cell division of a HSC and the generation of a MPP 10 (Fig. 1). This progenitor acts as a branching point for the myeloid and lymphoid lineages 11. Differentiation along the myeloid lineage is initiated by the common myeloid progenitor (CMP) that generates megakaryocytes, erythrocytes, granulocytes, and macrophages 12. The lymphoid lineage is marked by the surface expression of the tyrosine kinase Flt3 receptor that is first detected on the lymphoid-primed multipotent progenitor (LMPP). Although LMPPs have lost the megakaryocyte–erythroid lineage potential, they can give rise to granulocytes, macrophages, and lymphocytes 13,14. Reduced expression of the stem cell markers SCA-1 and c-Kit and upregulation of IL-7 receptor expression mark the next step in lymphopoiesis, represented by the common lymphoid progenitor (CLP) 15,16. Recently, the transmembrane protein Ly6D has been identified as an early B-cell marker that allows the subdivision of CLPs into Ly6D-negative all lymphoid progenitors (ALP) and B-cell-biased lymphoid progenitors (BLP) that express Ly6D on the surface 17. ALPs retain the potential for generating natural killer cells (NK), dendritic cells (DC), T and B cells, whereas BLPs show a markedly reduced T-cell potential and generate predominantly B cells 17. Pre-pro-B cells, also called fraction (Fr.) A in the nomenclature of Hardy and Hayakawa 1, are marked by the B220 isoform of the CD45 receptor but lack canonical B-cell markers including CD19 18. In the following pro-B-cell stages (Fr.B and Fr.C), the immunoglobulin heavy chain (IgH) genes are rearranged in a Rag-dependent manner. Successful rearrangement culminates in the pairing of heavy chain and surrogate light chains λ5 (Igll1) and VpreB1 (Vpreb1). Together with its signaling components, Igα (Cd79a) and Igβ (Cd79b), the pre-B-cell receptor (pre-BCR) is expressed on the surface of large pre-B-cells (Fr. C'). Passing this developmental checkpoint activates rearrangement of the immunoglobulin light genes in the small pre-B-cell stage (Fr.D). Productive recombination of the Igκ or Igλ locus results in the expression of the IgM B-cells receptor (BCR) on the surface of the immature B cell 19. Only a subset of these immature B cells leave the bone marrow and only a portion of them complete their development in the spleen and join the mature B-cell pool 20. Mature B cells circulate in blood and secondary lymphatic organs. After contact with a pathogen-derived antigen, mature B cells undergo class switch recombination (CSR) and somatic hypermutation (SHM) and differentiate into plasma cells that produce high affinity soluble antibodies 21.
Fig 1.
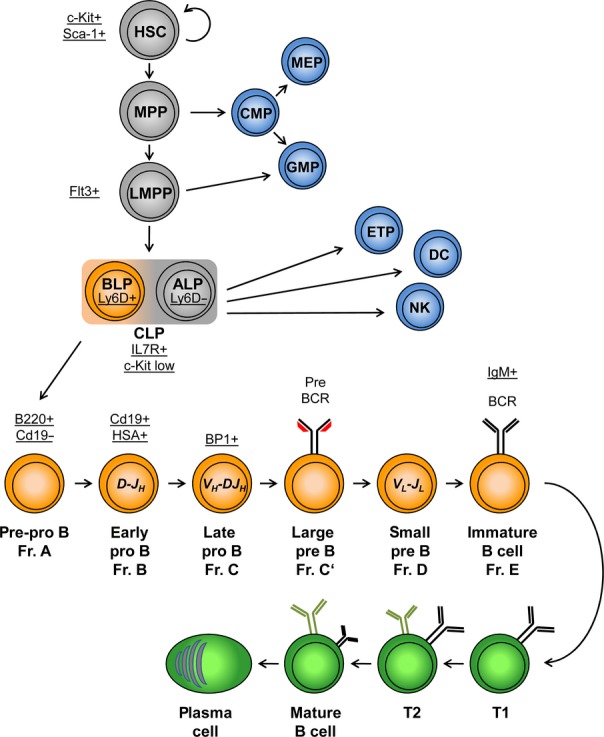
A schematic view of B-cell lymphopoiesis. Common developmental steps of B and non-B cells are colored in gray. Early B-cell development in the bone marrow is shown in orange, while late B-cell development in the periphery is depicted in green. Non-B cells are colored in blue. The developmental stages are marked by bold letters and the presence or absence of surface proteins, indicative of specific cell types, are underlined. Rearrangements of the heavy and light chains are written in italic letters. HSC, hematopoietic stem cell; MPP, multipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CLP, common lymphoid progenitor; ALP, all lymphoid progenitor; BLP, B-cell-biased lymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocytic/erythrocyte progenitor; GMP, ganulocyte/macrophage progenitor; ETP, early thymic progenitor; DC, dendritic cell; NK, natural killer cell; T1 and T2, transitional B cell 1 and 2. Adapted from Mandel and Grosschedl 2, Lai and Kondo 14, Roessler and Grosschedl 19, and Rolink, Andersson, and Melchers 20.
Early B-cell factor 1: protein structure and mechanism of action
Protein structure of EBF1
EBF1 is one of the key factors of B-cell differentiation. EBF1 was discovered as a factor with B lineage-specific DNA-binding activity to the Cd79a promoter 22. Because of its strong expression in early B cells, the factor was named EBF 22,23 which was later changed to EBF1. Purification of this factor from a transformed pre-B-cell line by sequence-specific DNA affinity chromatography characterized EBF1 as a dimer of two 65 kDa subunits that binds its palindromic DNA-binding motif 5′-TCCCNNGGGA with high affinity 24. Amino acid sequence analysis allowed for the molecular cloning of EBF1, which was also independently cloned as Olf1 in a yeast-one-hybrid screen, using the 5′ flanking region of the gene encoding olfactory marker protein (Omp) 23,25,26. Together with Collier, an Olf-1/EBF ortholog identified in Drosophila melanogaster, Olf-1 and EBF1 established a new family of transcription factors, which was named COE according to its founding members. EBF1 is highly conserved during metazoan evolution and shows strong sequence overlap with the three other members of the family, now termed EBF2, EBF3, and EBF4 27. All COE factors consist of four protein domains: an N-terminal DNA-binding domain (DBD), an IPT (Ig-like/plexins/transcription factors) domain, a helix-loop-helix (HLH) dimerization domain, and a C-terminal transactivation domain.
