Abstract
Diarrheal diseases cause illness and death among children younger than 10 years in developing countries. Conventional testing for the detection of hemorrhagic bacteria takes 2 to 5 days to yield complete information on the organism and its antibiotic sensitivity pattern. Hence, in the present study, we developed a molecular-based diagnostic assay that identifies common hemorrhagic bacteria in stool samples. A set of specific primers were designed for the detection of Salmonella spp., Shigella spp., enterohemorrhagic Escherichia coli (EHEC), and Campylobacter spp., suitable for use in a one-tube PCR assay. The assay in the present study simultaneously detected five genes, namely, ompC for the Salmonella genus, virA for the Shigella genus, eaeA for EHEC, 16S rRNA for the Campylobacter genus, and hemA for an internal control. Specific primer pairs were successfully designed and simultaneously amplified the targeted genes. Validation with 20 Gram-negative and 17 Gram-positive strains yielded 100% specificity. The limit of detection of the multiplex PCR assay was 1 × 103 CFU at the bacterial cell level and 100 pg at the genomic DNA level. Further evaluation of the multiplex PCR with 223 bacterium-spiked stool specimens revealed 100% sensitivity and specificity. We conclude that the developed multiplex PCR assay was rapid, giving results within 4 h, which is essential for the identification of hemorrhagic bacteria, and it might be useful as an additional diagnostic tool whenever time is important in the diagnosis of hemorrhagic bacteria that cause diarrhea. In addition, the presence of an internal control in the multiplex PCR assay is important for excluding false-negative cases.
INTRODUCTION
Enteric infections remain an important cause of morbidity in Malaysia. The incidence rate of diarrhea among Malaysian children was 23.6 episodes per 100 persons per year (1). Developing countries have the greatest diarrheal disease burden in both mortality and morbidity. In Malaysia, the most important causative pathogens for hemorrhagic diarrhea are Campylobacter spp., enterohemorrhagic Escherichia coli (EHEC), Shigella spp., Vibrio cholerae O139, and Salmonella spp. However, the presence of these pathogens is probably underestimated due to inappropriate diagnostic methods in clinical practice (1). Laboratory analyses to identify the causative pathogens of the diarrheal cases are not often performed, especially in outpatient clinics, due to the cost (1). However, the Infectious Diseases Society of America (IDSA) guidelines recommend stool cultures for severe, bloody, febrile, dysenteric, or persistent diarrheal illnesses (2). The laboratory results are critical for both individual patient care and for public health purposes. The lack of a specific diagnosis can hinder the clinician in provision of appropriate therapeutic and preventive measures. In the last decade, molecular methods for the identification of hemorrhagic pathogens became more important due to their rapidity, sensitivity, and reproducibility. These methods are an attractive alternative when conventional bacteriological techniques fail to identify microorganisms, particularly slow-growing, fastidious, or noncultivable organisms. Interfering factors such as antimicrobial therapy may cause false-negative culture results even in cases of infections due to easy-to-culture pathogens such as E. coli and Salmonella (3, 4). Rapid pathogen identification by molecular technology can reduce the costs associated with hospitalization and play a central role in detection and antibiotic susceptibility testing (5). Previous studies used a panel of parallel PCR assays or combined two multiplex PCR assays for the detection of the most common hemorrhagic bacteria causing gastroenteritis (6, 7). In the two conditions, multiple pipetting steps, repeated handling of PCR reagents in microvolumes, and processing variations are detrimental factors that can lead to false results (8). Here, we describe the application of a multiplex PCR design in a one-tube multiplex assay format for simultaneous detection of four bacteria (Salmonella spp., Shigella spp., EHEC, and Campylobacter spp.) which are involved in bloody diarrhea.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains used in this study for positive and negative control were obtained from the culture collections as listed in Table 1. These strains were inoculated onto Columbia blood agar (Merck, NJ) plates with 5% sheep blood for 24 h at 37°C. Campylobacter spp. were grown under a microaerobic atmosphere at 42°C for 48 h.
TABLE 1.
