Abstract
In this study, we investigated the performance of the FilmArray and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) in identifying microorganisms from blood culture (BC) bottles prior to positivity. First, we used simulated BacT/Alert FA Plus BC bottles with five each for Escherichia coli and Staphylococcus aureus isolates. The FilmArray identified all 10 isolates before BC positivity with 9/10 at 5 h and 1 at 7.5 h after incubation in the BC system. MALDI-TOF MS failed to identify the isolates prior to positivity. When the bottles were incubated for 2.5 h at room temperature (RT) before we put them into the BC system, the FilmArray identified 6/10 at 2.5 h and the remaining 4 at 5 h. Finally, we tested simulated BC bottles after incubation at RT. Interestingly, 9/10 isolates were identified with the FilmArray after 8 h of incubation at RT. Second, we studied clinical BC bottles in quadruplicate. When three-fourths of the parallel bottles signaled positive, the FilmArray was run on the fourth nonsignaled bottle and was found to be positive in 14/15 such cases. Third, we analyzed the performance of the FilmArray in the identification of microorganisms from clinical BC bottles before incubation in the system. Two milliliters of broth from 400 BC bottles was collected after arrival at the laboratory and stored at −70°C. Sixteen bottles later signaled positive in the system. When the frozen broth from these bottles was analyzed, the FilmArray identified all the microorganisms in 8/16 bottles prior to incubation in the BC system. This study shows that the FilmArray can identify microorganisms from BC bottles prior to positivity and in some cases even prior to incubation in the BC system.
INTRODUCTION
Early identification of microorganisms and their antimicrobial resistance properties in patients with bloodstream infections (BSI) has been shown to be crucial in the clinical management and initiation of the appropriate antimicrobial therapy (1). Blood culture is currently the gold standard for detecting and identifying microorganisms causing BSI (2). Despite being accurate and well established in the routine workflow, culture-based standard procedures may take up to 72 h (3, 4). Thus, there is an urgent need for methods that can rapidly identify microorganisms to the species level and the antimicrobial resistance properties.
Studies to date on BSI diagnostics have had three main areas of focus, namely, (i) non-culture based, (ii) blood culture based, and (iii) semi-culture-based methods, which are the focus area of this study. The idea of the non-culture-based methods is to bypass blood culturing and the associated time loss. While the obvious advantage is rapid identification, the disadvantages are diagnostically challenging and include a long hands-on time, specially trained personnel for performing the analysis, and a long processing time for identification (6 to 8 h) (5). Therefore, despite numerous investigations, the non-culture-based methods have not yet been established in routine diagnostics (5).
In recent years, culture-based methods experienced a revolution by the development of rapid identification methods that can be used directly on positive BC bottles. Methods that have been proven successful with this approach include direct matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) (6), the FilmArray (7), and fluorescence in situ hybridization using peptide nucleic acid probes (PNA-FISH) (8, 9). These methods have reduced the total time to identification of microorganisms significantly. However, they still require the use of blood cultures that have signaled positive (3).
The FilmArray blood culture identification (FA BCID) panel is a new molecular diagnostic method based on multiplex PCR (10). Identification of 24 common microorganisms and screening for the three antibiotic resistance genes (mecA, vanAB, and the KPC gene) takes 65 min in total from blood culture positivity. We have recently evaluated the FA BCID panel in a prospective clinical study where the method identified all microorganisms in 153/167 (91.6%) monomicrobial and 17/24 (71%) polymicrobial blood cultures (7).
The blood culture-based methods have decreased the time to identification after blood culture positivity from up to 72 h to <2 h (6–8). The next step in rapid diagnostics might be a semi-culture-based approach. Semi-culture-based identification is a new approach, and the term is defined as the identification and susceptibility testing of microorganisms from blood culture bottles before the bottles signal positive in the blood culture system. Few previous studies have explored this approach (11–13). The aim of this study was to evaluate the FilmArray and MALDI-TOF MS for (i) identification of bacteria, yeast, and antibiotic resistance genes in blood cultures before positivity in the blood culture systems and (ii) identification of bacteria and yeast after incubation of the blood cultures in room temperature.
MATERIALS AND METHODS
We conducted our investigation between June 2013 and November 2013 at Karolinska University Hospital, Huddinge, Sweden. The laboratory receives ca. 75,000 blood culture specimens per year from Karolinska University Hospital (Huddinge, Sweden), South General Hospital (Stockholm, Sweden), and Södertälje Hospital (Södertälje, Sweden). The total number of patient beds in these three tertiary hospitals are 1,569.