The N-terminal DNA-binding domain, spanning some 220 amino acids, shows the highest degree of sequence conservation, as the similarity between the evolutionarily most distantly related proteins still exceeds 80% 28,29. Biochemical analysis of the DBD demonstrated that its interaction with DNA is dependent on a zinc-coordination motif, H-X3-C-X2-C-X5-C, located between amino acids 157 and 170 29,30. Because of its difference to the canonical zinc finger structure, this atypical zinc finger motif was termed ‘zinc knuckle’ or ‘COE motif’ 29. Methylation interference assays showed that EBF1 contacts both the major and minor grooves of DNA 22. Recent determination of the crystal structures of EBF1 and an EBF1:DNA complex clarified the three-dimensional architecture of the DBD and elucidated the interaction between EBF and DNA at atomic resolution 31,32 (Fig. 2). The DBD folds into a β-sandwich, which consists of a four- and a five-stranded anti-parallel β-sheet. DNA interaction is mediated by three distinct DNA interaction modules, the GH loop, a central module, and the zinc knuckle, that extend from the β-sandwich 31. Most of the sequence specificity of DNA binding by EBF1 is provided by the central module. It is composed of small β-sheets and loops that reach deep into the major groove, contacting specific nucleotides of one half-site of the palindromic binding sequence. The zinc knuckle, which consists of short α-helices, contacts the other half-site in the minor groove. The large GH loop protrudes into the minor groove outside of the binding motif. Although the GH loop is important for DNA-binding affinity, it does not appear to contribute to the specificity of sequence recognition 31. The crystal structure confirmed the binding of DNA by a dimer of EBF1 22. Interestingly, the two monomers of EBF1 form a symmetric clamp over the entire binding motif. In this structure, each monomer contacts both half-sites of the palindromic consensus sequence. Although EBF1 shows no amino acid sequence similarity with other families of DNA-binding proteins, the three-dimensional architecture of the EBF1 DBD resembles the N-terminal half of the Rel-homology domain (RHD), which is found in NF-κB and NFAT 31,32.
Fig 2.
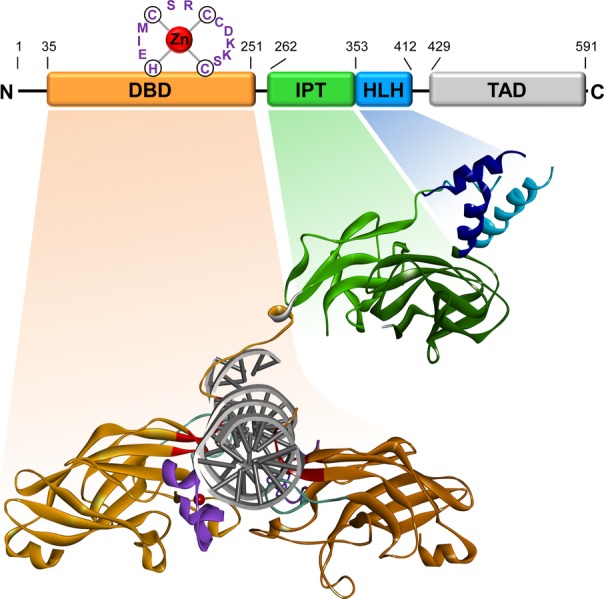
Structure of EBF1. A schematic presentation of the domain structure of murine EBF1 [modified after Hagman and Lukin 38] and the crystal structure of a DNA-bound EBF1 dimer that lacks the C-terminal transactivation domain [modified after Treiber et al. 31 using PDB file 3MLP are depicted]. The structure was modeled using Discovery Studio 3.5 Visualizer, Accelrys Inc., San Diego, CA. The DNA-binding domain (DBD) is colored in orange, the IPT domain in green, the helix-loop-helix (HLH) dimerization domain in blue and the C-terminal transactivation domain (TAD) in light gray. The DNA interacting modules within the DBD are colored in red (central motif), purple (zinc coordinating motif) and turquoise (GH loop). Zinc ions are represented by red spheres. The border amino acids of each domain are indicated in the schematic.
The DBD is followed by an IPT domain that extends from aa 262 to 345 33. The RRARR motif located between the DBD and the IPT domain was proposed as a putative nuclear localization signal (NLS) 25. As predicted by sequence comparison and underlined by the crystal structure, the IPT domain adopts an immunoglobulin-like fold. It resembles the C-terminal half of the RHD. The structural similarities of both DBD and IPT domain with the RHD strengthen the relationship between EBF1 and members of the Rel family 31,32. In contrast to NFAT and NF-κB, in which the IPT domain is involved in DNA binding, dimerization and protein–protein interaction 34,35, the function of the IPT domain of EBF, which is dispensable for DNA binding and dimerization 23, remains elusive.
EBF1 forms stable homo- and heterodimers via an HLH domain, consisting of two amphipathic helices 23,36,37. Dimerization of the four helices, two from each monomer, forms a helix bundle comparable to the dimerized basic HLH domains of other proteins like MyoD 31,38,39. The second helix is duplicated in vertebrates, resulting in a helix-loop-helix-loop-helix motif. However, the third helix is not essential for dimerization 37. Moreover, the crystal structure of EBF1 argues against an inclusion of the third helix in the HLH dimerization motif and raises the possibility that the third helix-like motif interacts with other proteins 31,32.
The C-terminal transactivation domain is only poorly conserved between the COE family members. Nevertheless, common properties are a strong enrichment of proline, serine, and threonine residues and a lack of predicted secondary structure. Despite the poor sequence conservation, the C-terminus of all four EBF members in mice contributes to the activation of gene expression in vector-based reporter assays 23,36,37. However, EBF4 shows a lower transactivation potential compared to the other family members 37. Aside from the C-terminal domain, an additional transactivating region may be located in the DBD because a truncated version of EBF lacking the C-terminus is still able to activate transcription 29.
The molecular mechanism of EBF1 function
EBF1 is the only COE factor that is expressed in the hematopoietic system, and it plays a critical role in B-cell development 2,6–8. Genome-wide chromatin immunoprecipitation analysis combined with deep sequencing (ChIP-seq) in pro-B cells identified approximately 5000 EBF1-occupied sites corresponding to some 3000 genes 40,41. These genes are strongly associated with B-cell function and encode many components of the B-cell receptor signal transduction cascade. However, EBF1 gain-of-function experiments in pre-pro-B cells and EBF1 loss-of-function studies in pro-B cells indicated that only a small fraction of EBF1-bound genes are regulated by EBF1 40. These studies also revealed that EBF1 can both activate and repress genes. Beside the presence of activated and repressed genes among EBF1-occupied targets, a third group of genes, termed ‘poised’ genes, are bound by EBF1 in early B-cell stages but expression is detected only at later stages of the B lineage 40. Notably, analysis of histone marks linked EBF1 binding to di-methylation of histone 3 at lysine 4 (H3K4me2). Ectopic expression of EBF1 in a pre-T-cell line resulted in H3K4me2 modification at B-cell-specific EBF1 targets, which was found to be independent of transcriptional activation or repression 40. Thus, binding of EBF1 to chromatin is associated with this histone modification. A strong coincidence of H3K4 di-methylation with EBF1 occupancy can also be detected in mature B cells, although some targets bound by EBF1 specifically in mature B cells show H3K4me2 modifications already in pro-B cells 42.
