Abstract
Chronic wasting disease (CWD), a transmissible spongiform encephalopathy of deer, elk, and moose, is the only prion disease affecting free-ranging animals. Since the disease was first identified in northern Colorado and southern Wyoming in 1967, new epidemic foci of the disease have been identified in 20 additional states, as well as two Canadian provinces and the Republic of South Korea. Identification of CWD-affected animals currently requires postmortem analysis of brain or lymphoid tissues using immunohistochemistry (IHC) or an enzyme-linked immunosorbent assay (ELISA), with no practical way to evaluate potential strain types or to investigate the epidemiology of existing or novel foci of disease. Using a standardized real-time (RT)-quaking-induced conversion (QuIC) assay, a seeded amplification assay employing recombinant prion protein as a conversion substrate and thioflavin T (ThT) as an amyloid-binding fluorophore, we analyzed, in a blinded manner, 1,243 retropharyngeal lymph node samples from white-tailed deer, mule deer, and moose, collected in the field from areas with current or historic CWD endemicity. RT-QuIC results were then compared with those obtained by conventional IHC and ELISA, and amplification metrics using ThT and thioflavin S were examined in relation to the clinical history of the sampled deer. The results indicate that RT-QuIC is useful for both identifying CWD-infected animals and facilitating epidemiological studies in areas in which CWD is endemic or not endemic.
INTRODUCTION
Chronic wasting disease (CWD) is an efficiently transmitted transmissible spongiform encephalopathy (TSE) of cervids (e.g., deer, elk, and moose) and is the only known prion disease affecting free-ranging nondomestic animals. As such, it is the only prion disease of animals whose control and eradication (through genotypic breeding schemes or herd reduction/depopulation efforts, for example) are problematic (1, 2). While the origins of CWD are uncertain, the disease has been present in wild cervid populations in northern Colorado and southern Wyoming for over 40 years (3, 4) and has now been identified in both captive and free-ranging cervids in 22 states, 2 Canadian provinces, and the Republic of Korea (5). With intensified national and international surveillance efforts, CWD continues to be identified in areas previously thought to be free of infection, including recent discoveries in Iowa, Texas, and Pennsylvania (www.promedmail.org, archive numbers 20120721.1210369, 20120711.1197183, and 20121014.1341794). The prevalence of CWD varies across North America but can be as high as 30% in some areas of Colorado, approaching 80% in captive populations (6).
Determination of prevalence rates in populations is dependent on a sensitive and specific “gold standard” diagnostic assay. Immunohistochemistry (IHC) was, until recently, considered the gold standard diagnostic test for chronic wasting disease and other prion diseases of animals and humans. For cervids, an enzyme-linked immunosorbent assay (ELISA) was recently approved by the US Department of Agriculture for primary diagnostic screening of field samples across the United States (7), although an amplification-based assay, similar to PCR, for the detection of CWD (or other TSEs) has been elusive to date. The true sensitivity and specificity of IHC or ELISA for the detection of infected individuals are unknown, although it is generally acknowledged that the assays underestimate the levels of prions in a given sample due to conventional proteolytic pretreatment steps to abolish cellular PrPC cross-reactivity (8–10). This limitation has led to increased interest in the development of assays that involve either amplification and detection of the protease-resistant prion protein (e.g., serial protein misfolding cyclic amplification [sPMCA] assay) (11) or fluorometric quantitation of seeded amplification activity (e.g., real-time [RT]-quaking-induced conversion [QuIC] assay) (12) or that otherwise avoid harsh proteolytic treatments (e.g., conformation-dependent immunoassay [CDI]) (13).
One component of chronic wasting disease field surveillance that has required additional research is the ability to distinguish prion strains in vitro. Because prion infections are devoid of agent nucleic acids and a host immune response, conventional infectious disease strain-typing methods (e.g., nucleic acid sequencing or antibody neutralization studies) are not possible. Despite this hurdle, at least two strains of CWD have been reported in natural isolates, with distinct pathological distributions in mouse bioassay results and biochemical traits in vitro (14–16). These strains may occasionally be found in the same individual, which, combined with the necessity for mouse bioassays, makes epidemiological studies difficult. Investigations into the unprecedented appearance of new epidemic foci across the United States (e.g., southeastern Wisconsin in 2002, central New York State in 2005, and north-central Missouri in 2010) have relied on anecdotal information regarding the origins of CWD infections in these areas, with no additional insight into strain identities or sources. The ability to distinguish strains in vitro has so far been limited to protease treatment or guanidine denaturation profiles, although fluorometric assays may hold promise in this area and could be useful in epidemiological investigations (17).