Bacterial species and strains used in this study and results of multiplex PCR assays
| Strain no. | Reference strain | Multiplex PCR result for: |
||||
|---|---|---|---|---|---|---|
| 16S rRNAa | ompCb | virAc | eaeAd | hemAe | ||
| 1 | Salmonella enterica (ATCC 25957) | − | + | − | − | + |
| 2 | Salmonella enterica (ATCC 14028) | − | + | − | − | + |
| 3 | Salmonella enterica clinical isolatesf | − | + | − | − | + |
| 4 | Salmonella enterica clinical isolatesg | − | + | − | − | + |
| 5 | Shigella flexneri (ATCC 12022) | − | − | + | − | + |
| 6 | Shigella sonnei (ATCC 25931) | − | − | + | − | + |
| 7 | Shigella boydii (ATCC 9207)h | − | − | + | − | + |
| 8 | Shigella dysenteriae | − | − | + | − | + |
| 9 | Escherichia coli (EHEC) (ATCC 43889) | − | − | − | + | + |
| 10 | Escherichia coli (ATCC 43890) | − | − | − | + | + |
| 11 | Campylobacter jejuni (ATCC 33560) | + | − | − | − | + |
| 12 | Campylobacter jejuni (ATCC 33559) | + | − | − | − | + |
| 13 | Campylobacter jejuni (ATCC 33291) | + | − | − | − | + |
| 14 | C. jejuni clinical isolatesf | + | − | − | − | + |
| 15 | Klebsiella pneumoniae (ATCC 13883)h | − | − | − | − | + |
| 16 | Proteus mirabilis (ATCC 29245)h | − | − | − | − | + |
| 17 | Pseudomonas aeruginosa (ATCC 27853)h | − | − | − | − | + |
| 18 | Enterococcus faecium LMG 16192i | − | − | − | − | + |
| 19 | Enterococcus faecalis (ATCC 29212)i | − | − | − | − | + |
| 20 | Vibrio cholerae (O1 classical)h | − | − | − | − | + |
| 21 | Yersinia enterocolitica (ATCC 23715)e | − | − | − | − | + |
| 22 | S. aureus (ATCC 33591) | − | − | − | − | + |
| 23 | S. aureus (ATCC 33592) | − | − | − | − | + |
| 24 | Staphylococcus saprophyticus (ATCC 13518) | − | − | − | − | + |
| 25 | S. aureus (ATCC 43300)h | − | − | − | − | + |
| 26 | S. aureus (ATCC 25923)f | − | − | − | − | + |
| 27 | S. aureus (ATCC 49775) clinical isolatesg | − | − | − | − | + |
| 28 | S. epidermidis (ATCC 14990)h | − | − | − | − | + |
| 29 | CoNS methicillin-resistante | − | − | − | − | + |
| 30 | Streptococcus sp. group A (ATCC 19615)g | − | − | − | − | + |
| 31 | Streptococcus sp. group B (ATCC 12401)g | − | − | − | − | + |
| 32 | Bacillus subtilis (ATCC 6633)e | − | − | − | − | + |
| 33 | Listeria monocytogenes (ATCC 7644)e | − | − | − | − | + |
| 34 | Corynebacterium spp.f | − | − | − | − | + |
| 35 | Citrobacter freundii (ATCC 8090)e | − | − | − | − | + |
| 36 | Gardnerella spp.e | − | − | − | − | + |
| 37 | Candida albicans (ATCC 10231)e | − | − | − | − | + |
Campylobacter genus.
Salmonella genus.
Shigella genus.
Escherichia coli genus.
Internal control.
Obtained from the Institute for Medical Research, Malaysia.
Obtained from the Department of Medical Microbiology, Institute for Medical Molecular Biotechnology.
Obtained from the Department of Medical Microbiology and Parasitology, School of Medical Sciences, Universiti Sains Malaysia.
Reference strain from Belgian Co-ordinated Collections of Microorganisms (BCCM), Ghent, Belgium.
Primer design for multiplex PCR assay.
Four different primer sets were designed based on sequence data obtained from GenBank databases (9). The ClustalW program in Vector NTI (version 9.0) software (Invitrogen, Carlsbad, CA) was used to align the DNA sequences. The conserved and nonconserved regions of the DNA sequence alignments were visualized using GeneDoc software (http://www.nrbsc.org/downloads). Based on the conserved regions of the alignment, specific primer pairs were designed to amplify the Salmonella spp., Shigella spp., EHEC, and Campylobacter genus genes. A primer pair based on the hemA gene was designed (759 bp) and was used as an internal control (Table 2). The five primer pairs (Research Biolabs, Kuala Lumpur, Malaysia) were designed in such a way that the PCR products ranged from 150 to 759 bp. The specificity of the designed primers was checked using BLAST, which is available at the GenBank website (http://www.ncbi.nlm.nih.gov/BLAST). The primer sequences for the five genes and expected PCR product sizes are shown in Table 2.
TABLE 2.