Further evidence for a function of EBF1 in modulating the epigenetic landscape comes from the analysis of the Cd79a promoter in plasmacytoma cells expressing ectopic EBF1 43. In pro-B, pre-B, and mature B cells, the Cd79a promoter is activated by the collaboration of several transcription factors including EBF1, RUNX1, E2A, and Pax-5 43–45. In hematopoietic progenitors, plasma cells and non-B cells, the Cd79a promoter is methylated at CpG dinucleotides, whereas DNA methylation decreases stepwise at the onset of B-cell differentiation. EBF1 expression in plasmacytoma cells was found to induce DNA demethylation at the Cd79a promoter 43. Recently, Tet2 has been linked to EBF1 in a study analyzing the hypermethylation status of certain tumors, including chondrosarcomas 46. Tet2 catalyzes the conversion of 5-methylcytosine to 5-hydroxmethylcytosine and thereby initiates demethylation of DNA. Although an interaction of Tet2 and EBF1 could explain the EBF1-linked DNA demethylation, a physical or functional interaction between these proteins still needs to be determined in normal B-lineage cells.
EBF1 function has also been linked to the chromatin-remodeling complexes SWI/SNF and Mi-2/NuRD. EBF1-mediated induction of chromatin accessibility at the Cd79a promoter is dependent on the SWI/SNF complex, whereas the Mi-2/NuRD complex is involved in chromatin compaction and DNA hypermethylation at the Cd79a promoter 47,48. However, no physical interaction between EBF1 and this complexes has been reported.
Finally, the zinc finger proteins ZNF423 and ZNF521 have been shown to interact directly with EBF1 and suppress EBF1 function 49,50. The interaction of ZNF423 (ROAZ, Ebfaz) was mapped to a 253 amino acid region of EBF1 (aa 240 to aa 492) including the HLH domain and was shown to inhibit EBF1-mediated transactivation in reporter assays 49. The DNA-binding motif of ZNF423 homodimers (5′-GCACCCNNGGGTGC) includes a perfect EBF1-binding site 51. Recently, aberrant ZNF423 expression has been linked to a B-cell maturation defect in B precursor acute lymphoblastic leukemia 52. The closely related ZNF521 was first identified in murine B-cell lymphoma cells and human hematopoietic progenitor cells 50,53. Alike ZNF423, it contains 30 zinc fingers of the C2H2 type and negatively regulates EBF1-mediated transcription. In the hematopoietic system, ZNF521 is abundantly expressed in early progenitors, but its expression drops rapidly upon differentiation 50. Knockdown of ZNF521 in hematopoietic progenitor cells leads to enhanced B-cell differentiation with an increased number of B cells 54. A functional role for ZNF521, which includes the physical interaction with EBF1, has also been shown in adipocytes 55.
Function of EBF1 in the regulatory network of B-cell differentiation
Priming of B-cell development
Ebf1 is expressed in B-lineage cells, in adipocytes, and specific neuronal cell types 23,26,28,56. EBF1 has been proposed to act as a ‘pioneer’ transcription factor of B-cell differentiation as it is able to induce epigenetic changes and to initiate locus activation. Increased chromatin accessibility allows for further regulation by other transcription factors and modifiers of chromatin structure. However, target site recognition by EBF1 requires a ‘permissive’ chromatin context. Chromatin immunoprecipitation experiments to detect binding of ectopically expressed EBF1 demonstrated that EBF1 can bind B cell-specific targets in T-lineage cells, but not in non-hematopoietic cells, such as fibroblasts 40. Moreover, genome-wide analysis of EBF1 binding in B-lineage cells and pre-adipocytic cells revealed only a small set of shared EBF1-occupied sites, indicating that only few B-cell target genes are bound in other tissues and vice versa (57, S.B. & R.G., unpublished data). A permissive hematopoietic-specific chromatin context could be established by the promiscuous transcription associated with multi-lineage priming in HSCs 58–60. The mechanism of multi-lineage priming is still obscure, but it could involve transcription factors that are expressed in HSCs 59. In addition, lymphoid-specific changes in chromatin structure may contribute to the establishment of an epigenetic landscape upon which B-lymphoid transcription factors, such as EBF1 and Pax-5, can act. Lymphoid-specific priming of chromatin and enhancers has been associated with a set of hematopoietic factors including Ikaros, Pu.1, and E2A.
Ikaros, encoded by Ikzf1, has multiple functions in hematopoiesis, including the regulation of self-renewal capacity of HSCs and regulation of lymphopoiesis 61,62. This zinc finger protein can act both as an activator or a repressor of transcription by recruiting chromatin-remodeling complexes, including Mi-2/NuRD and SWI/SNF 63,64. A knockout of Ikzf1 leads to an arrest in LMPPs, as the cells fail to upregulate Flt3 and differentiate into CLPs 61,65. Ikaros also contributes to the rearrangement of immunoglobulin genes by regulating the expression of Rag genes and mediating chromatin accessibility at the IgH and Igκ locus 66–68. Forced expression of EBF1 can partially rescue Ikaros-deficient progenitors. Although EBF1 can overcome the developmental block and promote differentiation into CD19-positive pro-B cells, it fails to restore Rag-dependent rearrangement of the IgH locus 66. Recently, a genome-wide binding study in pre-B cells broadened our understanding of Ikaros function in B-cell development as many of the identified target genes are associated with activation of the B-cell program. In particular, Ikaros-regulated genes are highly enriched for pre-BCR signaling, cell cycle progression, and VDJ recombination 69,70. Interestingly, Ikaros represses the expression of PU.1 by direct binding to the Spi1 promoter 71, and indirectly by promoting Gfi1 expression. As Gfi1 competes with PU.1 binding at the Spi1 promoter, Ikaros-mediated upregulation of Gfi1 reduces PU.1 expression by interrupting an auto-regulatory loop 72.