In the present study, we have applied a standardized RT-QuIC seeded amplification assay with two different fluorophores (thioflavin T [ThT] and thioflavin S [ThS]) to examine, in a blinded manner, retropharyngeal lymph node (RLN) samples collected at necropsy from white-tailed and mule deer (Odocoileus virginianus and Odocoileus hemionus, n = 1201) and moose (Alces alces, n = 42), during routine CWD surveillance in Colorado, Illinois, Nebraska, New York, and Texas. We analyzed various aspects of amplification in positive animals, i.e., (i) time to threshold, (ii) slope, and (iii) peak fluorescence, and correlated our amplification results with several a priori variables, including age, sex, species, genotype, and harvest location. We hypothesized that RT-QuIC results would correlate with ELISA and IHC results reported by contributing state agencies and that there would be amplification characteristics unique to either genotype or geographical regions of endemicity. Our results demonstrate that the RT-QuIC assay is comparable to conventional assays for CWD detection, in terms of sensitivity, and they predict that seeded amplification using various fluorophores may eventually prove to be a useful, rapid, and inexpensive tool for advanced epidemiological studies in ongoing and newly identified foci of chronic wasting disease.
MATERIALS AND METHODS
Study population.
The study areas included distinct geographic regions in Colorado, Illinois, Nebraska, New York, and Texas. Animals were harvested during routine surveillance through the course of the 2010-2011 (New York, n = 100; Nebraska, n = 280) or 2012 (Texas, n = 126) big-game hunting seasons, as part of targeted CWD surveillance outside big-game hunting seasons in 2013 (Illinois, n = 695), or as road kill in 2004 to 2010 (Colorado, n = 42). Lymph node samples were initially tested by either ELISA (at New York State Department of Environmental Conservation/New York State Veterinary Diagnostic Laboratory, Nebraska Game and Parks Commission, or University of Nebraska Veterinary Services Laboratory) or IHC (at Texas Department of Wildlife and Parks/Texas State Veterinary Laboratory, Illinois Department of Natural Resources/Illinois Department of Agriculture, or Colorado Division of Parks and Wildlife/Colorado State University Veterinary Diagnostic Laboratory), with results withheld until prion seeding assays were complete.
Tissue collection and processing.
Retropharyngeal lymph node samples (i.e., those specific tissues described above) were collected during postmortem examinations and were submitted frozen to the Prion Research Center (PRC) at Colorado State University. Each sample was assigned a unique numerical designation, which was recorded to allow blinded evaluations. Samples were initially prepared as 2% (wt/vol) homogenates in RT-QuIC dilution buffer (phosphate-buffered saline [PBS] with 0.05% sodium dodecyl sulfate [SDS]) by using a BulletBlender (NextAdvance) with 0.5-mm zirconium oxide beads and 1.5-ml conical screw-cap tubes. Samples were homogenized using three 5-min cycles of homogenization (at a speed setting of 10) and were then kept at −80°C until RT-QuIC analysis.
RT-QuIC procedure.
RT-QuIC assays were performed using a truncated form of the Syrian hamster recombinant PrP (SHrPrP) (residues 90 to 231) in pET41b, expressed and purified as described previously (18, 19). In brief, 1-liter cultures of lysogeny broth (LB) containing autoinduction supplements (EMD Biosciences) were inoculated with SHrPrP-expressing Rosetta strain Escherichia coli, grown overnight, and harvested when optical density (OD) at 600 nm of ∼3 was reached. Cells were lysed with Bug Buster reagent with supplemented Lysonase (EMD Biosciences), and inclusion bodies (IBs) were harvested by centrifugation of the lysate at 15,000 × g. IB pellets were washed twice and stored at −80°C until purification (typically ≤24 h). IB pellets were solubilized with 8 M guanidine hydrochloride in 100 mM NaPO4 and 10 mM Tris (pH 8.0), clarified by centrifugation at 15,000 × g for 15 min, and added to Super Flow nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) preequilibrated with denaturing buffer (6.0 M guanidine hydrochloride, 100 mM NaPO4, 10 mM Tris [pH 8.0]). Denatured SHrPrP and Ni-NTA resin were incubated for 45 min at room temperature with rotation and then were added to an XK fast protein liquid chromatography column (GE Healthcare). Refolding was achieved on the column by using a linear refolding gradient of denaturing buffer to refolding buffer (100 mM NaPO4, 10 mM Tris [pH 8.0]) over 340 ml at 0.75 ml/min. SHrPrP was eluted with a linear gradient of refolding buffer to elution buffer (100 mM NaPO4, 10 mM Tris [pH 8.0], 500 mM imidazole [pH 5.5]) over 100 ml at 2.0 ml/min. Fractions were pooled and dialyzed against two changes of 4.0 liters of dialysis buffer (20 mM NaPO4 [pH 5.5]). Recovered SHrPrP was adjusted to a final concentration of ∼0.5 mg/ml.