Sequences of primers used for multiplex PCR assays
| Bacteria | Gene | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) | Product size (bp) | GenBank accession no. |
|---|---|---|---|---|---|
| Internal control | hemA | GAGCAGCGTCCATTGTGAGATC | ATTCTCAGATATGTGTGGTGGACTA | 759 | AF227752 |
| Shigella | virA | AAGTACATACTATTACAGCTCCGG | GCAGTGTCGTTTTTAGGGACAACT | 600 | D26468.1 |
| Campylobacter | 16S rRNA | AGTTGGAAACGACTGCTAATACTC | TTAATGGTTAAGCCATTAGATTTCAC | 450 | HQ864829.1 |
| Salmonella spp. | ompC | CGTATCGGCTTCAAAGGCGAAAC | GAAGTCGGTGTTACGGTAGGTAG | 299 | AF039309.1 |
| Escherichia coli (EHEC) | eaeA | CGGCTGGCATGAGTCATACAATAA | AAACAAAGCAACATTATCACCATAATA | 150 | JQ350704.1 |
Multiplex PCR assay.
The monoplex PCR for each gene and the multiplex PCR assay were standardized using genomic DNA extracted from reference strains. A mixture of DNAs from four reference strains, namely, Salmonella enterica (ATCC 25957), Shigella flexneri (ATCC 12022), Escherichia coli (ATCC 43889), and Campylobacter jejuni (ATCC 33560), which contained the four genes of interest, was used as a positive control (Fig. 1). DNase-free distilled water was used as a negative control. In addition, a plasmid (pCR 2.1-TOPO; Invitrogen) that contained the hemA gene (1 pg) was used as a template for the internal control. To rule out false-negative results, an internal control (primer pair and template) was incorporated in every reaction mixture, including negative controls.
FIG 1.
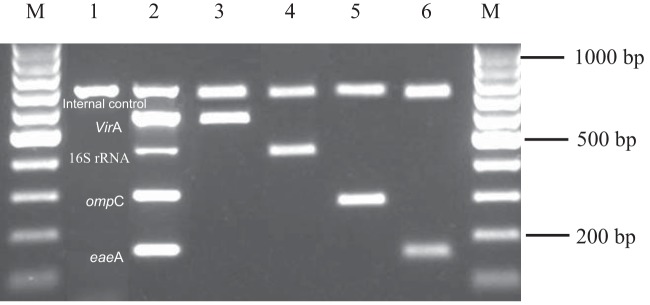
Multiplex PCR assay profile with reference strains. Lanes M, 100-bp markers; lane 1, negative control; lane 2, positive control; lane 3, Shigella flexneri ATCC 12022 (virA); lane 4, Campylobacter jejuni ATCC 33560 (16S rRNA); lane 5, Salmonella enteric ATCC 25957 (ompC); lane 6, EHEC 43889 (eaeA).
Diagnostic evaluation of the multiplex PCR was done using the lysates from 223 bacterium-spiked stool specimens. Stool specimens obtained from healthy children were spiked with hemorrhagic bacteria to imitate clinical specimens that were obtained from patients with diarrhea. Stool suspensions were prepared using a previous method (10). Briefly, 10 g stool was added to 1× phosphate-buffered saline (PBS) (pH 7.4) to make a 10% (wt/vol) stool suspension. A volume of 200 μl of the stool suspension was aliquoted into different 1.5-ml tubes and spiked with a single colony of the study bacteria. The whole spiked stool suspension was inoculated into 5 ml of alkaline peptone water (Oxoid, Basingstoke, United Kingdom) and incubated for 4 h at 37°C. Thereafter, 1 ml of the alkaline peptone water culture was boiled for 5 min and pelleted by centrifugation at 5000 × g for 10 min. The resultant supernatant was used as the template for the PCR assay. Then, 2 μl of the supernatants (lysates) was used in the multiplex PCR assays.
The optimized concentration of primer for each gene (0.2 pmol 16S rRNA, 1 pmol virA, 1 pmol ompC, 0.6 pmol eaeA, and 0.8 pmol hemA) was used in the multiplex PCR. The other components used in the PCR were 400 μM dNTPs (Fermentas, Vilnius, Lithuania), 3.125 mM MgCl2, 1× PCR buffer, and 0.75 U Taq DNA polymerase (Fermentas). The PCR was carried out using a Mastercycler Gradient (Eppendorf, Hamburg, Germany) with one cycle of initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 30 s, annealing for 30 s at 70°C, and extension at 72°C for 30 s, followed by an extra cycle of annealing at 70°C for 30 s, and a final extension at 72°C for 5 min. The PCR products were analyzed by electrophoresis on 1.5% low electroendosmosis (EEO) agarose gels (Promega, Madison, WI), with SYBR Safe at 100 V for 75 min. PCR products were visualized under UV illumination and photographed using an image analyzer ((Bio-Rad, Hemel Hempstead, United Kingdom).