The Ets transcription factor PU.1 is widely expressed in the hematopoietic system and performs distinct roles in the myeloid and lymphoid lineages 73. In accordance with its broad function in the hematopoiesis, PU.1-deficient mice are born alive but die within 24 h. They lack mature macrophages, neutrophils, B, and T cells, while erythrocytes and megakaryocytes are present 74. Of particular interest is the concentration-dependent function of PU.1 in the fate decision between myeloid and lymphoid lineage. High levels of PU.1 promote macrophage development, whereas low expression levels support B-cell development 75. Myeloid lineage differentiation is supported by PU.1-dependent upregulation of developmentally important cytokine receptors including Csf1r 76. In lymphopoiesis, a crucial step is the activation of Il7r by PU.1 in the CLP stage 77. In line with this finding, ectopic expression of Il7r in PU.1-deficient fetal liver progenitors is able to rescue early B-cell development 78. Interestingly, ectopic expression of EBF1 also overcomes the block of B-cell differentiation in PU.1-deficient progenitors 79. The kinetics of the EBF1-mediated rescue of differentiation is even faster than the rescue by Il7r expression, indicating that the developmental block in PU.1-deficient cells is only partly due to misregulation of Il7r and is mainly a consequence of impaired Ebf1 expression 79. Aside from direct activation of target genes, a recent genome-wide study linked the binding of PU.1 to nucleosome remodeling and the deposition of mono-methylation at lysine 4 of histone 3 80.
Tcf3 encodes the basic helix-loop-helix transcription factor E2A that exists in two different splice variants, termed E12 and E47. E2A-deficient mice develop normally but display increased postnatal death 81. E2A-deficient mice lack B cells due to a developmental block at the pre-pro-B-cell stage 81,82. A more detailed analysis revealed that E2A is also needed for the maintenance of the HSC pool by restricting entry into the cell cycle 83,84. Furthermore, E2A plays a role in lymphoid priming by supporting LMPP development from HSCs 85. This function is reflected in the reduced numbers of LMPPs in E2A-deficient mice. Strikingly, this effect seems to be dose-dependent as heterozygote animals show an intermediate reduction in LMPPs. It has been shown that E2A initiates expression of a subset of lymphoid-associated genes including IL7r. Interestingly, many of these E2A-dependent genes display potential binding sites for PU.1 and Ikaros in their regulatory regions, indicating a synergistic regulation 85.
The complex mechanisms of lymphoid cell priming are far from being understood and may require additional transcription factors, such as Tal1, Fli1, and Runx1 that define HSCs 60. Runx1 is of special interest as it has been linked to chromatin remodeling at the Spi1 promoter, eventually leading to the expression of PU.1 86. Furthermore, Runx1 was shown to recruit Tal1 and Fli1 to Runx1-binding sites 60. Miz and Myb may contribute to lymphoid priming by modulating the sensitivity to IL-7 receptor signaling 87–89, and Bcl11a appears to act prior to the expression of Ebf1 90. Thus, cooperation of several transcription factors and chromatin modifiers may generate conditions that are permissive for the B-lineage program and the action of the lineage-restricted transcription factors EBF1 and Pax-5.
Transcriptional regulation of Ebf1 expression
The transition from a pluripotent lymphoid precursor to B-lineage cells is marked by the expression of Ebf1 and Pax-5. In the hematopoietic system, these two transcription factors are expressed solely in the B-cell lineage and therefore, they can be considered as crucial activators of the B-cell program. Ebf1-deficient mice display a complete block of B lymphopoiesis at the pre-pro-B-cell stage, slightly earlier than that observed in Pax-5-deficient mice 91,92. This and other observations indicate that Ebf1 expression precedes Pax-5 expression. Further evidence for a sequential activation came from single cell analysis of CLPs. This cell population is heterogeneous and differential expression of Ly6d distinguishes ALPs that still have B- and T-cell potential and BLPs that have lost T-cell potential 17. Moreover, fractionation of this heterogeneous population according to the expression of an Igll1 promoter-controlled reporter gene and Rag1 allowed the identification of three stages with different lineage potentials. Expression analysis of those sub-fractions at the single cell level indicated that many cells express Ebf1 but lack Pax-5 expression, underlining the sequential expression of EBF1 and Pax-5 93.
Transcriptional activation of Ebf1 depends on E2A encoded by Tcf3, which acts in concert with FoxO1 and IL-7 receptor signaling to establish expression of Ebf1 in BLPs 94. Ectopic expression of EBF1 can, at least partially, rescue the developmental block in E2A-deficient mice 95. This observation confirms that the initiation of Ebf1 expression is one of the major functions of E2A. Splice variant-specific deletion of E2A isoforms revealed that only E47- but not E12-deficient mice display a developmental block at the pre-pro-B-cell stage, comparable to that observed in Ebf1-deficient mice, suggesting that Ebf1 is specifically activated by the E47 isoform of E2A 96.
The expression of Ebf1 is mediated by two distinct promoters, the distal α-promoter and the stronger proximal β-promoter 97,98. Transcription from these promoters results in the expression of two EBF1 isoforms, EBF1α and β. Due to alternative splicing, transcripts from the distal promoter lack the start codon that is used for translation of EBF1β, generating an isoform that lacks 14 N-terminal amino acids of the EBF1β isoform. However, no functional difference between the two EBF1 isoforms has yet been described. The distal α-promoter is regulated by E47 and EBF1, and it contains two consensus Ikaros-binding sites 97. Moreover, the α-promoter is regulated by IL-7 receptor signaling 98. In mice, B-cell differentiation requires IL7 receptor signaling. IL-7R-deficient mice show a developmental arrest at the pre-pro-B-cell stage and impaired Ebf1 expression 99. Forced expression of EBF1 can partially rescue B-cell development in IL-7R-deficient mice 99. Furthermore, Ebf1 expression can be restored by a constitutive active form of STAT5 that mediates a transcriptional response independent of IL-7 signaling 99. STAT5 has been shown to preferentially activate Ebf1 from the distal promoter although no STAT5 binding can be detected at this promoter 98. Thus, IL-7 signaling may activate Ebf1 expression indirectly by providing permissive conditions for Ebf1 activation by other factors. Expression from the proximal β-promoter is driven by Ets1, Pax-5, PU.1, and the RUNX1/CBF-β complex 98. In the absence of Runx1, increased accumulation of the repressive histone mark H3K27me3 can be detected at the proximal Ebf1 promoter, suggesting that Runx1 changes the epigenetic landscape at the proximal Ebf1 β-promoter 100. Recent studies focusing on the epigenetic regulation of Ebf1 expression identified the SWI/SNF-like BAF complex, which facilitates transcription from the proximal promoter, and MYSM1, which appears to act on both promoters 101,102.
