Abstract
The species Corynebacterium pyruviciproducens was described in 2010 based on the features of a single strain. In this report, we describe the chemotaxonomic and phenotypic characteristics of 11 C. pyruviciproducens clinical strains isolated in Switzerland and Canada in comparison to the strain 06-17730T. Heterogeneities within the type strain were found in the 16S rRNA gene and in biochemical markers. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) identification of this species could not be achieved since currently this bacterial species is not included in the corresponding database. Reliable identification is obtained only with sequence-based identification tools. Results of antimicrobial susceptibility testing of this species with an extended panel of antimicrobials are presented here for the first time.
INTRODUCTION
Corynebacterium pyruviciproducens was first described in 2010, based on studies of the strain 06-17730T (= WAL 19168T = CCUG 57046T = ATCC BAA-1742T = DSM 45585T) cultured from a groin abscess (1). This bacterial species has now been isolated from several clinical specimens in laboratories in Switzerland and Canada. During routine bacteriological identification procedures, we observed a number of differences in the phenotypic and other properties between these and the published description of C. pyruviciproducens strain 06-17730T (1). These observations as well as heterogeneities in the 16S rRNA sequences are presented in this report. In the species description publication, Tong and colleagues (1) reported that the C. pyruviciproducens type strain was highly sensitive to ampicillin, ceftriaxone, clindamycin, and erythromycin. Since no other information concerning the antimicrobial susceptibility of C. pyruviciproducens has been published to date, we present in this report detailed data on the antimicrobial susceptibility testing of isolates from our laboratories, including new susceptibility data for the strain 06-17730T using an extended panel of antimicrobials.
MATERIALS AND METHODS
Strains and type strain.
Four C. pyruviciproducens strains were isolated from in-patients of the University Hospital Basel, Switzerland, during the years 2011 and 2012 (laboratory number [source], 718260/2011 [urine], 713182/2012 [urine], 707471/2012 [urine], and 607685/2011 [bone biopsy specimen]). Seven Canadian isolates were referred to the National Microbiology Laboratory (NML number [source], 94-0264 [synovial fluid], 95-0358 [blood culture], 96-0085 [scapular abscess], 00-0179 [cesarean section incision fluid], 00-A-091 [chest abscess], 100222 [skin infection], and 130606 [breast abscess fluid]). All strains were identified as C. pyruviciproducens using sequence-based methods. Data for all strains were amalgamated for this study, with comments about differences in underlying methods added as required. C. pyruviciproducens strain 06-17730T was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) (accession no. DSM 45585T).
Phenotypic methods.
Swiss isolates were tested for lipophilia after observation for growth on normal blood plates compared with growth on blood plates containing 1% Tween 80. Canadian isolates were tested for lipophilia by comparing growth in brain heart infusion broth (BHIB) (Becton, Dickinson) with growth in BHIB enriched with 1% Tween 80 (2).
Biochemical testing of all strains was performed using the API Coryne panel (bioMérieux) according to the manufacturer's instructions. Enzyme production of the Canadian isolates was analyzed using the API ZYM panel (bioMérieux).
The Canadian strains except NML 130606 were tested for products of glucose fermentation using conventional tube biochemical tests (2). Cellular fatty acid composition was determined with the Sherlock system (MIDI) as described previously (3) using version 4.5 of the MIDI operating system and MIDI library generation software.
MALDI-TOF mass spectrometry analysis.
Colonies were smeared onto a steel target plate and overlaid with 1 μl of matrix solution consisting of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile-2.5% trifluoroacetic acid (normal protocol) and of 1 μl of 70% formic acid was added to the smears before use of the matrix solution (short protocol). Measurements were performed using a Microflex LT matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) system and analyzed with Biotyper 3.0 SR software (MALDI Biotyper; Bruker Daltonics GmbH).
Molecular phylogenetic analysis.
16S rRNA gene sequencing was done as described previously, generating >1,400 bp (4) with partial (∼400 bp) rpoB gene sequencing of Canadian isolates being done as outlined by Khamis et al. (5). ClustalW software and the neighbor-joining algorithm found in MEGA 5.2 were used for phylogenetic analysis.
Determination of MICs of different antimicrobials.