Two percent RLN homogenates were diluted 1:100 in RT-QuIC dilution buffer, and 5 μl of this resultant 0.02% homogenate was added to 95 μl of RT-QuIC reaction buffer (350 mM NaCl, 10 μM EDTA, 10 μM ThT, and 0.1 mg/ml SHrPrPC), yielding a final lymph node homogenate concentration of 0.002% in RT-QuIC reaction buffer. These 100-μl preparations were evaluated in parallel with both positive- and negative-control tissues in adjacent wells of a 96-well plate, along with unspiked controls spiked with 5 μl of RT-QuIC buffer alone. Control homogenates consisted of a pooled preparation of 6 CWD-positive white-tailed deer (CBP6) or tissue-matched negative-control samples collected in a CWD-negative area of New York State. Plates were then subjected to 96 cycles of shaking and incubation at 42°C (approximately 24 h), with cycles of 1 min of shaking (700 rpm, double orbital) and 1 min of rest. ThT fluorescence readings (450-nm excitation and 480-nm emission, bottom read, 20 flashes per well) were taken following each 15-min cycle, using a gain setting of 1,200. Positive samples were defined as those that crossed a threshold of fluorescence, a threshold determined by the average fluorescence of the three tissue-matched negative-control samples over the course of the experiment plus five standard deviations. The time to positivity was defined as the time at which a sample fluorescence emission crossed the cycle threshold (CT). The time to positive fluorescence of test samples was then compared with that of positive-control tissue samples, typically yielding values between 0 and 1; samples with earlier times to threshold fluorescence thus had values closer to or greater than 1. The slope of the amplification curve was determined as the increase in relative fluorescence over time. These analyses were performed using MARS analytical software.
Positive samples were evaluated separately in triplicate in three separate experiments, using two different protocols, the first using ThT, as described above, and the second using thioflavin S (ThS) (10 μm) in place of ThT. Thioflavin S fluorescence readings (480-nm excitation and 510-nm emission, bottom read, 20 flashes per well) were taken following each 15-min cycle, using a gain setting of 1,400. Criteria for identification of seeding activity in both protocols were used as described above for ThT. Values for the time to positive threshold, slope, and fluorescence plateau were averaged across the 9 replicates for each sample and each fluorophore.
Cervid PRNP PCR amplification and sequence analysis.
DNA was extracted from frozen CWD-positive RLNs using a commercial kit (Qiagen), following the manufacturer's instructions. Consensus primer pairs specific for amplification of PRNP in mule deer and white-tailed deer were described previously by O'Rourke and colleagues (20). PRNP sequences were amplified using HotStart DNA polymerase (Qiagen) with forward primer 223 (5′-ACACCCTCTTTATTTTGCAG-3′) and reverse primer 224 (5′-AGAAGATAATGAAAACAGGAAG-3′), which yielded an approximately 830-bp product. PCR conditions were as follows: 95°C for 5 min, 35 cycles of denaturation at 95°C for 60 s, annealing at 54°C for 60 s, and extension at 72°C for 60 s, and then extension at 7°C for 7 min, under standard buffer conditions with 2.5 mM MgCl2 (Qiagen). PCR products were analyzed on 1.5% agarose EZ-vision-stained gels.
PCR products were purified using a commercial kit (Qiagen) to remove unincorporated deoxynucleoside triphosphates (dNTPs) and primers and then were bidirectionally sequenced using forward primer 223 and reverse primer 224 (GeneWiz Inc., South Plainfield, NJ). Chromatographic data were aligned using CLC Main Workbench 6.8.4 software. All sequences were individually analyzed for conflicts and secondary peaks, in order to create all necessary contigs and associated consensus sequences. DNA and amino acid sequences were aligned using ClustalW (codons) in Mega 5.2 to determine amino acid polymorphisms, focusing on amino acid residues 95 and 96 (white-tailed deer) and 225 (mule deer).
Analysis of RT-QuIC metrics.
Our analyses had two major foci, i.e., (i) to determine whether there was a relationship between ELISA scores and RT-QuIC metrics (ThT and ThS scores, amplitudes, and slopes) for CWD-positive deer and (ii) to evaluate the best predictors (location/state, species, sex, age, and PrP genotype at amino acid position 96) of RT-QuIC metrics for CWD-positive mule deer and whitetail deer. All analyses were conducted using the program R (www.r-project.org), using the stats package. To determine if there was a relationship between ELISA scores and RT-QuIC metrics, we performed Spearman correlations. ELISA scores were available only for deer from Nebraska, and we evaluated the relationships for all deer and for mule deer and whitetail deer independently. To evaluate how well the RT-QuIC metrics were predicted by the harvest location, deer species, sex, age, and PrP genotype at position 96, we used an information theoretic approach (21) in which all single predictor variables were evaluated using linear regression, based on a Gaussian distribution, and ranked based on the Akaike information criterion (AIC) corrected for small sample size (see Table S1 in the supplemental material) (21). This approach is of value because it enables determination of the most parsimonious model or set of models to explain RT-QuIC metric scores, as well as calculation of variable importance weights to determine the relative importance of one predictor variable over others (21). Because information theory departs from frequency-based statistical approaches, which are dependent on P values, we also calculated the coefficient of variation (r2) so that the relative fits of models to the data could be assessed.
RESULTS
Conventional detection of PrPres in RLN tissues.