Study design.
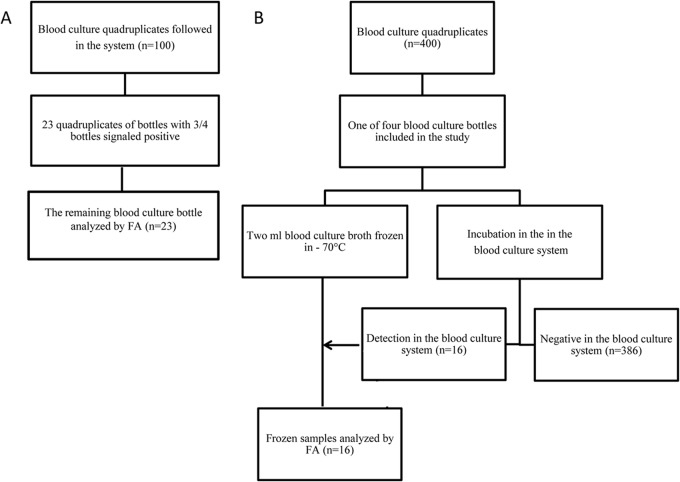
We evaluated the performance of the FilmArray (BioFire Diagnostics, Inc., Salt Lake City, UT, USA) and MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) in identification of microorganisms from blood culture bottles prior to positivity in three different settings, (i) in vitro (controlled analysis with spiked blood culture bottles), (ii) ex vivo (clinical samples analyzed during incubation in the blood culture system), and (iii) ex vivo (clinical samples analyzed after transport and before incubation in culture system). Figures 1 and 2 depict the study design.
FIG 1.
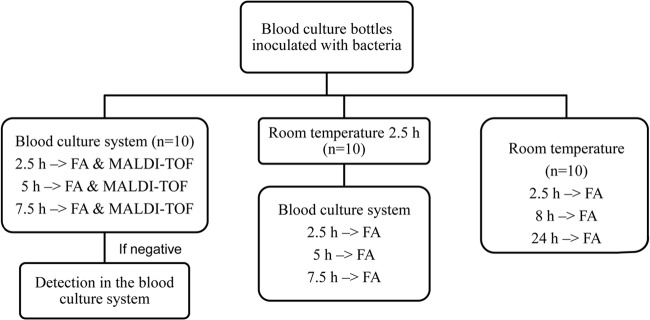
Flow chart of in vitro studies of the FilmArray (FA) and MALDI-TOF MS on BacT/Alert FA Plus blood culture bottles inoculated with Escherichia coli (n = 5) and Staphylococcus aureus (n = 5).
FIG 2.
Flow chart of clinical blood culture bottles tested with the FilmArray (FA) prior to positivity in the blood culture system (A) and prior to incubation in the blood culture system (B).
Blood culture bottles and blood culture system.
BacT/Alert aerobic/FA Plus and anaerobic/FN Plus (bioMérieux, Durham, NC, USA) blood culture bottles were used for the investigation. Bottles were incubated in the BacT/Alert 3D (bioMérieux) automated blood culture system until positivity or for a maximum of 5 days.
FilmArray BCID.
The FilmArray is a closed in vitro diagnostic system that combines nucleic acid extraction from clinical specimens, high-order nested multiplex PCR, and post-PCR DNA melt curve analysis. The FA BCID panel covers 19 bacteria and 5 yeasts: Enterobacteriaceae, Escherichia coli, Enterobacter cloacae complex, Klebsiella oxytoca, Klebsiella pneumoniae, Serratia marcescens, Proteus spp., Acinetobacter baumannii, Haemophilus influenzae, Neisseria meningitidis, Pseudomonas aeruginosa, Staphylococcus spp., Staphylococcus aureus, Streptococcus spp., Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Enterococcus spp., Listeria monocytogenes, Candida albicans, Candida glabrata, Candida krusei, Candida parapsilosis, and Candida tropicalis. In addition, the FA BCID panel also tests for the three antimicrobial resistance genes (vanAB, mecA, and the KPC gene). Briefly, 1 ml hydration buffer was injected into the pouch using the FA product syringe. Then, 100 μl of the blood culture broth was diluted in FA dilution buffer. A diluted sample (300 μl) was injected into the FA pouch before loading into the FilmArray system for analysis. Extraction, amplification, detection, and control analyses were automated within the pouch. Results for the analysis were provided by the software automatically after use of endpoint melting curve data. Each pouch included two internal run controls for both the primary amplification and the analyte-specific detection stages.