MICs were determined for the Swiss strains with Etest strips (bioMérieux) on Mueller-Hinton agar with blood (Heipha) using inocula of 1 McFarland unit incubated in an atmosphere of 5% CO2 at 37°C for 48 h. The Clinical and Laboratory Standards Institute (CLSI) breakpoints for corynebacteria (6) were used for the interpretation of the MICs of all tested antimicrobial agents except for amoxicillin-clavulanic acid, piperacillin-tazobactam, cefuroxime, moxifloxacin, and tigecycline, which were interpreted by using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) non-species-related breakpoints (7). Canadian isolates were analyzed using CLSI-described broth microdilution methods (8) and then interpreted (6) using Trek Sensititre plates and cation-adjusted Mueller-Hinton broth with lysed horse blood (Thermo Scientific Oxoid).
Nucleotide sequence accession numbers.
Nearly the entire 16S rRNA gene sequence is deposited in GenBank under the following strain designations and accession no.: NML 94-0264, GU304653; NML 95-0358, GU304655; NML 00-0179, GU304659; NML 96-0085, GU304657; 718260/2011, JX501770; 607685/2011, JX501771; 713182/2012, KF372872; and 707471/2012, KF372873. Strain names and accession nos. of rpoB gene sequences are the following: NML 94-0264, GU304654; NML 95-0358, GU304656; NML 96-0085, GU304658; and NML 00-0179, GU304660.
RESULTS AND DISCUSSION
All isolates, after 24 to 48 h of incubation in an atmosphere containing 5% CO2, were observed to be small, translucent to grayish, catalase-positive colonies, visible on Columbia agar supplemented with 5% sheep blood (BD Diagnostic Systems). The Canadian strains tended to grow better in 5% CO2 than with incubation in air or anaerobically. Gram staining revealed irregular, granulated, and long Gram-positive rods that did not show the typical “palisade” appearance of corynebacteria. The CAMP reaction performed on blood plates in the presence of Staphylococcus aureus ATCC 25923 showed variable results (Table 1). All Swiss isolates were found to be nonlipophilic, including the type strain. Five of seven Canadian isolates were lipophilic. The results of the biochemical tests are summarized in Table 1. Using the API Coryne panel, we identified isolates as Corynebacterium glucuronolyticum with scores consistent with good to excellent identification profiles where esculin was hydrolyzed but as the Corynebacterium renale group with doubtful identification profiles if esculin was negative (Table 1). Enzyme production was found to give essentially the same results as those found by Tong et al. (1) when Canadian isolates were tested. Like C. glucuronolyticum, C. pyruviciproducens is the only other Corynebacterium species to date which consistently produces β-glucuronidase, and so these species may be misidentified for each other if identification is made based solely on the use of phenotypic assays where production of this enzyme is a heavily weighted test.
TABLE 1.
Biochemical traits of C. pyruviciproducens isolates
| Biochemical trait | Results fora: |
|||
|---|---|---|---|---|
| Strain 06-17730Tb | Strain 06-17730Tc | Swiss strains (n = 4) (no. positive/no. tested [%])d | Canadian strains (n = 7) (no. positive/no. tested [%])d | |
| CAMP reaction | − | + | + | V (3/6 [50], 1 ND) |
| Lipophilia | + | − | − | + (5/5 [100], 2 ND) |
| Reduction in nitrates | − | − | − | V (2/7 [29]) |
| β-Glucosidase (esculin) | − | − | V (3/4 [75]) | V (4/7 [57]) |
| Fermentation of: | ||||
| d-Xylose | + | + | − | V (2/7 [29]) |
| d-Maltose | + | − | − | V (4/7 [57]) |
| d-Sucrose | + | − | V (1/4 [25]) | V (6/7 [86]) |
| Glycogen | − | − | − | V (2/7 [29]) |
| Fructose | − | ND | ND | V (5/7 [71]) |
API Coryne codes: for strain 06-17730T, 2200725; for strain 06-17730T, 2200(5/7)04; for Swiss strains, 2200304 (1), 2240304 (2), and 2240305 (1); and for Canadian strains, 0200304 (1), 2200705 (1), 2240305 (2), 2240705 (1), 3240105 (1), and 3240305 (1). +, positive; −, negative.
Data as described by Tong et al. (1).
As observed in this study for DSM 45585T.
All Swiss or Canadian patient isolates were positive or weakly positive for catalase, for fermentation of glucose and ribose, and for the production of pyrazinamidase and β-glucuronidase. Using conventional tube sugars, Canadian isolates were also variable for maltose, galactose, glycerol, mannose, trehalose, and salicin, but maltose was negative for all strains when the API Coryne panel was used. Conventional or API Coryne reactions were negative for urease and gelatin hydrolysis, mannitol, and lactose. Pyrrolidonyl arylamidase, alkaline phosphatase, β-galactosidase, α-glucosidase, and N-acetyl-β-glucosaminidase were not detected in any strain. V, variable, that is, at least 1 strain expressed that result; ND, not done.