Retropharyngeal lymph node specimens were analyzed by referring state agencies, using either conventional ELISAs (New York and Nebraska) or IHC tests (Colorado, Illinois, and Texas). The Colorado State University Veterinary Diagnostic Laboratory did not detect CWD infection in 42 moose from Colorado, while the University of Nebraska Veterinary Diagnostic Center identified 5 mule deer and 5 white-tailed deer as positive for CWD by ELISA. Ages ranged from 1 to 3 years, with both sexes and only homozygous 95Q/Q and 96G/G (white-tailed deer) and 225S/S (mule deer) animals represented. ELISA scores ranged from 2.961 to 3.292. The Illinois Department of Agriculture identified 12 of 695 white-tailed deer samples as CWD positive by IHC. Ages ranged from <1 to 3 years of age, with both sexes and both 96G/G and 96G/S genotypes represented; all animals were homozygous for glycine at amino acid position 95. The Texas Veterinary Services Laboratory identified one of 126 mule deer samples as CWD positive by IHC (225S/S), while the New York State Veterinary Diagnostic Laboratory did not identify any CWD-positive animals among the 100 white-tailed deer samples submitted to the PRC for analysis (Table 1).
TABLE 1.
Characteristics of CWD-positive deer samples from Nebraska, Illinois, and Texas, as determined by RT-QuIC analysis
| Animal identification no. | State | County | Speciesa | Sexb | Age (yr)c | ELISA score | Amino acid |
ThT |
ThS |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 95 | 96 | 225 | Score | Amplitudee | Slope | Score | Amplitudee | Slope | |||||||
| 1059 | NE | Box Butte | MD | F | 3 | 3.143 | GG | SS | 0.885 | 10,818 | 1.514 | 1.080 | 6,584 | 0.429 | |
| 1088 | NE | Sioux | MD | M | 3 | 3.079 | GG | SS | 0.445 | 6,243 | 1.878 | 0.619 | 6,730 | 0.376 | |
| 1127 | NE | Phelps | WTD | F | 2 | 3.292 | GG | SS | 0.768 | 10,329 | 1.722 | 0.964 | 6,778 | 0.407 | |
| 1128 | NE | Buffalo | WTD | M | 1 | 3.292 | GG | SS | 0.702 | 10,047 | 1.017 | 0.865 | 6,973 | 0.335 | |
| 1154 | NE | Furnas | WTD | M | 2 | 2.961 | GG | SS | 0.506 | 8,594 | 1.797 | 0.589 | 6,895 | 0.274 | |
| 1187 | NE | Sioux | MD | M | 2 | 3.151 | GG | SS | 0.844 | 11,146 | 1.716 | 1.025 | 7,162 | 0.434 | |
| 1218 | NE | Webster | WTD | M | 2 | 3.104 | GG | SS | 1.060 | 11,409 | 1.704 | 1.037 | 5,879 | 0.322 | |
| 1254 | NE | Custer | WTD | M | 3 | 2.966 | GG | SS | 1.025 | 12,314 | 1.753 | 0.996 | 5,800 | 0.285 | |
| 1267 | NE | Scottsbluff | MD | M | 1 | 3.11 | GG | SS | 0.811 | 10,293 | 2.250 | 0.718 | 5,831 | 0.283 | |
| 1268 | NE | Sioux | MD | F | NA | 2.983 | GG | SS | 0.818 | 11,170 | 1.228 | 0.826 | 5,890 | 0.289 | |
| 1344 | TX | El Paso | MD | M | 4.5 | NA | GG | SS | 0.666 | 10,233 | 2.691 | 0.966 | 5,126 | 0.375 | |
| 1468 | IL | Kane | WTD | M | 2 | NA | GG | SS | 0.691 | 8,561 | 2.247 | 0.684 | 5,376 | 0.452 | |
| 1475 | IL | McHenry | WTD | M | 3 | NA | GG | SS | 0.676 | 8,805 | 3.167 | 0.659 | 5,013 | 0.610 | |
| 1543 | IL | Stephenson | WTD | F | 2 | NA | GG | SS | 0.751 | 11,172 | 0.861 | 0.761 | 6,928 | 0.363 | |
| 1563 | IL | Boone | WTD | F | <1 | NA | GG | SS | 0.479 | 7,725 | 0.810 | 0.984 | 5,886 | 0.378 | |
| 1606 | IL | DeKalb | WTD | F | 2 | NA | GSd | SS | 0.846 | 11,165 | 0.901 | 0.929 | 6,277 | 0.357 | |
| 1617 | IL | DeKalb | WTD | F | 1 | NA | GG | SS | 0.818 | 11,187 | 0.768 | 0.955 | 6,191 | 0.340 | |
| 1761 | IL | McHenry | WTD | M | 1 | NA | GG | SS | 0.979 | 11,999 | 0.846 | 1.164 | 5,765 | 0.400 | |
| 1771 | IL | Grundy | WTD | F | 2 | NA | GG | SS | 0.784 | 11,086 | 1.179 | 1.005 | 6,288 | 0.340 | |
| 1801 | IL | Kane | WTD | M | 1 | NA | GG | SS | 0.879 | 6,784 | 0.773 | 0.766 | 7,243 | 0.365 | |
| 1912 | IL | Ogle | WTD | F | <1 | NA | GG | SS | 1.022 | 10,855 | 2.771 | 0.912 | 7,636 | 0.371 | |
| 1918 | IL | DeKalb | WTD | M | 1 | NA | GG | SS | 0.824 | 9,278 | 1.386 | 0.725 | 7,139 | 0.373 | |
| 1923 | IL | Ogle | WTD | M | 3 | NA | GS | SS | 0.848 | 10,352 | 1.516 | 0.521 | 5,684 | 0.875 | |
MD, mule deer; WTD, whitetail deer.