Once initiated, Ebf1 expression is enforced and maintained by multiple positive regulatory feedback loops (Fig. 3). E2A and FoxO1 are both connected with EBF1 in a reciprocal positive feedback loop 103,104. This regulatory unit is established by a feed-forward loop, in which E2A promotes the expression of FoxO1 105. Moreover, both E2A and FoxO1 regulate the expression of the IL-7 receptor and therefore indirectly modulate Ebf1 expression 85,106. Moreover, expression of Ebf1 is under a positive auto-regulatory feedback via the EBF1-binding site in the distal promoter. Consistent with this autoregulation, ectopic expression of EBF1 enhances to the activity of the distal but not proximal promoter 98. In Ebf1 heterozygous mutant mice, however, Ebf1 transcription from both promoters is reduced suggesting that the proximal promoter is indirectly regulated by EBF1, possibly via Pax-5 98. Pax-5 binds multiple sites in the Ebf1β promoter, and a reciprocal positive feedback loop between EBF1 and Pax-5 is established by the EBF1-mediated activation of Pax-5 via binding to an intragenic enhancer in the Pax-5 locus 98,107.
Fig 3.
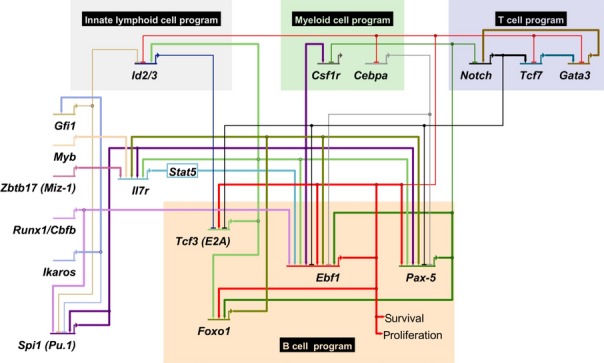
Regulatory network governing early B-cell development. Key factors involved in lineage priming are depicted on the left while major transcription factors regulating B-cell specification and commitment are shown in the orange box. Genes promoting alternative lineage decisions are highlighted in gray (innate lymphoid lineage), light green (myeloid lineage), and light purple (T-cell lineage). Positive regulation is depicted with thick lines that end in an arrow, while repression is represented by thin, barred lines.
Specification of the B-cell lineage
EBF1 is a key regulator of B-cell lineage specification. Expression of EBF1 can be detected at the earliest developmental stage represented by the BLP, and it continues until the onset of plasma cell differentiation 108,109. A conventional knockout of Ebf1 results in a complete lack of B cells, as their development is blocked at the transition from Fr. A pre-pro-B cells to Fr. B pro-B cells 91. No rearrangement at the Igμ locus is detected in vivo, whereas Ebf1-deficient cells cultured in vitro show D to JH but not VH to DJH recombination 110. In heterozygous Ebf1 knockout mice, the number of B-cell progenitors (Fr. B and C) is reduced by 50% 91. The important role of EBF1 in the specification of B-cell development was further shown by adoptive transfer experiments in which forced expression of EBF1 in wildtype bone marrow cells enriched for HSCs skewed differentiation along the B-cell pathway at the expense of other hematopoietic lineages like T cells, NK cells, and lymphoid-derived DC 111.
Early biochemical studies indentified numerous key genes for B-cell differentiation, including Cd79a, Cd79b, Cd19, Blk, Igll1, and Vpreb1 as EBF1 targets (22,112–115). Microarray analysis of EBF1-deficient CLPs validated Pax-5 as a direct target and revealed additional EBF1 targets like FoxO1 and Pou2af1 116,117. Recent genome-wide ChIP-seq analysis in pro-B cells confirmed these target genes and extended the list to some 3000 EBF1-occupied gene loci that are enriched for genes determining B-cell identity and genes regulating the (pre-) BCR signaling cascade 40,41. However, the regulation of EBF1 target genes requires the collaboration of EBF1 with other transcription factors. Many EBF1-regulated genes, including Vpreb1,Igll1, and Cd79a, also contain binding sites for E2A, and ectopic expression of EBF1 and E2A in non-B-cell lines results in a synergistic activation of the endogenous Igll1 and Vpreb1 surrogate light chain genes and Igll1 reporter constructs 115,118,119. Moreover, a collaboration of EBF1 and E2A is underlined by the analysis of Ebf1+/−E2a+/− double heterozygous mutant mice. These mice show an impaired differentiation of Fr.B pro-B cells and reduced expression of Pax-5,Rag1,Rag2, and Cd79a, whereas these phenotypes are not observed in either single-heterozygous mouse 116. Similarly, Ebf1+/−Runx1+/− double heterozygous mice show more severe defects in B cell-specific gene expression and differentiation than those observed in to single-heterozygous mice 120.
Recent ChIP-seq data allowed further insight into the regulatory network. A genome-wide ChIP analysis of Ikaros in pre-B cells identified binding sites of EBF1, E2A, Pax-5, and FoxO1 in the vicinity of Ikaros-bound regions 69. In addition, PU.1 occupancy is associated with C/EBP- and AP1-binding sites in macrophages, whereas PU.1-bound regions are flanked by EBF1-, E2A-, Oct-, and NF-κB-binding sites in splenic B cells 80. Notably, a comparison of PU.1-bound regions in sequential developmental stages of B lymphopoiesis revealed a change in the composition of neighboring factor binding sites 80. In Ebf1-deficient pre-pro-B cells, PU.1-bound regions are primarily associated with E2A- and Runx-binding motifs, whereas PU.1 ChIP peaks are additionally flanked by EBF1- and Oct-binding sites in Rag-deficient pro-B cells. In mature splenic B cells, the association of PU.1-bound regions with Runx is reduced and an association with the NF-κB motif is gained. These observations suggest that PU.1 achieves different tasks at specific developmental stages by cooperation with different neighboring factors 80. A similar conclusion was reached by ChIP-seq analysis of E2A binding. In CLPs, a large number of E2A-occupied genes contain Ikaros- and/or PU.1-binding sites in their regulatory regions 85. Genome-wide binding assays of E2A in Ebf1-deficient pre-pro-B cells and Rag1-deficient pro-B cells revealed that E2A-bound regions are predominantly flanked by Runx-binding sites 41. In pro-B cells, E2A-bound regions were additionally flanked by EBF1- and FoxO1-binding motifs. Finally, genome-wide ChIP experiments for EBF1-bound regions in pro-B cells suggested that the EBF1-occupied regions are strongly enriched in binding sites for E2A, Pax-5, Runx, NF-κB, Stat1, Ets, and Nrf1 40,41. In mature B cells, many EBF1-occupied regions overlap with EBF1-bound regions in pro-B cells. However, additional EBF1-occupied sites are gained while others are lost during differentiation, resulting in a shift in the composition of neighboring binding sites between pro-B and mature B cells 42.