Evaluation of the multiplex PCR assay.
Analytical specificity was evaluated using DNA lysates prepared from pure cultures of 20 Gram-negative and 17 Gram-positive strains obtained from different sources (Table 1). The limit of detection (LoD) was evaluated using various concentrations of genomic DNA starting from 1 μg to 10 pg and lysate starting from 107 to 102 CFU/ml obtained from reference strains. Double-blinded diagnostic evaluation of the multiplex PCR was carried out. The assay was validated quantitatively and qualitatively with 223 randomly selected bacterium-spiked stool specimens. The PCR result is considered a valid negative when only the PCR internal control amplicon is observed. If all amplicons are absent, the PCR is defined as inhibited, and the test should be repeated. The results were compared with the conventional methods, which were considered the gold standard (11).
RESULTS
In the present study, the multiplex PCR was optimized successfully to identify the Salmonella genus (ompC), Shigella genus (virA), Escherichia coli (eaeA), and Campylobacter genus (16S rRNA) genes simultaneously. Stepwise optimization of primer concentration, annealing temperature, MgCl2, dNTP, and Taq polymerase was carried out. The multiplex PCR gave the best results when 3.125 mM MgCl2, 400 μM dNTP, 0.75 U Taq polymerase, and 70°C annealing temperature were used. Based on the analysis of a collection of 37 well-defined bacterial strains, the newly developed multiplex PCR exhibited a high specificity (Table 1). None of the primers showed cross-reactions in the detection channels of the other multiplex assays. No other bacterial species except Salmonella spp., Shigella spp., Escherichia coli, and Campylobacter spp. revealed positive results (Table 1). Overall, the analytical specificity of the multiplex PCR assay was 100% for the detection of hemorrhagic bacteria reference strains. The LoD of the multiplex PCR at the DNA level was found to be 100 pg DNA (data not shown), whereas at the bacterial level, it was found to be 1 × 103 CFU/ml (data not shown). All of the reference strains of Salmonella spp. were positive for the ompC gene by multiplex PCR, and all of the Shigella spp. were positive for the virA gene; moreover, both Escherichia coli strains were positive for the eaeA gene and the all Campylobacter strains were positive for the 16S rRNA gene. The remaining reference strains (23 out of 37; 6 Gram-negative and 17 Gram-positive) were negative (Table 1). Upon completion of the standardization of the multiplex PCR assay with reference strains, the assay was validated with a randomly selected 223 bacteria-spiked stool specimens. Among these, 14 specimens contained a single bacterial gene, 9 specimens had 2 different genes, 18 specimens included 3 genes, and 59 specimens had 4 genes (Table 3). The diagnostic accuracy of a multiplex PCR for detection of hemorrhagic bacteria was found to have 100% sensitivity and specificity.
TABLE 3.
Performance of multiplex PCR assay for detecting hemorrhagic bacteria
| Targeted gene(s) | Multiplex PCR assay results |
||||
|---|---|---|---|---|---|
| No. of bacterium-spiked stool specimens | Positive PCR bandsb | Isolate(s) found | Sensitivity (%) | Specificity (%) | |
| ompC and ICa | 14 | ++ | Salmonella spp. | 100 | 100 |
| virA and IC | 14 | ++ | Shigella spp. | 100 | 100 |
| eaeA and IC | 14 | ++ | E. coli | 100 | 100 |
| 16S rRNA and IC | 14 | ++ | Campylobacter | 100 | 100 |
| ompC, virA and IC | 9 | +++ | Salmonella spp., Shigella spp. | 100 | 100 |
| ompC, eaeA and IC | 9 | +++ | Salmonella spp., E. coli | 100 | 100 |
| ompC, 16S rRNA and IC | 9 | +++ | Salmonella spp., Campylobacter | 100 | 100 |
| virA, eaeA and IC | 9 | +++ | Shigella spp., E. coli | 100 | 100 |
| virA, 16S rRNA and IC | 9 | +++ | Shigella spp., Campylobacter | 100 | 100 |
| eaeA, 16S rRNA and IC | 9 | +++ | E. coli, Campylobacter | 100 | 100 |
| ompC, virA, eaeA and IC | 18 | ++++ | Salmonella spp., Shigella spp., E. coli | 100 | 100 |
| virA, eaeA, 16S rRNA and IC | 18 | ++++ | Shigella spp., E. coli, Campylobacter | 100 | 100 |
| ompC, eaeA, 16S rRNA and IC | 18 | ++++ | Salmonella spp., E. coli, Campylobacter | 100 | 100 |
| ompC, virA, eaeA, 16S rRNA and IC | 59 | +++++ | Salmonella spp., Shigella spp., E. coli, Campylobacter | 100 | 100 |
IC, internal control.