F, female; M, male.
Individuals <1 year of age were assigned values of 0.5 for analyses. NA, data were not available for that sample.
Heterozygosities are indicated with bold type.
Amplitude is given in relative fluorescence units.
RT-QuIC analysis of RLN tissues.
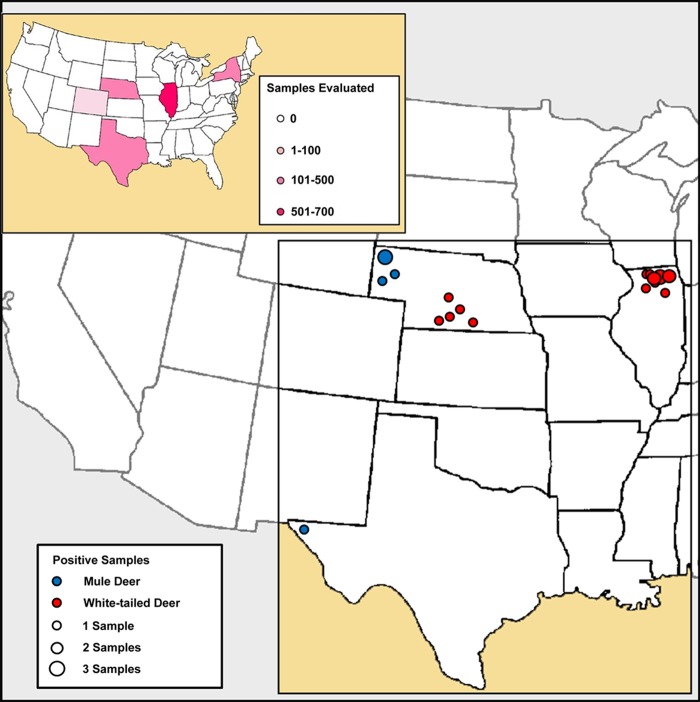
RT-QuIC analysis of RLN samples from Illinois revealed seeded amplification in 12/695 samples, corresponding to IHC-positive samples. Analysis of samples from Nebraska showed seeded amplification in 10/280 samples, corresponding to ELISA-positive lymph nodes, while 1/126 samples submitted by the Texas Department of Parks and Wildlife demonstrated seeded amplification in RT-QuIC analysis; the RLN specimen from this deer was also positive by IHC. Forty-two RLN samples from moose in Colorado and 100 white-tailed deer in New York remained negative by RT-QuIC (Fig. 1 and 2). RT-QuIC positivity correlated 100% with IHC and ELISA positivity (i.e., 100% sensitivity and specificity), with amplification scores ranging from 0.479 to 1.06 using ThT and from 0.521 to 1.16 using ThS. The RT-QuIC results for CWD-positive animals are summarized in Table 1.
FIG 1.
Summary of retropharyngeal lymph node samples evaluated and positive sample locations. Samples included 100 white-tailed deer lymph nodes from New York State, 695 white-tailed deer lymph nodes from Illinois, 280 white-tailed and mule deer lymph nodes from Nebraska, 126 mule deer lymph nodes from Texas, and 42 moose lymph nodes from Colorado. Of 1,243 samples evaluated, 11 RT-QuIC assay-positive deer were identified in Illinois, 10 in Nebraska, and one in Texas.
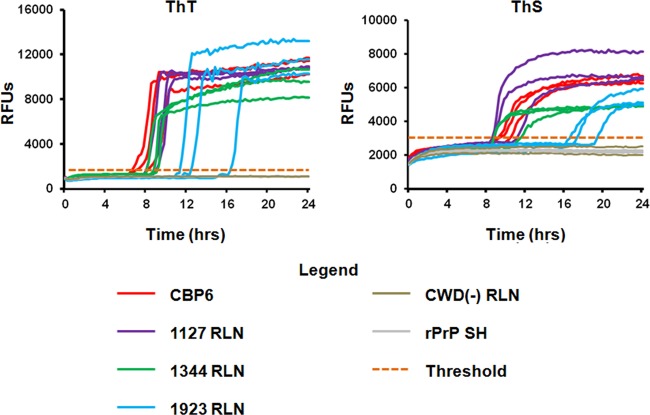
FIG 2.
RT-QuIC results from CWD-positive deer. Samples that were positive in the initial screening were reanalyzed in triplicate in three separate experiments, using either thioflavin T (ThT) or thioflavin S (ThS). Positive controls (CBP6) and multiple negative controls (CWD-negative lymph nodes and untreated Syrian hamster [SH] recombinant PrP) were included on each experimental plate. The threshold for amplification (orange dotted line) was determined by averaging the relative fluorescence units (RFUs) of negative-control samples over the course of the experiment and adding five standard deviations. Seeded amplification is demonstrated by increases in ThT and ThS fluorescence over time in the positive-control sample as well as each of three positive lymph nodes from study deer; negative-control samples do not show seeded amplification.