In addition to the integration of inputs from multiple transcription factors at cis-acting regions, the regulatory network underlying B lymphopoiesis also involves extensive cross-regulation of transcription factors. As outlined above, EBF1 is involved in multiple cross-regulatory feedback loops with E2A, FoxO1, and Pax-5 that help to stabilize the developmental decision of B-cell specification.
Commitment to the B-cell lineage
Besides its role in B-cell specification, EBF1 also participates in the repression of alternative cell fates, termed B-cell commitment. Early studies identified Pax-5 as a commitment factor for the B lineage because Pax-5−/− cells, which are arrested at the late pro-B-cell stage (Fr. B to Fr. C) and show rearrangement of proximal but not distal VH segments 92,121,122, have acquired lineage plasticity 123,124. Upon depletion of IL-7, Pax-5-deficient pro-B cells give rise in vitro to nearly all hematopoietic cell lineages except for B cells 123,124. In addition, adoptive transfer of Pax-5−/− B-cell progenitors into Rag2−/− mice that lack lymphocytes allowed for a reconstitution of the T-cell compartment 125. Even mature B cells can be converted into T cells after conditional deletion of Pax-5 via de-differentiation to uncommitted progenitors 126. Insight into the mechanism by which Pax-5 antagonizes alternative cell fates was provided by the molecular analysis of Pax-5-bound and regulated genes, which indicated that Pax-5 represses a large number of non-B cell-specific genes, including Csf1r and Notch1, which are major determinants of macrophage and T-cell development, respectively 123,127,128.
Several recent studies suggested an additional role for EBF1 in B-cell commitment. Analysis of the developmental potential of Ebf1-deficient lymphoid progenitors that were injected into lethally irradiated mice indicated that the Ebf1-deficient progenitors give rise to several lineages including myeloid, dendritic, NK, and T cells 110. Moreover, they can differentiate into T cells and myeloid cells in vitro, depending on the supportive conditions 110. In contrast, ectopic expression of EBF1 restricts their alternative developmental potential and promotes the generation of B cells. The lineage restriction by EBF1 is independent of Pax-5 because EBF1 expression in Pax-5-deficient fetal liver progenitors inhibits their myeloid and T-lineage potential in vivo 110. Furthermore, the myeloid differentiation capacity of Pax-5-deficient pro-B cells is repressed by ectopic expression of EBF1 in vitro 110. Likewise, enforced expression of EBF1 impedes T-cell development in Pax-5-deficient progenitors cultured under T-cell promoting conditions 129. Together, these results provide strong evidence for a role of EBF1 in preventing alternative lineage development in progenitors, independent of Pax-5.
Evidence for a role of EBF1 in the maintenance of B-cell identity came from experiments analyzing the developmental plasticity of Ebf1-deficient pro-B cells in vivo 130. Tamoxifen-induced deletion of floxed alleles in late pro-B cells that were adoptively transferred into alymphoid Rag2−/−Il2rg−/− double-deficient mice allowed for the generation of CD4/CD8 double-positive T cells in the thymus and single-positive T cells in the spleen 130. Importantly, single cell analysis indicated that the peripheral T cells carried rearrangements of both the B-cell and T-cell receptor genes. In particular, the frequent detection of rearrangements using distal VH segments confirmed that the cells were generated from Pax-5-expressing late pro-B cells because Pax-5 is required for the rearrangement of distal VH segments. The transferred pro-B cells also carried a Bcl2 transgene to enhance survival of cells during the process of lineage conversion. However, the limited number of B cell-specific gene rearrangements indicates that only few of the injected cells were able to convert into T cells. Aside from the conversion into T cells, Ebf1-deficient pro-B cells convert into innate lymphoid cells of type 2 and 3 (ILC2 and ILC3). Conversion into the myeloid lineage, however, was only rarely detected 130. Interestingly, in the bone marrow of recipient mice, a relatively large population of CD19-positive cells expressing Pax-5 but lacking Ebf1 expression was detected. Microarray analysis of this cell population indicated that these cells resemble lymphoid progenitors, and suggested that lineage conversion of EBF1-deleted cells occurs via de-differentiation to an intermediate progenitor-like state 130.
EBF1 antagonizes alternative cell fates by direct repression of several genes specific for alternative lineages, including Tcf7, Gata3, Cebpa,Id2, and Id3. Tcf7 and Gata3 are expressed in the earliest T-cell stages and are critical regulators of T-cell development 94. Upon induction via Notch signaling, Tcf1, encoded by Tcf7, promotes T-cell differentiation by the activation of T-cell fate determinants including Bcl11b and Gata3 131. The zinc finger protein Gata3 has been proven to be essential for early T-cell development, at least in part, by regulating Notch1 expression 132,133. Both genes, Tcf7 and Gata3, are bound by EBF1, and they show increased expression upon loss of EBF1 129,130. EBF1 induces the repressive histone mark H3K27me3 at the Gata3 locus 129. A direct regulation of Gata3 by EBF1 was underlined by the expression of a synthetic zinc finger protein that blocks binding of EBF1 to a regulatory site of Gata3, which resulted in increased Gata3 expression and restoration of T-lineage potential 129.
EBF1 also promotes B-cell development by repression of Id2 and Id3 110,134. ID proteins are antagonists of E proteins, including E2A, and heterodimer formation between ID and E proteins inhibits the DNA-binding ability of E proteins 135. ID2 is a major regulator of all innate lymphoid cell types 136,137, and the lineage conversion of Ebf1-deficient pro-B cells into ILCs is likely accounted for by the de-repression of Id2 130.
Finally, EBF1 also represses a key determinant of myeloid differentiation, Cebpa, and thereby antagonizes myeloid differentiation 110. However, CEBPα also antagonizes the expression of Ebf1 and Pax-5, and ectopic expression of CEBPα in a pre-B-cell line efficiently converts the cells into a macropahage-like cell type 138. Although Ebf1 and Pax-5 both regulate B-cell lineage commitment, they appear to repress distinct genes. Notably, EBF1 mainly represses genes encoding transcription factors that determine alternative cell fates, whereas Pax-5 inhibits genes encoding receptors that promote alternative cell fates and thereby renders cells unresponsive to alternative lineage signals. By repressing distinct genes, EBF1 and Pax-5 antagonize alternative cell fates by a ‘double-lock mechanism’.