MALDI-TOF MS.
MALDI-TOF MS was performed directly from blood culture bottles as previously described by Saffert et al. (14). Briefly, 1.5 ml blood culture broth was centrifuged for 5 min at 110 relative centrifugal forces (rcf). Following the centrifugation, 1 ml supernatant was transferred to an Eppendorf tube and centrifuged for 2 min at high speed (20,800 rcf). The pellet was washed with 1 ml molecular-grade water (Sigma-Aldrich, St. Louis, MO, USA) and centrifuged for 2 min at high speed. Extraction was performed by exposing the pellet to 70% formic acid (30 μl) (Fluka, St. Louis, MO, USA) and 30 μl acetonitrile (Fluka). After a 2-min centrifugation at high speed, 1 μl supernatant was spotted on a steel 96-spot MALDI plate (Bruker Daltonics, Bremen, Germany) and allowed to dry before application of the MALDI matrix (1 μl) (Bruker Daltonics). MALDI-TOF MS (Bruker Daltonics) was used to perform the analysis. The Bruker Biotyper 3.0 software and library (Bruker Daltonics) were used for spectra analysis. According to the manufacturer's instructions, scores of >2.0 were considered identification at the species level and scores of >1.7 were considered identification at the genus level.
Standard clinical microbiological methods.
Gram staining was done immediately after blood cultures signaled positive. The results of the Gram stains directed further subculturing onto relevant agar plates. Observed growth on agar plates was used for identification of microorganisms by using MALDI-TOF MS (Bruker Daltonics, Bremen, Germany), the Vitek 2 XL system (bioMérieux, France), and various validated desktop spot tests. These tests included catalase, oxidase, and indole spot tests and agglutination for Staphylococcus aureus (Staphaurex latex test; Remel Europe Ltd., Dartford, United Kingdom), group A streptococci, and Streptococcus pneumoniae (Oxoid, Basingstoke, United Kingdom). The susceptibility testing was performed by disc diffusion according to the EUCAST method.
Simulated blood culture bottles. (i) Preparation of simulated blood culture bottles.
Escherichia coli and S. aureus, two of the most common causes of bacterial BSIs, were used in the in vitro experiments (15). Eight previous blood culture isolates, four each from S. aureus and E. coli and two reference strains, S. aureus (ATCC 29213) and E. coli (ATCC 25922), were included in the study. The frozen isolates were thawed, cultured onto blood agar plates, and then incubated at 37°C for 24 h. Inoculum was prepared by suspending colonies from agar plates in 0.01 M phosphate buffer solution (pH 7.3 to 7.4) to a 0.5 McFarland standard (1.5 × 108 CFU/ml) in DensiCheck (bioMérieux, l'Etoile, France). Inoculum was diluted, and each bottle was spiked with 100 μl final inoculum containing 1, 000 CFU of the individual bacteria. Finally, 10 ml of 37°C defibrinated horse blood (Håtunalab AB, Bro, Sweden) was added to each BacT/Alert FA Plus blood culture bottle. Similarly, 100 μl of the final suspension was simultaneously cultured on three blood agar plates and incubated at 36°C for 24 h to control the bacterial concentration that was inoculated in the respective blood culture bottle.
(ii) Identification of bacteria with the FilmArray and MALDI-TOF MS prior to positivity in the blood culture system.
Bottles were prepared as described above and then were immediately incubated in the blood culture system. Blood culture broth from the incubated bottles was taken at the time intervals of 2.5 h, 5 h, and 7.5 h and analyzed by the FilmArray and direct MALDI-TOF MS. The bottles were kept in the blood culture system until they signaled positive. The times to detection (TTD) of blood cultures in the blood culture system were documented. The positive blood cultures were then analyzed by direct MALDI-TOF MS since the method could not identify bacteria before blood culture positivity. MALDI-TOF MS was not included in the subsequent experiments due to the observed failure of identification of bacteria during culture.
(iii) Simulated transport time before loading the bottles in the blood culture system.
In this experiment, we investigated the role of transport time in identification of bacteria prior to culture positivity. We investigated the effect of transport time by preparing spiked blood cultures as previously described and incubating 10 spiked bottles (five each of S. aureus and E. coli) for 2.5 h at room temperature before loading them in the blood culture system. The FilmArray analyses were performed at the time intervals of 2.5 h, 5 h, and 7.5 h after incubation of the bottles in the blood culture system. Finally, bottles were left in the blood culture system until signaling positive.