Correlation between RT-QuIC amplification analyses and clinical variables.
In a comparison of ELISA versus RT-QuIC analysis, the directions of relationships between ELISA scores and RT-QuIC metrics were generally similar among deer species (Table 2). The notable exception to this was ThT slope, with positive and negative slopes for mule deer and whitetail deer, respectively. However, correlation values indicated that the positive relationship observed for mule deer was only weakly supported, whereas the negative correlation between ELISA scores and ThT slopes was a moderately strong trend. Overall and for white tail deer, there were significant positive correlations between ELISA scores and ThS slopes.
TABLE 2.
Spearman correlations between ELISA scores and QuIC metrics for CWD-positive deer
| Parameter | All deer (n = 10) |
Mule deer (n = 5) |
Whitetail deer (n = 5) |
|||
|---|---|---|---|---|---|---|
| ρ | P | ρ | P | ρ | P | |
| ThT score | −0.024 | 0.947 | 0.600 | 0.350 | 0.154 | 0.805 |
| ThT amplitude | −0.116 | 0.751 | 0.000 | 1.000 | 0.051 | 0.935 |
| ThT slope | −0.377 | 0.283 | 0.200 | 0.783 | −0.821 | 0.089 |
| ThS score | 0.413 | 0.235 | 0.600 | 0.350 | 0.154 | 0.805 |
| ThS amplitude | 0.456 | 0.185 | 0.500 | 0.450 | 0.308 | 0.614 |
| ThS slope | 0.699 | 0.024a | 0.700 | 0.233 | 0.975 | 0.005a |
Significant result. ρ values indicate the strength of relationships for individual deer species, but the statistical power to detect significant relationships is decreased, owing to the small sample sizes in these cases.
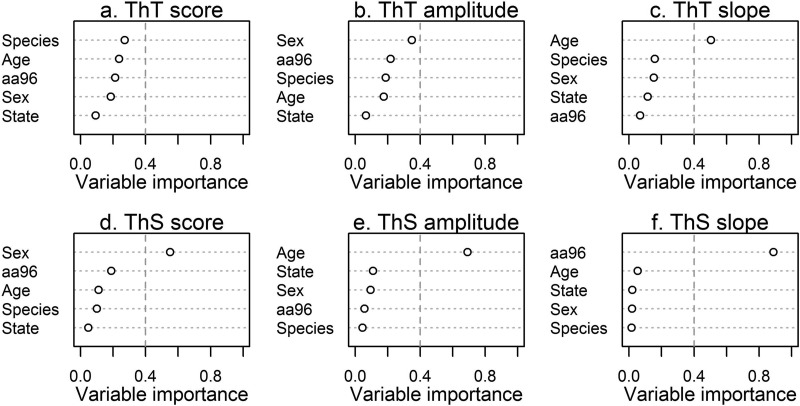
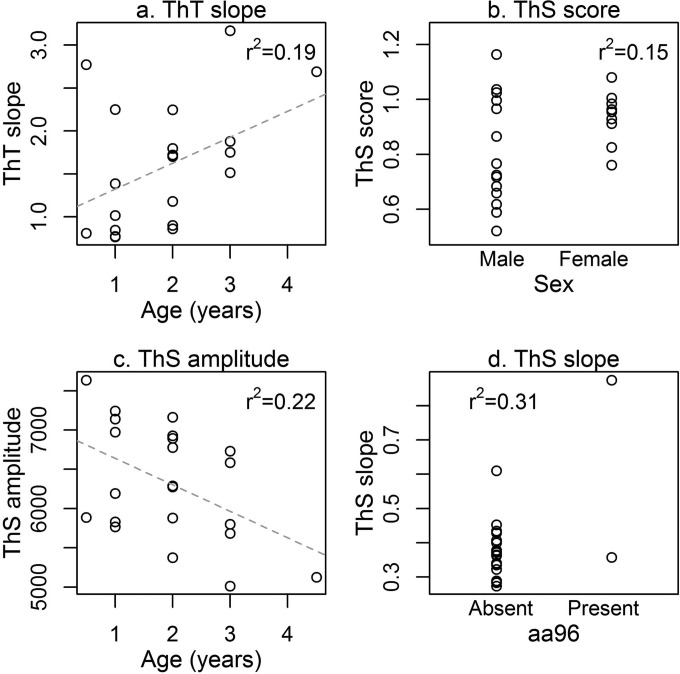
When RT-QuIC metrics were evaluated as predictors of a priori variables, there was generally one predictor variable that was a distinctly better/more important predictor (variable importance weight of >0.4) than the rest (Fig. 3 and Table 2). This was particularly distinct for ThT slope, ThS score, ThS amplitude, and ThS slope (Fig. 3c to f). However, there was model uncertainty regarding the best predictor of ThT score and ThT amplitude (Fig. 3a and b). For RT-QuIC metrics with one predictor variable better than the rest (variable importance weight of >0.4), we plotted that association with the RT-QuIC metric (Fig. 4). ThT slope and ThS amplitude exhibited positive and negative relationships with deer age, respectively, while female deer tended to have higher and less variable ThS scores. Interestingly, ThS slopes were higher in the limited number of individual deer with the 96G/S genotype present.