The Canadian strains, except NML 130606, showed trace to small volumes of succinic, propionic, and lactic acids, in addition to acetic and pyruvic acids as described by Tong et al. (1). The cellular fatty acid profiles for the 7 Canadian strains were found to be consistent with those described by Tong et al., including the observation that tuberculostearic acid (TBSA) could not be detected (1) (Canadian data not shown).
Identification of C. pyruviciproducens could not be achieved with MALDI-TOF mass spectrometry since this bacterial species is not included in the current Bruker MALDI Biotyper database.
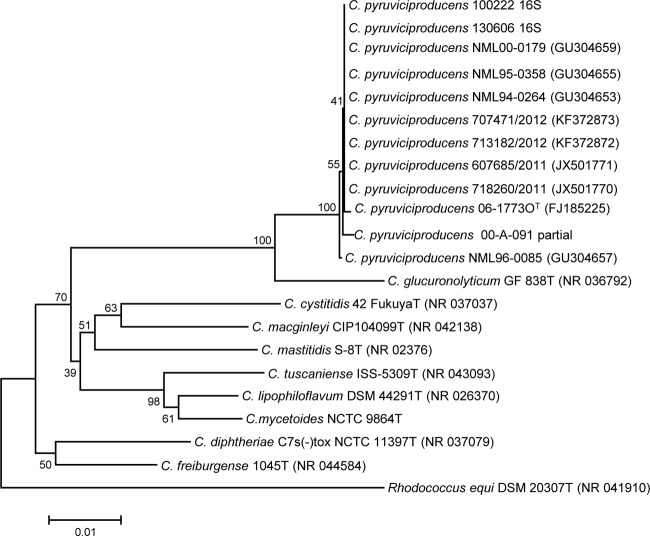
We demonstrate the nearly identical alignment between the C. pyruviciproducens type strain and the clinical isolates described in this study when partial rpoB gene sequences were analyzed (see the partial rpoB gene sequences with GenBank accession numbers assigned to these isolates as shown in Fig. 2). On the other hand, when the full 16S rRNA gene sequences of the strains of this study were aligned with the C. pyruviciproducens strain 06-17730T, 11 polymorphisms (99.22% identity) were detected, mainly in the first 200-bp fragment of the gene (Fig. 1).
FIG 2.
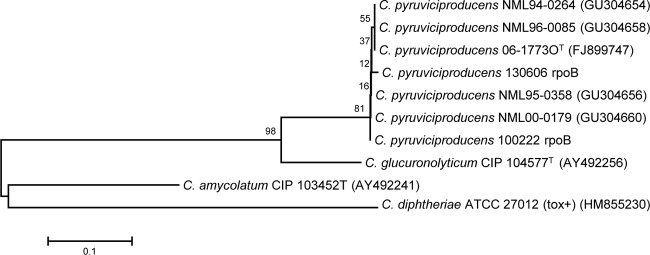
Relationship of partial rpoB gene sequences inferred by the neighbor-joining method for Canadian study strains of C. pyruviciproducens. The numbers at the nodes are bootstrap values (percentage based on 1,000 replications); only values ≥50% shown. GenBank accession numbers are given in parentheses. Bar, 0.1 substitution per nucleotide position, with Corynebacterium diphtheriae used as the outgroup.
FIG 1.
Relationship of nearly full-length 16S rRNA gene sequences inferred by the neighbor-joining method for all study strains identified as Corynebacterium pyruviciproducens. The numbers at the nodes are bootstrap values (percentage based on 1,000 replications); only values ≥50% shown. GenBank accession numbers are given in parentheses. Bar, 0.01 substitution per nucleotide position, with Rhodococcus equi used as the outgroup.
The combined antimicrobial susceptibility profiles for C. pyruviciproducens isolates and a review for the C. pyruviciproducens type strain are presented in Table 2. The susceptibility testing of the type strain and aggregated analysis of the strains in this study showed that this species was always susceptible to tigecycline, gentamicin, vancomycin, rifampin, linezolid, quinupristin-dalfopristin, chloramphenicol, and daptomycin, whereas the susceptibilities to beta-lactams, erythromycin, clindamycin, fluoroquinolones, tetracycline, and trimethoprim-sulfamethoxazole were variable (Table 2). The NML skin isolate 100222 and bone isolate 607685/2011 were found to be more profoundly resistant than other strains. Both isolates were susceptible to vancomycin, daptomycin, gentamicin, rifampin, and linezolid, whereas isolate 100222 was additionally susceptible to carbapenems and quinupristin-dalfopristin and 607685/2011 was susceptible to tetracycline, tigecycline, ciprofloxacin, and moxifloxacin.
