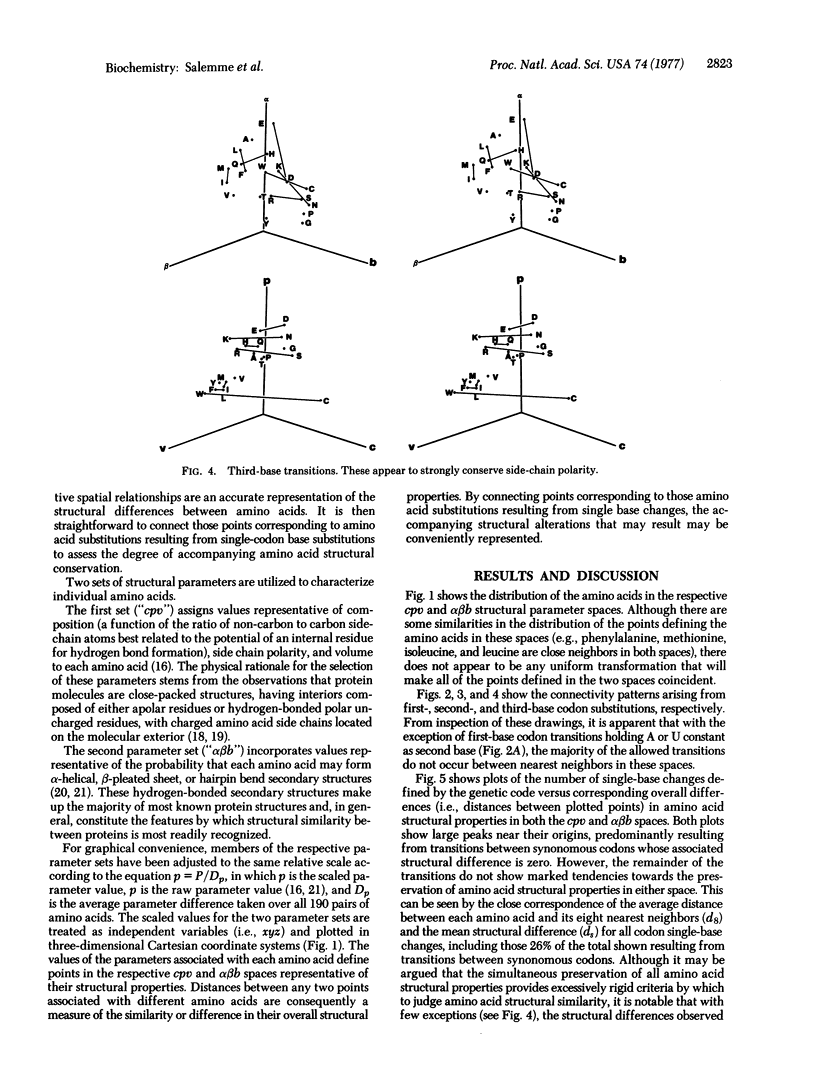
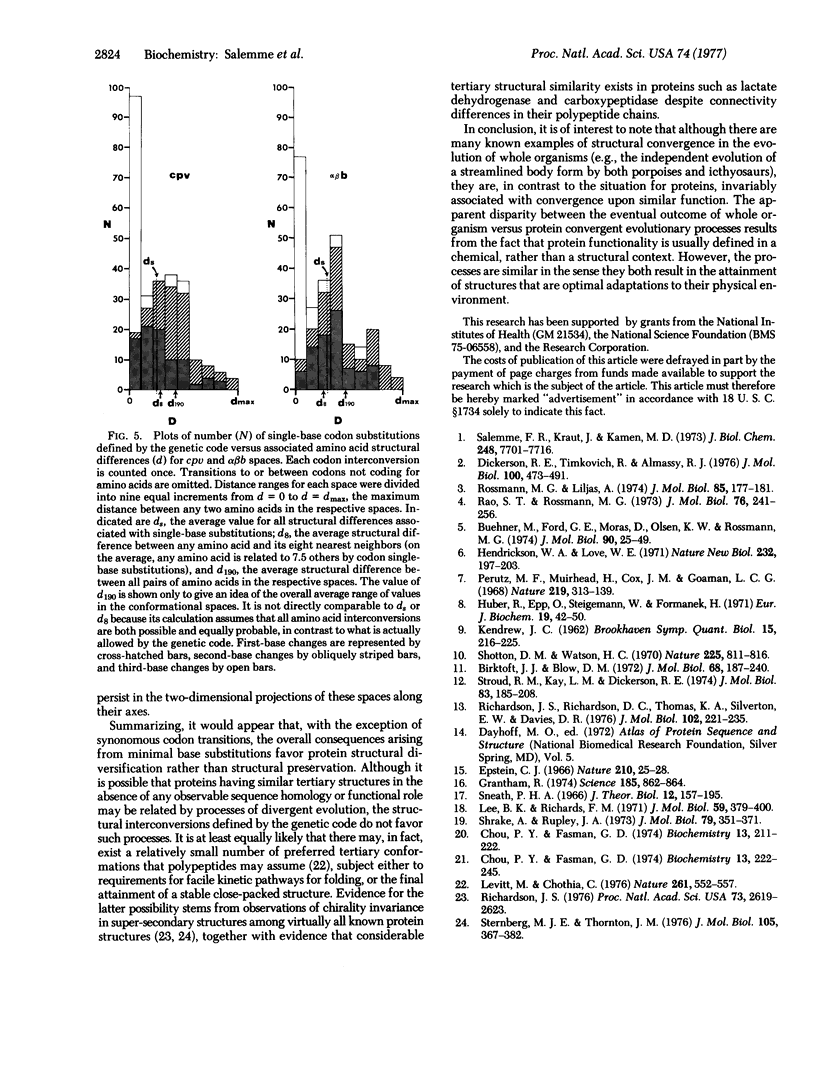
Abstract
Several recent protein crystallographic structure determinations have demonstrated the existence of considerable tertiary structural similarity among proteins otherwise having little similarity in either amino acid sequence or biological function. In order to assess the possibility that such proteins may have arisen through processes of divergent evolution from a common ancestor, a graphical presentation is given which correlates the pattern of allowed single base substitutions defined by the genetic code with the associated changes in the structural properties of the encoded amino acids. The results show that while a large degree of structural conservation is evident due to codon synonomy, there is, in general, little tendency for the code to be structurally conservative in the majority of the cases where codon single-base changes result in amino acid substitutions. The possible consequences of this pattern of potential amino acid substitutions are discussed in relation to protein evolutionary processes.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Buehner M., Ford G. C., Olsen K. W., Moras D., Rossman M. G. Three-dimensional structure of D-glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1974 Nov 25;90(1):25–49. doi: 10.1016/0022-2836(74)90254-x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Timkovich R., Almassy R. J. The cytochrome fold and the evolution of bacterial energy metabolism. J Mol Biol. 1976 Feb 5;100(4):473–491. doi: 10.1016/s0022-2836(76)80041-1. [DOI] [PubMed] [Google Scholar]
- Epstein C. J. Role of the amino-acid "code" and of selection for conformation in the evolution of proteins. Nature. 1966 Apr 2;210(5031):25–28. doi: 10.1038/210025a0. [DOI] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974 Sep 6;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Love W. E. Structure of lamprey haemoglobin. Nat New Biol. 1971 Aug;232(33):197–203. doi: 10.1038/newbio232197a0. [DOI] [PubMed] [Google Scholar]
- Huber R., Epp O., Steigemann W., Formanek H. The atomic structure of erythrocruorin in the light of the chemical sequence and its comparison with myoglobin. Eur J Biochem. 1971 Mar 1;19(1):42–50. doi: 10.1111/j.1432-1033.1971.tb01285.x. [DOI] [PubMed] [Google Scholar]