FIG 3.
Variable importance weights of location, species, sex, age, and amino acid predictors of QuIC metrics. Where there was clearly a best predictor variable, variable importance weights were >0.4 (vertical dashed line). Of the correlates analyzed, likely predictors of age included ThT slope and ThS amplitude, while ThS score was correlated with sex and ThS slope seemed to be a good predictor of amino acid 96 (aa96) identity (for full model comparison tables, see Table S1 in the supplemental material).
FIG 4.
Distribution of QuIC metric data associated with the best predictor variable in each case (see Fig. 3). Coefficients of variation are given in each panel. These results show a positive correlation between ThS slope and cervid PrP amino acid 96 identity (for full model comparison table, see Table S1 in the supplemental material). The dashed line represents the trend line determined from the data.
DISCUSSION
Amplification-based assays, long used in clinical settings for the detection of viral and bacterial agents in clinical samples, have to date not been available for use in postmortem screening for transmissible spongiform encephalopathies. The slow evolution of prion amplification assays, which take advantage of the propensity of the abnormal PrPres isoform to convert mammalian and, more recently, recombinant PrPC in vitro, has made it increasingly practical to employ these approaches in clinical settings. The real-time (RT)-quaking-induced conversion (QuIC) seeded amplification assay offers the additional advantage of avoiding the proteolytic or acidic pretreatments commonly required for both conventional TSE detection assays (e.g., immunohistochemical analyses and ELISAs) and serial protein misfolding cyclic amplification (sPMCA) assays. The RT-QuIC assay has been reported to amplify PrPres seeds present in brain dilutions in the femtogram range, comparable to bioassays (19, 22); in the present study, however, our goal was to compare seeded amplification directly to conventional IHC and ELISA with samples collected postmortem.
Our findings, through blinded analysis of over 1,200 field samples collected from various cervid species across the United States, demonstrate that RT-QuIC is capable of accurately identifying IHC- and ELISA-positive retropharyngeal lymph node specimens. With continued development of the RT-QuIC assay, it may be possible to identify subclinically positive, TSE-affected individuals that are not positive by conventional detection systems, which may represent a significant number of animals in areas in which CWD is endemic. It remains to be determined whether the RT-QuIC assay could be used for antemortem detection of CWD infection, using rectal biopsy specimens or other clinical samples that are available antemortem (e.g., blood or cerebrospinal fluid [CSF] samples) (18, 23, 24).
Apart from enhanced sensitivity, conventional amplification assays for viral and bacterial pathogens offer a second distinct advantage, i.e., the ability to identify pathogen-derived nucleic acid sequences or specific antibody responses, facilitating epidemiological investigations. It is commonly accepted that infectious prions lack both a nucleic acid component and a specific host immune response but still exhibit distinct strain properties; therefore, alternative methodologies for identifying TSE strains in vitro are necessary. A number of fluorophores have been shown to bind prion aggregates; indeed, this finding has been incorporated into the RT-QuIC assay, providing visual evidence of seeded amplification through cumulated binding of one of these fluorophores (thioflavin T). Little is known about how or where in the prion protein structure this binding may occur, although strain discrimination has been reported using luminescent conjugated polymers (LCPs), fluorophores that emit conformation-dependent fluorescence spectra (17). Our analysis of CWD-positive lymph nodes using two conventional fluorophores revealed that some components of the RT-QuIC analysis scheme, including thioflavin S score, amplitude, and slope, may be predictors of a CWD-positive cervid's background or possibly CWD strain traits. No definitive evidence of geographic grouping was observed across ThT or ThS metrics, although it is possible that incorporation of LCPs into the RT-QuIC assay may allow more-precise discrimination of clinical TSE isolates and eventual strain correlation.
In summary, we report the first deployment of an amplification-based assay for the detection of CWD in cervid field cases. Blinded RT-QuIC analysis yielded results correlating directly with those of conventional IHC analyses and ELISAs. Further work is needed to assess whether RT-QuIC analysis may contribute to CWD strain distinction, antemortem detection, and advanced epidemiological studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01NS061902 and NCRR K01OD010994 and grant DO D12ZO-045 from the Morris Animal Foundation. Moose sampling was funded in part by Colorado Parks and Wildlife and the US Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We sincerely thank Byron Caughey, Jason Wilham, and Christina Orrú for their assistance in the development of the RT-QuIC assay for use with cervid samples. We also thank all of the state and federal agencies that provided archived and ongoing sample collections, as well as the private parties, hunters, and professional harvestmen without whose conservation efforts these samples would not have been available for analysis.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01258-14.