TABLE 2.
Antimicrobial susceptibility testing results
| Antimicrobial agenta | Results for DSM 45585T = 06-17730T: |
MIC range (μg ml−1) |
No. susceptible/total no. (%) | No. of intermediate or resistant/immune/total no. (%) | No. with lower MICs/total no. (%) | No. with higher MICs/total no. (%) | ||
|---|---|---|---|---|---|---|---|---|
| MIC | Interpretationb | Swiss isolates (n = 4) | Canadian isolates (n = 6) | |||||
| Penicillin | 0.094 | S | 0.064–32 | ≤0.06–4 | 8/10 (80) | 2/10 (20) | ||
| Amoxicillin-clavulanic acidc | 0.094 | NB | 0.047–32 | ≤2/1 to >16/8 | 4/6 (67) | 2/6 (33) | ||
| Piperacillin-tazobactamc,d | 0.5 | S | 0.25–32 | NDe | 3/4 (75) | 1/4 (25) | ||
| Cefuroximec | 0.125 | NB | 0.19–3 | ≤0.5–1 | 10/10 (100) | 0 (0) | ||
| Ceftriaxone | 1.5 | I | 1–16 | 0.25 to >2 | 6/10 (60) | 4/10 (40) | ||
| Cefepime | 0.25 | S | 0.25–16 | ≤0.5–4 | 8/10 (80) | 2/10 (20) | ||
| Imipenemd | 0.064 | S | 0.064–12 | ND | 3/4 (75) | 1/4 (25) | ||
| Erythromycin | 2 | R | 0.023–32 | >4 | 2/10 (20) | 8/10 (80) | ||
| Clindamycin | >256 | R | 0.125 to >256 | >2 | 1/10 (10) | 9/10 (90) | ||
| Tetracycline | 0.75 | S | 0.25–1 | ≤1 to >16 | 5/10 (50) | 5/10 (50) | ||
| Tigecyclinec | 0.032 | NB | 0.032–0.19 | 0.03–0.5 | 10/10 (100) | 0 (0) | ||
| Gentamicin | 0.25 | S | 0.094–0.38 | ≤2 | 10/10 (100) | 0 (0) | ||
| Ciprofloxacin | 0.19 | S | 0.25 to >32 | ≤0.5 to >2 | 7/10 (70) | 3/10 (30) | ||
| Moxifloxacinc | 0.064 | NB | 0.19–6 | ≤1–2 | 7/10 (70) | 3/10 (30) | ||
| Vancomycin | 0.38 | S | 0.38–0.5 | ≤0.5–1 | 10/10 (100) | 0 (0)4 | ||
| Rifampin | <0.002 | S | 0.002–0.003 | ≤0.5 | 10/10 (100) | 0 (0) | ||
| Linezolid | 0.38 | S | 0.25–0.38 | 0.5 | 10/10 (100) | 0 (0) | ||
| Daptomycin | 0.032 | S | 0.023–0.032 | ≤0.06–0.12 | 10/10 (100) | 0 (0) | ||
| Ampicillinc,d | ND | ND | ≤0.12–16 | 5/6 (83) | 1/6 (17) | |||
| Azithromycinc,d | ND | ND | >2 | 0/2 (0) | 2/2 (100) | |||
| Cefotaximed | ND | ND | ≤0.12–2 | 5/6 (83) | 1/6 (17) | |||
| Chloramphenicolc,d | ND | ND | 4 | 6/6 (100) | 0 (0) | |||
| Ertapenamc,d | ND | ND | ≤0.5 to >2 | 4/6 (67) | 2/6 (33) | |||
| Gatifloxacinc,d | ND | ND | ≤1–4 | 5/6 (83) | 1/6 (17) | |||
| Levofloxacinc,d | ND | ND | ≤0.25 to >8 | 5/6 (83) | 1/6 (17) | |||
| Meropenemc,d | ND | ND | ≤0.25–0.5 | 6/6 (100) | 0/6 (0) | |||
| Quinupristin-dalfopristind | ND | ND | ≤0.12–0.25 | 6/6 (100) | 0/6 (0) | |||
| Trimethoprim-sulfamethoxazoled | ND | ND | ≤0.5/9.5–4/76 | 5/6 (83) | 1/6 (17) | |||
The antimicrobial agents shown were tested by both laboratories either by Etest (Swiss, 4 patient strains and the type strain) or broth microdilution (Canadian, 6 patient strains; NML 00A091 did not grow well enough in broth to perform testing).