Role of EBF1 in mature B cells
EBF1 is expressed not only at the onset of B-cell development but throughout B-cell differentiation until the plasma cell stage. The block in early B-cell development of Ebf1-deficient mice obscured an analysis of EBF1 function in mature B cells but two recent studies, using a conditional knockout of Ebf1, provided insight into the role of EBF1 in later stage B cells 42,109. Tamoxifen-induced deletion of Ebf1 in pre-B cells revealed an increased cell death and a defect in cell cycle progression as the mutant cells accumulate at the G1 phase 42. Notably, transformation of primary Ebf1-deficient pro-B cells with Abelson murine leukemia virus (A-MuLV) overcame the proliferation defect but did not rescue the survival defect. However, survival and proliferation of Ebf1-deficient pro-B cells is observed upon transformation by A-MuLV and ectopic expression of Myb 42.
Conditional deletion of Ebf1 in peripheral B cells generates a strong reduction in marginal zone (MZ) B cells in the spleen, as well as B1 cells in the peritoneum 42,109. In contrast, EBF1 seems to be dispensable for the generation and/or maintenance of follicular B (FoB) cells. However, surface expression of CD19 and CD21 on Ebf1-deficient follicular B cells is strongly decreased. Stimulation of Ebf1-deficient FoB cells showed impaired calcium mobilization and BCR signaling, including reduced phosphorylation of CD79a, CD19, and Akt 42. Moreover, BCR stimulation of Ebf1-deficient FoB cells results in augmented apoptosis and impaired proliferation 42,109. Finally, Ebf1 deletion affects germinal center (GC) B-cell development. Immunization of mice with sheep red blood cells (SRBC) initiates the differentiation of mature splenic B cells into GC B cells that perform CSR and SHM 42,109. Although GCs are formed after immunization of mice deficient for EBF1 in mature B cells, the number of GC B cells is strongly reduced, indicating that EBF1 is required for the maintenance of GC B cells 42,109. Thus, EBF1 plays multiple roles in the later stages of B-cell differentiation that may help to coordinate the processes of differentiation, cell proliferation, and survival.
Conclusion
Efforts aimed at understanding the regulatory circuitry that underlies B lymphopoiesis have identified several transcription factors that generate a permissive chromatin context in progenitor cells and/or determine a lineage-specific pattern of gene expression. Notably, the regulatory determinants of B lymphopoiesis are interconnected via feedback loops that stabilize lineage decisions and coordinate their action in establishing and maintaining cell type-specific patterns of gene expression. Genome-wide analysis of transcription factor occupancy has provided us an interesting insight into the combinatorial regulation of target genes. Future studies will involve single cell analysis to elucidate the heterogeneity of cell populations and the developmental capacity of individual cells. Moreover, clarification of the causal relationships of transcription factor binding, epigenetic regulation, and changes in higher order chromatin structure will be necessary to obtain a comprehensive understanding of this complex developmental process.
Acknowledgments
We are grateful to Peter Nielsen for critically reading the manuscript and Marika Rott for help in finalizing the figures and manuscript. Research in the Grosschedl lab is supported by funds from the Max Planck Society and the German Research Foundation. The authors have no conflict of interest to declare.
References
- 1.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 2.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22:161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Welner RS, Pelayo R, Kincade PW. Evolving views on the genealogy of B cells. Nat Rev Immunol. 2008;8:95–106. doi: 10.1038/nri2234. [DOI] [PubMed] [Google Scholar]
- 5.Panaroni C, Wu JY. Interactions between B lymphocytes and the osteoblast lineage in bone marrow. Calcif Tissue Int. 2013;93:261–268. doi: 10.1007/s00223-013-9753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22:177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zandi S, Bryder D, Sigvardsson M. Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol Rev. 2010;238:47–62. doi: 10.1111/j.1600-065X.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 9.Mercer EM, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–425. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Agosto JA, et al. The hematopoietic stem cell and its niche: a comparative view. Genes Dev. 2007;21:3044–3060. doi: 10.1101/gad.1602607. [DOI] [PubMed] [Google Scholar]
- 11.Arinobu Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Akashi K, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 13.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 16.Mansson R, et al. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- 17.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allman D, Li J, Hardy RR. Commitment to the B lymphoid lineage occurs before DH-JH recombination. J Exp Med. 1999;189:735–740. doi: 10.1084/jem.189.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roessler S, Grosschedl R. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin Immunol. 2006;18:12–19. doi: 10.1016/j.smim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Rolink AG, Andersson J, Melchers F. Molecular mechanisms guiding late stages of B-cell development. Immunol Rev. 2004;197:41–50. doi: 10.1111/j.0105-2896.2004.0101.x. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z, et al. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann NY Acad Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagman J, Travis A, Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 1991;10:3409–3417. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagman J, et al. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 24.Travis A, et al. Purification of early-B-cell factor and characterization of its DNA-binding specificity. Mol Cell Biol. 1993;13:3392–3400. doi: 10.1128/mcb.13.6.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 26.Kudrycki K, et al. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol Cell Biol. 1993;13:3002–3014. doi: 10.1128/mcb.13.5.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois L, Vincent A. The COE–Collier/Olf1/EBF–transcription factors: structural conservation and diversity of developmental functions. Mech Dev. 2001;108:3–12. doi: 10.1016/s0925-4773(01)00486-5. [DOI] [PubMed] [Google Scholar]
- 28.Liberg D, Sigvardsson M, Akerblad P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol Cell Biol. 2002;22:8389–8397. doi: 10.1128/MCB.22.24.8389-8397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagman J, et al. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields S, et al. The ‘zinc knuckle’ motif of Early B cell Factor is required for transcriptional activation of B cell-specific genes. Mol Immunol. 2008;45:3786–3796. doi: 10.1016/j.molimm.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treiber N, et al. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev. 2010;24:2270–2275. doi: 10.1101/gad.1976610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siponen MI, et al. Structural determination of functional domains in early B-cell factor (EBF) family of transcription factors reveals similarities to Rel DNA-binding proteins and a novel dimerization motif. J Biol Chem. 2010;285:25875–25879. doi: 10.1074/jbc.C110.150482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bork P, et al. Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci. 1999;24:261–263. doi: 10.1016/s0968-0004(99)01416-4. [DOI] [PubMed] [Google Scholar]
- 34.Muller CW, Harrison SC. The structure of the NF-kappa B p50:DNA-complex: a starting point for analyzing the Rel family. FEBS Lett. 1995;369:113–117. doi: 10.1016/0014-5793(95)00541-g. [DOI] [PubMed] [Google Scholar]
- 35.Cramer P, Muller CW. A firm hand on NFkappaB: structures of the IkappaBalpha-NFkappaB complex. Structure. 1999;7:R1–R6. doi: 10.1016/s0969-2126(99)80002-1. [DOI] [PubMed] [Google Scholar]
- 36.Wang SS, Tsai RY, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang SS, Betz AG, Reed RR. Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol Cell Neurosci. 2002;20:404–414. doi: 10.1006/mcne.2002.1138. [DOI] [PubMed] [Google Scholar]
- 38.Hagman J, Lukin K. Early B-cell factor ‘pioneers’ the way for B-cell development. Trends Immunol. 2005;26:455–461. doi: 10.1016/j.it.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Ma PC, et al. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 40.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyory I, et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012;26:668–682. doi: 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier H, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 44.Sigvardsson M, et al. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier H, et al. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23:1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao H, et al. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramirez J, et al. MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol. 2012;32:5078–5088. doi: 10.1128/MCB.00819-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bond HM, et al. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood. 2004;103:2062–2070. doi: 10.1182/blood-2003-07-2388. [DOI] [PubMed] [Google Scholar]