- KENDREW J. C. Side-chain interactions in myoglobin. Brookhaven Symp Biol. 1962 Dec;15:216–228. [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F., Muirhead H., Cox J. M., Goaman L. C. Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 A resolution: the atomic model. Nature. 1968 Jul 13;219(5150):131–139. doi: 10.1038/219131a0. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. Handedness of crossover connections in beta sheets. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2619–2623. doi: 10.1073/pnas.73.8.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C., Thomas K. A., Silverton E. W., Davies D. R. Similarity of three-dimensional structure between the immunoglobulin domain and the copper, zinc superoxide dismutase subunit. J Mol Biol. 1976 Apr 5;102(2):221–235. doi: 10.1016/s0022-2836(76)80050-2. [DOI] [PubMed] [Google Scholar]
- Rossman M. G., Liljas A. Letter: Recognition of structural domains in globular proteins. J Mol Biol. 1974 May 5;85(1):177–181. doi: 10.1016/0022-2836(74)90136-3. [DOI] [PubMed] [Google Scholar]
- Salemme F. R., Kraut J., Kamen M. D. Structural bases for function in cytochromes c. An interpretation of comparative x-ray and biochemical data. J Biol Chem. 1973 Nov 25;248(22):7701–7716. [PubMed] [Google Scholar]
- Shotton D. M., Watson H. C. Three-dimensional structure of tosyl-elastase. Nature. 1970 Feb 28;225(5235):811–816. doi: 10.1038/225811a0. [DOI] [PubMed] [Google Scholar]
- Shrake A., Rupley J. A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol. 1973 Sep 15;79(2):351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Sneath P. H. Relations between chemical structure and biological activity in peptides. J Theor Biol. 1966 Nov;12(2):157–195. doi: 10.1016/0022-5193(66)90112-3. [DOI] [PubMed] [Google Scholar]
- Sternberg M. J., Thornton J. M. On the conformation of proteins: the handedness of the beta-strand-alpha-helix-beta-strand unit. J Mol Biol. 1976 Aug 15;105(3):367–382. doi: 10.1016/0022-2836(76)90099-1. [DOI] [PubMed] [Google Scholar]
- Stroud R. M., Kay L. M., Dickerson R. E. The structure of bovine trypsin: electron density maps of the inhibited enzyme at 5 Angstrom and at 2-7 Angstron resolution. J Mol Biol. 1974 Feb 25;83(2):185–208. doi: 10.1016/0022-2836(74)90387-8. [DOI] [PubMed] [Google Scholar]