(iv) Spiked blood cultures incubated at room temperature and analyzed with the FilmArray.
Ten blood culture bottles (five each of S. aureus and E. coli) were spiked as previously described and incubated at room temperature. The FilmArray analyses were then performed at the time intervals of 2.5 h, 8 h, and 24 h of room temperature incubation. The chosen time intervals were based on our previously reported observation on transport times of blood cultures at Karolinska University Hospital (16). After the microorganism identification with the FilmArray, the blood cultures were immediately incubated in the blood culture system. The times to detection for the blood culture bottles in the system were documented.
Clinical blood culture bottles. (i) Identification of bacteria from clinical samples prior to positivity in the blood culture system.
Blood culture bottles in quadruplicate were investigated for inclusion in this part of the study. The criteria for inclusion in the study were (a) BacT/Alert aerobic/FA Plus and anaerobic/FN Plus blood culture bottles, (b) samples for four blood culture bottles taken at the same time, and (c) pending blood culture bottle results. When three out of the four samples signaled positive in the blood culture system, the fourth blood culture was tested by the FilmArray before signaling for blood culture positivity. In order to analyze the specificity of the FilmArray, we tested 8 blood culture bottles that were negative at the end of 5 days of incubation in the blood culture system.
(ii) Identification of bacteria from clinical samples prior to incubation in the blood culture system.
Two-milliliter blood culture broth from one bottle from each of the 400 blood culture bottles were taken from the clinical samples after arrival at the laboratory. These samples were immediately put in a −70°C freezer. Simultaneously, the blood culture bottles were loaded in the blood culture system. Blood cultures were routinely followed for positivity or until the end of the incubation time of 5 days. When the blood culture bottle signaled positive, the respective frozen broth was thawed and analyzed with the FilmArray. If the initial analysis was FilmArray negative, 1 ml broth was centrifuged at 21,000 rcf for 5 min. The supernatant (850 μl) was discarded, and 100 μl of the pellet was analyzed again by The FilmArray. Transport times of the respective sample and times to detection of microorganisms in the blood culture system were documented. In order to analyze the specificity of the FilmArray in this setting, we tested frozen broth from 4 blood culture bottles that were negative at the end of 5 days of incubation in the blood culture system.
RESULTS
Identification Prior to Detection in Spiked Blood Culture Bottles. (i) Identification of bacteria prior to detection in the blood culture system.
The FilmArray identified 10/10 (100%) bacteria in spiked samples before detection in the blood culture system. Nine out of 10 (five S. aureus and four E. coli) isolates were identified at 5 h, and the remaining one E. coli isolate was identified at 7.5 h of incubation in the blood culture system (Table 1). The median time to detection (TTD) of growth for the blood culture bottles in the automated blood culture system was 11.1 h (range, 9.12 to 25 h). In contrast, MALDI-TOF MS did not detect any of the isolates prior to their signaling positive in the blood culture system. Identification with the direct MALDI-TOF MS method was, however, successful in 10/10 (100%) samples after blood culture bottles signaled positive with an average MALDI-TOF MS score of 2.1 (data not shown).
TABLE 1.
In vitro study results of blood culture bottles inoculated with Escherichia coli (n = 5) and Staphylococcus aureus (n = 5)a
| Time in blood culture system (h) | No. of bottles after direct incubation in the indicated blood culture system with: |
No. of bottles after incubation in the FilmArray at RTb for 2.5 h with: |
||||
|---|---|---|---|---|---|---|
|
E. coli |
S. aureus |
E. coli | S. aureus | |||
| FilmArray | MALDI-TOF MS | FilmArray | MALDI-TOF MS | |||
| 2.5 | 0 | 0 | 0 | 0 | 3c | 3c |
| 5 | 4 | 0 | 5 | 0 | 2d | 2d |
| 7.5 | 1 | 0 | NAe | 0 | NA | NA |
| Until positive signal | NA | 5 | NA | 5 | NA | NA |
At specified time points in the blood culture system, blood culture broth was aspirated and subjected to testing with the FilmArray and MALDI-TOF MS. When a bottle tested positive with a method, no additional testing with that method was performed.
RT, room temperature.
The total time to identification was 5 h after inoculation in the blood culture bottles.
The total time to identification was 7.5 h after inoculation in the blood culture bottles.
NA, not assessed, as microorganisms in all bottles were identified by the assay.