REFERENCES
- 1.Williams ES, Miller MW, Kreeger TJ, Kahn RH, Thorne ET. 2002. Chronic wasting disease of deer and elk: a review with recommendations for management. J. Wildl. Manage. 66:551–563. 10.2307/3803123. [DOI] [Google Scholar]
- 2.Nodelijk G, van Roermund HJ, van Keulen LJ, Engel B, Vellema P, Hagenaars TJ.2011. Breeding with resistant rams leads to rapid control of classical scrapie in affected sheep flocks. Vet. Res. 42:5. 10.1186/1297-9716-42-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams ES, Young S. 1980. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J. Wildl. Dis. 16:89–98. 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 4.Williams ES, Young S. 1982. Spongiform encephalopathy of Rocky Mountain elk. J. Wildl. Dis. 18:465–471. 10.7589/0090-3558-18.4.465. [DOI] [PubMed] [Google Scholar]
- 5.Saunders SE, Bartelt-Hunt SL, Bartz JC. 2012. Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg. Infect. Dis. 18:369–376. 10.3201/eid1803.110685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keane DP, Barr DJ, Bochsler PN, Hall SM, Gidlewski T, O'Rourke KI, Spraker TR, Samuel MD. 2008. Chronic wasting disease in a Wisconsin white-tailed deer farm. J. Vet. Diagn. Invest. 20:698–703. 10.1177/104063870802000534. [DOI] [PubMed] [Google Scholar]
- 7.USDA. 2013. Chronic wasting disease: revised program standards. USDA, Washington, DC. [Google Scholar]
- 8.Haley NJ, Mathiason C, Carver S, Telling GC, Zabel MC, Hoover EA. 2012. Sensitivity of protein misfolding cyclic amplification versus immunohistochemistry in ante-mortem detection of chronic wasting disease. J. Gen. Virol. 93:1141–1150. 10.1099/vir.0.039073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safar JG, Geschwind MD, Deering C, Didorenko S, Sattavat M, Sanchez H, Serban A, Vey M, Baron H, Giles K, Miller BL, Dearmond SJ, Prusiner SB. 2005. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U. S. A. 102:3501–3506. 10.1073/pnas.0409651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haley N, Mathiason C, Zabel MD, Telling GC, Hoover E. 2009. Detection of sub-clinical CWD infection in conventional test-negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS One 4:e7990. 10.1371/journal.pone.0007990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saborio GP, Permanne B, Soto C. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810–813. 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 12.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. 2007. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 4:645–650. 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 13.Safar JG, Scott M, Monaghan J, Deering C, Didorenko S, Vergara J, Ball H, Legname G, Leclerc E, Solforosi L, Serban H, Groth D, Burton DR, Prusiner SB, Williamson RA. 2002. Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat. Biotechnol. 20:1147–1150. 10.1038/nbt748. [DOI] [PubMed] [Google Scholar]
- 14.Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, Balachandran A, McKenzie D, Castilla J, Soto C, Jewell J, Graham C, Hoover EA, Telling GC. 2010. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328:1154–1158. 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrott MR, Sigurdson CJ, Mason GL, Hoover EA. 2012. Evidence for distinct chronic wasting disease (CWD) strains in experimental CWD in ferrets. J. Gen. Virol. 93:212–221. 10.1099/vir.0.035006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymond GJ, Raymond LD, Meade-White KD, Hughson AG, Favara C, Gardner D, Williams ES, Miller MW, Race RE, Caughey B. 2007. Transmission and adaptation of chronic wasting disease to hamsters and transgenic mice: evidence for strains. J. Virol. 81:4305–4314. 10.1128/JVI.02474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Polymenidou M, Schwarz P, Leclerc M, Hammarstrom P, Wuthrich K, Aguzzi A. 2007. Prion strain discrimination using luminescent conjugated polymers. Nat. Methods 4:1023–1030. 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- 18.Haley NJ, Van de Motter A, Carver S, Henderson D, Davenport K, Seelig DM, Mathiason C, Hoover E. 2013. Prion-seeding activity in cerebrospinal fluid of deer with chronic wasting disease. PLoS One 8:e81488. 10.1371/journal.pone.0081488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. 2010. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 6:e1001217. 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. 2004. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J. Gen. Virol. 85:1339–1346. 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- 21.Burnham KP, Anderson DR. 2002. Model selection and inference: a practical information-theoretic approach. Spring-Verlag, New York, NY. [Google Scholar]
- 22.Peden AH, McGuire LI, Appleford NE, Mallinson G, Wilham JM, Orru CD, Caughey B, Ironside JW, Knight RS, Will RG, Green AJ, Head MW. 2012. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking-induced conversion. J. Gen. Virol. 93:438–449. 10.1099/vir.0.033365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elder AM, Henderson DM, Nalls AV, Wilham JM, Caughey BW, Hoover EA, Kincaid AE, Bartz JC, Mathiason CK. 2013. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS One 8:e80203. 10.1371/journal.pone.0080203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfe LL, Spraker TR, Gonzalez L, Dagleish MP, Sirochman TM, Brown JC, Jeffrey M, Miller MW. 2007. PrPCWD in rectal lymphoid tissue of deer (Odocoileus spp.). J. Gen. Virol. 88:2078–2082. 10.1099/vir.0.82342-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.