S, sensitive; I, intermediate; R, resistant (or immune); NB, no breakpoints by CLSI.
CLSI does not describe breakpoints for this antimicrobial agent, so MICs were not interpreted by the Canadian laboratory when done. For this study, both Swiss and Canadian strains were interpreted using European Committee on Antimicrobial Susceptibility Testing (EUCAST) non-species-related breakpoints (6) for amoxicillin-clavulanic acid, piperacillin-tazobactam, cefuroxime, moxifloxacin, and tigecycline. Data for amoxicillin-clavulanic acid and azithromycin were obtained only for 2 Canadian strains (NML 100222 and NML 130606).
Data from the Swiss or Canadian laboratories, but not both.
ND, not done.
This report describes variations in several phenotypic traits from those described in the C. pyruviciproducens species novum description publication. Interestingly, some discrepant characteristics were observed by reinvestigating the type strain, demonstrating differences in the results for the CAMP reaction and for lipophilia testing; data shown here suggest that at least some discrepant traits may have been caused by different methodology and interpretation criteria (e.g., different results were observed, depending on whether Tween incorporated into plates rather than broths was used for demonstrating lipophilia). The laboratory identification of C. pyruviciproducens can be achieved only by 16S rRNA or rpoB gene sequencing, since this species is not present in the API Coryne database or in the current version of the Bruker MALDI-TOF Biotyper database. Even identification by 16S rRNA gene sequencing was more difficult than expected, as the GenBank deposit for the type strain (accession no. FJ185225) is labeled as Corynebacterium sp. 06-17730, rather than as C. pyruviciproducens 06-17730T when BLASTed against the nr/nt database, and this strain is not otherwise found in GenBank's curated refseq database. The Swiss and Canadian strains showed significant 16S rRNA sequence heterogeneities (up to 11 mismatches) compared to those of the type strain sequence, but identification was confirmed by demonstrating a close relationship to the type strain of C. pyruviciproducens with partial sequencing of rpoB (Fig. 2).
In conclusion, identification of C. pyruviciproducens can, to date, only be achieved by sequence-based methods. The phenotypic and genotypic characterization of 11 C. pyruviciproducens isolates from Canada and Switzerland shows differences from the type strain on both the chemotaxonomic and 16S rRNA gene sequence levels.
ACKNOWLEDGMENTS
The technical services of T. Burdz and D. Wiebe in performance of the 16S rRNA and rpoB gene sequencing are gratefully acknowledged.
Footnotes
Published ahead of print 20 June 2014
REFERENCES
- 1.Tong J, Liu C, Summanen P, Xu H, Finegold SM. 2010. Corynebacterium pyruviciproducens sp. nov., producing pyruvic acid. Int. J. Syst. Evol. Microbiol. 60:1135–1140. 10.1099/ijs.0.011783-0. [DOI] [PubMed] [Google Scholar]
- 2.Bernard KA, Munro C, Wiebe D, Ongsansoy E. 2002. Characteristics of rare or recently-described Corynebacterium species recovered from human clinical material in Canada. J. Clin. Microbiol. 40:4375–4381. 10.1128/JCM.40.11.4375-4381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard KA, Bellefeuille M, Ewan EP. 1991. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J. Clin. Microbiol. 29:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard K, Shuttleworth L, Munro C, Forbes-Faulkner JC, Pitt D, Norton JH, Thomas AD. 2002. Propionibacterium australiense sp. nov., derived from granulomatous bovine lesions. Anaerobe 8:41–47. 10.1006/anae.2000.0408. [DOI] [Google Scholar]
- 5.Khamis A, Raoult D, La Scola B. 2004. rpoB gene sequencing for identification of Corynebacterium species. J. Clin. Microbiol. 42:3925–3931. 10.1128/JCM.42.9.3925-3931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved 2nd ed. CLSI document M45-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibility Testing. 2013. Breakpoint tables for interpretation of MICs and zone diameters, version 3.1. European Committee on Antimicrobial Susceptibility Testing, Vaxjo, Sweden. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI document M100–S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]