(ii) Effect of transport time on the FilmArray performance.
We analyzed the effect of a 2.5-h room temperature incubation before loading of the spiked blood culture bottles in the blood culture system. Interestingly, 6/10 (60%) spiked samples were identified by the FilmArray after 2.5 h of incubation in the blood culture system (Table 1). These isolates were three each of S. aureus and E. coli. The remaining four samples (two each of S. aureus and E. coli) were identified after 5 h of incubation in the blood culture system. The median time to detection in the blood culture system was 9.1 h (range, 8.2 to 10.8 h). In this setting, the identification of S. aureus and E. coli by the FilmArray was done in a total of 5 and 7.5 h after the inoculation of bacteria in the blood culture bottles.
(iii) FilmArray analysis prior to incubation in the blood culture system.
Direct identification prior to incubation of the spiked blood culture bottles in the blood culture system was successful in 9/10 (90%) isolates with the FilmArray after 8 h of incubation at room temperature. These were five E. coli and four S. aureus. The remaining one S. aureus isolate was identified after 24 h of incubation at room temperature. The median TTD of spiked samples was 5.5 h (range, 3.5 to 6.7 h) in the blood culture system after the simulated transport time of 8 h.
Identification prior to detection in clinical samples. (i) Identification of bacteria with the FilmArray prior to detection in the blood culture system.
In total, 100 blood culture bottles in quadruplicate were investigated for inclusion. Twenty-three bottles were included for analysis in the study. Fifteen of 23 (65%) bottles became positive, and 8/23 (35%) were blood culture negative. The FilmArray identified 14/15 (93%) microorganisms in positive bottles prior to detection in the blood culture system (Table 2). These were four S. aureus, five coagulase-negative staphylococci, two E. coli, and one each of Klebsiella pneumoniae and Enterococcus faecium. The FilmArray-negative and blood culture-positive sample was one S. pneumoniae isolate that signaled positive after 74 h in the blood culture system. Eight blood culture bottles that did not signal positive until the end of the incubation time of 5 days in the blood culture system were also negative on the FilmArray analysis.
TABLE 2.
Identification of microorganisms from blood culture bottles by the FilmArray during incubation in the systema
| System results | No. of bottles | Microorganism(s) and gene identified by: |
|
|---|---|---|---|
| FilmArray | Standard method after signal in blood culture system | ||
| FilmArray positive/culture positive (n = 14) | 4 | Staphylococcus aureus | Staphylococcus aureus |
| 4 | Staphylococcus spp. and mecA | Staphylococcus epidermidis and mecA | |
| 1 | Staphylococcus spp. and mecA | Coagulase negative staphylococci and mecA | |
| 1 | Enterococcus spp. | Enterococcus faecium | |
| 1 | Enterococcus spp. | Enterococcus faecalis | |
| 2 | Escherichia coli | Escherichia coli | |
| 1 | Klebsiella pneumoniae | Klebsiella pneumoniae | |
| FilmArray negative/culture positive (n = 1) | 1 | None detected | Streptococcus pneumoniae |
| FilmArray negative/culture negative (n = 8) | 8 | None detected | No growthb |
Twenty-three blood culture bottles were analyzed in quadruplicate. One of the 4 bottles was removed from the system and analyzed when the 3 parallel blood culture bottles signaled positive in the blood culture system.
Incubation in the blood culture system for 5 days.
(ii) Identification of microorganisms with the FilmArray before incubation in the blood culture system.
Sixteen of the 400 blood culture bottles included in the study signaled positive for growth. In total, 8 out of 16 (50%) samples were correctly identified by the FilmArray (Table 3). The microorganisms in five samples were directly identified by the FilmArray. These were one each of E. coli, Streptococcus pyogenes, S. pneumoniae, and Candida albicans and one polymicrobial blood culture containing Klebsiella oxytoca and Citrobacter freundii. The remaining 11 samples were centrifuged, and the pellet was analyzed with the FilmArray. The FilmArray then identified microorganisms in an additional three samples, each containing S. aureus. Eight out of 16 (50%) samples remained FilmArray negative. These were two each of Klebsiella pneumoniae and E. coli, one each of Staphylococcus epidermidis, Enterococcus faecalis, and S. aureus and one polymicrobial sample with E. coli and α-streptococci. Four blood culture bottles that did not signal positive until the end of the incubation time of 5 days in the blood culture system were also negative on the FilmArray analysis.
TABLE 3.