- 51.Tsai RY, Reed RR. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol Cell Biol. 1998;18:6447–6456. doi: 10.1128/mcb.18.11.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harder L, et al. Aberrant ZNF423 impedes B cell differentiation and is linked to adverse outcome of ETV6-RUNX1 negative B precursor acute lymphoblastic leukemia. J Exp Med. 2013;210:2289–2304. doi: 10.1084/jem.20130497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warming S, et al. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Kruppel-like zinc finger protein. Blood. 2003;101:1934–1940. doi: 10.1182/blood-2002-08-2652. [DOI] [PubMed] [Google Scholar]
- 54.Mega T, et al. Zinc finger protein 521 antagonizes early B-cell factor 1 and modulates the B-lymphoid differentiation of primary hematopoietic progenitors. Cell Cycle. 2011;10:2129–2139. doi: 10.4161/cc.10.13.16045. [DOI] [PubMed] [Google Scholar]
- 55.Kang S, et al. Regulation of early adipose commitment by Zfp521. PLoS Biol. 2012;10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jimenez MA, et al. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Griffin MJ, et al. Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem. 2013;288:35925–35939. doi: 10.1074/jbc.M113.491936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 59.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 60.Lichtinger M, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nichogiannopoulou A, et al. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 2010;584:4910–4914. doi: 10.1016/j.febslet.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 63.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 64.O'Neill DW, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida T, et al. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynaud D, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heizmann B, Kastner P, Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med. 2013;210:2823–2832. doi: 10.1084/jem.20131735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiang Y, et al. A multifunctional element in the mouse Igkappa locus that specifies repertoire and Ig loci subnuclear location. J Immunol. 2011;186:5356–5366. doi: 10.4049/jimmunol.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferreiros-Vidal I, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–1782. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- 70.Schwickert TA, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zarnegar MA, Rothenberg EV. Ikaros represses and activates PU.1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene. 2012;31:4647–4654. doi: 10.1038/onc.2011.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spooner CJ, et al. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 74.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 75.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 76.Zhang DE, et al. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeKoter RP, et al. Regulation of the interleukin-7 receptor alpha promoter by the Ets transcription factors PU.1 and GA-binding protein in developing B cells. J Biol Chem. 2007;282:14194–14204. doi: 10.1074/jbc.M700377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 79.Medina KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 82.Bain G, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 83.Semerad CL, et al. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Q, et al. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dias S, et al. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoogenkamp M, et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol. 2008;20:247–256. doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Greig KT, et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kosan C, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 90.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 91.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 92.Urbanek P, et al. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 93.Mansson R, et al. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 94.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beck K, et al. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- 98.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kikuchi K, et al. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seo W, et al. Runx1-Cbfbeta facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209:1255–1262. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choi J, et al. The SWI/SNF-like BAF complex is essential for early B cell development. J Immunol. 2012;188:3791–3803. doi: 10.4049/jimmunol.1103390. [DOI] [PubMed] [Google Scholar]
- 102.Jiang XX, et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity. 2011;35:883–896. doi: 10.1016/j.immuni.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mansson R, et al. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci USA. 2012;109:21028–21033. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Welinder E, et al. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci USA. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Decker T, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 108.Hagman J, Ramirez J, Lukin K. B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1. Curr Top Microbiol Immunol. 2012;356:17–38. doi: 10.1007/82_2011_139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vilagos B, et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med. 2012;209:775–792. doi: 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Z, et al. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 2003;22:4759–4769. doi: 10.1093/emboj/cdg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akerblad P, et al. The B29 (immunoglobulin beta-chain) gene is a genetic target for early B-cell factor. Mol Cell Biol. 1999;19:392–401. doi: 10.1128/mcb.19.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gisler R, Akerblad P, Sigvardsson M. A human early B-cell factor-like protein participates in the regulation of the human CD19 promoter. Mol Immunol. 1999;36:1067–1077. doi: 10.1016/s0161-5890(99)00092-9. [DOI] [PubMed] [Google Scholar]
- 114.Akerblad P, Sigvardsson M. Early B cell factor is an activator of the B lymphoid kinase promoter in early B cell development. J Immunol. 1999;163:5453–5461. [PubMed] [Google Scholar]
- 115.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 116.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 117.Zandi S, et al. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- 118.Sigvardsson M. Overlapping expression of early B-cell factor and basic helix-loop-helix proteins as a mechanism to dictate B-lineage-specific activity of the lambda5 promoter. Mol Cell Biol. 2000;20:3640–3654. doi: 10.1128/mcb.20.10.3640-3654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gisler R, Sigvardsson M. The human V-preB promoter is a target for coordinated activation by early B cell factor and E47. J Immunol. 2002;168:5130–5138. doi: 10.4049/jimmunol.168.10.5130. [DOI] [PubMed] [Google Scholar]
- 120.Lukin K, et al. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc Natl Acad Sci USA. 2010;107:7869–7874. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hesslein DG, et al. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nutt SL, et al. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 123.Nutt SL, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. [PubMed] [Google Scholar]
- 124.Zandi S, et al. Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc Natl Acad Sci USA. 2012;109:15871–15876. doi: 10.1073/pnas.1210144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rolink AG, et al. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–606. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 126.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 127.Delogu A, et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 128.Souabni A, et al. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17:781–793. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]
- 129.Banerjee A, et al. Transcriptional repression of Gata3 is essential for early B cell commitment. Immunity. 2013;38:930–942. doi: 10.1016/j.immuni.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nechanitzky R, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 131.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hendriks RW, et al. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 133.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thal MA, et al. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc Natl Acad Sci USA. 2009;106:552–557. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 136.Klose CS, et al. Transcriptional control of innate lymphocyte fate decisions. Curr Opin Immunol. 2012;24:290–296. doi: 10.1016/j.coi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 137.Spits H, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 138.Bussmann LH, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]