Identification of microorganisms by FilmArray directly from blood culture bottles before incubation in the blood culture systema
| System result | Microorganism(s) identified by: |
|
|---|---|---|
| FilmArray prior to incubation in blood culture system | Standard blood culturing methods | |
| FilmArray positive/culture positive (n = 8) | Klebsiella oxytoca and Enterobacteriaceae | Klebsiella oxytoca and Citrobacter freundii |
| Streptococcus pneumoniae | Streptococcus pneumoniae | |
| Streptococcus pyogenes | Streptococcus pyogenes | |
| Escherichia coli | Escherichia coli | |
| Candida albicans | Candida albicans | |
| Staphylococcus aureusb | Staphylococcus aureus | |
| Staphylococcus aureusb | Staphylococcus aureus | |
| Staphylococcus aureusb | Staphylococcus aureus | |
| FilmArray negative/culture positive (n = 8) | None detected | Escherichia coli and Streptococcus spp. |
| None detected | Staphylococcus epidermidis | |
| None detected | Staphylococcus aureus | |
| None detected | Enterococcus faecalis | |
| None detected | Klebsiella pneumoniae | |
| None detected | Klebsiella pneumoniae | |
| None detected | Escherichia coli | |
| None detected | Escherichia coli | |
| FilmArray negative/culture negative (n = 4) | None detected | No growth (5 days) |
FilmArray analysis was performed on the previously stored broth after the respective blood culture bottle signaled positive. Broth samples with negative FilmArray results were retested with FA after a centrifugation step.
FilmArray positive after centrifugation.
DISCUSSION
Rapid identification of microorganisms in patients with BSI is critical for tailoring the appropriate antimicrobial therapy (17). In this study, we investigated a new potential use of the FilmArray and MALDI-TOF MS for identifying microorganisms from blood cultures prior to signaling positive in the automated blood culture system. In the in vitro study, we showed that the FilmArray could identify microorganisms in 9/10 (90%) spiked blood cultures at 5 h and in 1/10 (10%) at 7.5 h after incubation in the blood culture system. In contrast, the median time to detection in the blood culture system was 11.1 h (range, 9.1 to 25 h). In parallel, we investigated a direct MALDI-TOF MS method for identifying S. aureus and E. coli after up to 8 h in the blood culture system. Unfortunately, the present direct MALDI-TOF MS method showed results similar to those of the Loonen et al. (11) investigation, and it did not identify any of the 10 strains after 8 h of culturing in the blood culture system. The method did, however, identify all strains after blood culture positivity.
The promising results of the in vitro investigation on the FilmArray encouraged further exploration of the method for testing clinical blood cultures. In the first part of the clinical investigation, identification of microorganisms before blood culture positivity was evaluated. The FilmArray identified all microorganisms in 14/15 (93%) blood cultures, including one blood culture that contained two different bacterial species, before blood culture positivity. The remaining 1/15 (7%) FilmArray-negative sample had a long incubation time in the blood culture system (72 h).
We simulated a short transport time of 2.5 h to analyze the effect of transport time on the FilmArray results. Following the additional transport time of 2.5 h, the FilmArray identified 6/10 (60%) of the microorganisms in spiked blood cultures at 2.5 h and the remaining 4/10 (40%) at 5 h after incubation. The median time to detection for the same samples in the blood culture system was, however, 9.1 h (range, 8.2 to 10.8 h). This suggests that the short transport time is beneficial for the performance of the FilmArray but has no significant effect on the time to detection in the blood culture system.
In the final part of the in vitro study, the FilmArray was investigated for identification of microorganisms in spiked blood cultures before incubation in the blood culture system. Interestingly, the FilmArray identified 9/10 (90%) of the microorganisms at 8 h in spiked blood cultures without the use of the blood culture system. This proposes a new and, to our knowledge, not previously explored use of blood cultures. To investigate this approach on clinical samples, we froze culture broth from 400 clinical blood cultures after arrival at the laboratory. The 16 blood cultures that later signaled positive were analyzed by the FilmArray, and all microorganisms in 8/16 (50%) samples were identified from the frozen blood culture broth. The freezing of the clinical samples might potentially have a negative effect on the FilmArray performance, and it is important that the samples be analyzed directly after transport in the future. Interestingly, the FilmArray identified one C. albicans isolate from the frozen sample, suggesting that the method can identify even yeast before incubation of the blood in the blood culture system. This might be compared to standard diagnostic methods that took 48 h to identify the C. albicans isolate. The present study also included one polymicrobial clinical blood culture bottle where the microorganisms were identified by the FilmArray before incubation of the bottle in the blood culture system. To put this finding in perspective: the semi-culture approach results in identification of the microorganisms in a polymicrobial blood culture in 70 min after arrival at the laboratory. In contrast, when processed by standard microbiological methods, the same polymicrobial sample would first require incubation in the blood culture system for detection and then an additional 24 to 48 h for identification. This finding might also be compared to direct MALDI-TOF MS methods where several studies have shown that MALDI-TOF MS is unreliable for identifying microorganisms in polymicrobial blood cultures (18, 19). Recently, we showed that the FilmArray identified all microorganisms in 17/24 (71%) clinical polymicrobial blood culture bottles after they signaled positive in the blood culture system (7). The semi-culture approach of identifying microorganisms before incubating the blood culture in the blood culture system might equip both the laboratory and the physician with a new possible “urgent analysis” if required (i.e., prolonged transport times for hospitals that are at long distances from the laboratory and/or a patient who is rapidly deteriorating). Blood cultures taken according to standard procedures might be analyzed by the FilmArray before incubation in the blood culture system.
There were no vanAB- or KPC gene-positive samples in the present study. Interestingly, all six MecA-positive Staphylococcus spp. detected by the FilmArray were resistant to cefoxitin in susceptibility testing, indicating that the assay might detect resistance genes in a semi-culture-based approach.
The present results indicate that the FilmArray applied to blood culture fluid after 5 h of incubation in the blood culture system might be a useful diagnostic strategy for routine use in order to detect bloodstream infection. Thus, including hands-on time and time in the FA system, the total time from the sampling to the identification result is approximately 6 h. This should be compared to the times for non-culture-based molecular diagnostic methods that are performed directly on blood samples. These methods are aimed to bypass the need for culturing in an attempt to quickly identify microorganisms. The LightCycler SeptiFast test, the Magicplex real-time PCR test, and the VYOO test are three commercial PCR assays that are used for detecting microorganisms directly from blood samples without the need for culturing. Despite bypassing the need for culturing, these methods take 6 to 8 h to identify microorganisms. Compared to non-culture-based methods, the total time to identification using the present semi-culture approach with the FilmArray is ca. 6 h. This short time to identification suggests that the FilmArray might be used in a selected number of patients in a manner similar to that of the non-culture-based methods. While the FilmArray only requires 5 min of hands-on time and almost no personnel training to perform the analysis, the non-culture-based methods require considerably longer hands-on time for extraction and PCR in addition to specially trained staff (5). The simplicity of the FilmArray is an obvious advantage as it can be performed by non-laboratory-trained personnel and can be implemented in 24/7 clinical settings (i.e., intensive care units). The semi-culture approach has the added benefit of early identification as well as simultaneous culturing for further processing in situations where an extended evaluation of the microorganisms is also needed (i.e., sequencing, collecting, and freezing isolates, etc). Moreover, there is still an obvious need for antimicrobial susceptibility testing of the collected strains, which is currently not possible with the non-culture-based methods.
The specificity of the assay is essential in implementation of the FilmArray in a semi-culture-based approach. In the present study, we tested a total of 12 blood culture bottles that were negative in the blood culture system. In our previous FilmArray study, we tested another 12 negative blood culture samples (7). In all 24 negative blood culture bottles, the FilmArray results were also negative. In line with our findings, Blaschke et al. (20) reported that 18 blood culture bottles that were culture negative were also negative in the FilmArray. The present and previously published data indicate that the FilmArray has high specificity for negative blood culture bottles.
It is important to note that we analyzed the performance of the FilmArray only in BacT/Alert FA Plus in the simulated bottles and in BacT/Alert FA Plus and BacT/Alert FN Plus in the clinical samples. It is not possible to generalize the present data to all types of blood culture bottles in the clinical routine, and the performance of the FilmArray may vary in other types of bottles.
There are a wide range of bacteria and yeast that can cause BSI. A limitation of the present study is that we analyzed only E. coli and S. aureus in the simulated blood cultures. Further studies analyzing other microorganisms, including fastidious bacteria yeasts and polymicrobial BC samples, are warranted.
This study proposes a unique application of the FilmArray that might open new possibilities for the rapid identification of microorganisms in patients with BSI. The present data indicate that the FilmArray can be used in identification of microorganisms from blood culture bottles before they signal positive and has the potential of identifying the microorganisms in blood cultures directly after arrival at the laboratory. This might be an important supplement to traditional blood culturing methods by providing rapid microorganism and antimicrobial resistance gene identification. Further studies are warranted to investigate the performance of the semi-culture-based FilmArray method in clinical settings.
ACKNOWLEDGMENTS
We thank BioFire Diagnostics Inc. and Biotech-IgG AB for providing the equipment and reagents for the FilmArray assay.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 20 June 2014
REFERENCES
- 1.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 244:379–386. 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 2.Beekmann SE, Diekema DJ, Chapin KC, Doern GV. 2003. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J. Clin. Microbiol. 41:3119–3125. 10.1128/JCM.41.7.3119-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellinghausen N, Kochem AJ, Disque C, Muhl H, Gebert S, Winter J, Matten J, Sakka SG. 2009. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 47:2759–2765. 10.1128/JCM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee A, Mirrett S, Reller LB, Weinstein MP. 2007. Detection of bloodstream infections in adults: how many blood cultures are needed? J. Clin. Microbiol. 45:3546–3548. 10.1128/JCM.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber J, Nierhaus A, Braune SA, de Heer G, Kluge S. 2013. Comparison of three different commercial PCR assays for the detection of pathogens in critically ill sepsis patients. Med. Klin. Intensivmed. Notfmed. 108:311–318. 10.1007/s00063-013-0227-1. [DOI] [PubMed] [Google Scholar]
- 6.Fothergill A, Kasinathan V, Hyman J, Walsh J, Drake T, Wang YF. 2013. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J. Clin. Microbiol. 51:805–809. 10.1128/JCM.02326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol. 51:4130–4136. 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azevedo NF, Jardim T, Almeida C, Cerqueira L, Almeida AJ, Rodrigues F, Keevil CW, Vieira MJ. 2011. Application of flow cytometry for the identification of Staphylococcus epidermidis by peptide nucleic acid fluorescence in situ hybridization (PNA FISH) in blood samples. Antonie Van Leeuwenhoek 100:463–470. 10.1007/s10482-011-9595-9. [DOI] [PubMed] [Google Scholar]
- 9.Harris DM, Hata DJ. 2013. Rapid identification of bacteria and Candida using PNA-FISH from blood and peritoneal fluid cultures: a retrospective clinical study. Ann. Clin. Microbiol. Microb. 12:2. 10.1186/1476-0711-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DH, Ririe KM. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047. 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loonen AJ, Jansz AR, Stalpers J, Wolffs PF, van den Brule AJ. 2012. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 31:1575–1583. 10.1007/s10096-011-1480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang MC, Lin WH, Yan JJ, Fang HY, Kuo TH, Tseng CC, Wu JJ. 2013. Early identification of microorganisms in blood culture prior to the detection of a positive signal in the BACTEC FX system using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J. Microbiol. Immunol. Infect., in press. 10.1016/j.jmii.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Lim SH, Mix S, Xu Z, Taba B, Budvytiene I, Berliner AN, Queralto N, Churi YS, Huang RS, Eiden M, Martino RA, Rhodes P, Banaei N. 2014. Colorimetric sensor array allows fast detection and simultaneous identification of sepsis-causing bacteria in spiked blood culture. J. Clin. Microbiol. 52:592–598. 10.1128/JCM.02377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saffert RT, Cunningham SA, Mandrekar J, Patel R. 2012. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagn. Microbiol. Infect. Dis. 73:21–26. 10.1016/j.diagmicrobio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Biedenbach DJ, Moet GJ, Jones RN. 2004. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). Diagn. Microbiol. Infect. Dis. 50:59–69. 10.1016/j.diagmicrobio.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Rönnberg C, Mildh M, Ullberg M, Ozenci V. 2013. Transport time for blood culture bottles: underlying factors and its consequences. Diagn. Microbiol. Infect. Dis. 76:286–290. 10.1016/j.diagmicrobio.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Seifert H. 2009. The clinical importance of microbiological findings in the diagnosis and management of bloodstream infections. Clin. Infect. Dis. 48(suppl 4):S238−S245. 10.1086/598188. [DOI] [PubMed] [Google Scholar]
- 18.Buchan BW, Riebe KM, Ledeboer NA. 2012. Comparison of the MALDI Biotyper system using Sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J. Clin. Microbiol. 50:346–352. 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagacé-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ. 2012. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J. Clin. Microbiol. 50:3324–3328. 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn. Microbiol. Infect. Dis. 74:349–355. 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]