++, one bacterial gene with an internal control; +++, two bacterial genes with an internal control; ++++, three bacterial genes with an internal control; +++++, four bacterial genes with an internal control.
DISCUSSION
Bloody diarrhea is a communicable disease that occurs in developed countries as sporadic cases and occasional outbreaks of various magnitudes and causes epidemics and disease in developing countries and endemic areas. The WHO predicts that there will still be about 5 million deaths in children younger than 5 years by 2025; most of these will be in developing countries, and most will be due to infectious diseases such as diarrhea (12). Diarrheal disease deaths are largely preventable if adequate treatment is sought early in the course of the illness (13). Recent studies suggest that rapid detection systems can decrease the costs associated with hospitalization and refine the application of antibiotic treatment (5). Implementation of the molecular tools increased the detection frequency of enteric pathogens 3-fold (19.2% compared to 6.4% with conventional methods) (7). Moreover, a considerable proportion of positive fecal samples previously confirmed as negative by culture methods have been detected by using molecular tools (14). The present study validated a multiplex PCR assay for the detection of hemorrhagic bacteria in stool samples. The performance of this assay was comparable to that of conventional methods for the detection of Salmonella spp., Shigella spp., Escherichia coli, and Campylobacter spp. simultaneously in stool, but results were seen within 4 h in comparison to 3 to 5 days by conventional methods. Although there are numerous reports on PCR assays for the detection of hemorrhagic bacteria (15, 16), only a few of these assays have incorporated internal controls (14, 17). The presence of PCR inhibitors (like bilirubin, bile salts, and heme in the feces) in stool samples may inhibit amplification and limit the usefulness of the PCR technique (18, 19). Therefore, inclusion of an internal control in the reaction is mandatory for the diagnostic test to rule out false-negative results and check for PCR inhibitors in samples (11). Various internal controls, i.e., plasmids or human genes, which have to be added to the sample, are used in diagnostic PCR assays. The multiplex PCR assay developed in our study successfully amplified all five target genes from a single reaction tube, and the primers did not interact with each other to produce false results. To avoid biases, the multiplex PCR assay was evaluated with a randomly selected 223 bacterium-spiked stool specimens, and the diagnostic performance was determined. Among the 223 specimens, 56 were positive for single bacteria, 54 showed 2 bacterial types, another 54 revealed 3 bacteria, and 59 samples contained 4 bacterial types. The overall analysis of the multiplex PCR showed 100% sensitivity, specificity, positive predictive value, and negative predictive value. The high positive predictive value (100%) suggested high test reliability and accuracy, while the 100% negative predictive value showed that the PCR assay could discriminate nonhemorrhagic bacteria. These results suggest that the primers designed were highly specific and no cross-reactivity with DNA from other organisms was observed. The LoD of this PCR assay was found to be 100 pg of DNA at the genomic DNA level and 1 × 103 CFU/ml at the bacterial cell level, which was below the bacterial burden (103 to 109 CFU/g) often reported in symptomatic patients as measured by culture (20) and also comparable with LoDs previously described (6, 7).
The mixing of primers in a single tube decreases costs and time and increases the ease of the assay compared to the more expensive real-time PCR. Theron et al. (21) and Thong et al. (22) reported that the enrichment procedure prior to the PCR enhances the total number of bacteria present and lower LoDs might be achieved; however, this procedure is time-consuming and minimizes the time-saving advantages of direct stool sample PCRs. In developing countries, mixed infections are common, especially in diarrheal samples (23), making it difficult to determine which pathogen is responsible for symptoms. In this study, the multiplex PCR assay successfully identified the mixed infections simultaneously in a single stool sample with 100% sensitivity and specificity, showing that the presented multiplex PCR assay is robust and effective. Previously, several individual PCR assays have generally used to detect a broad range of enteric bacteria (24, 25). Currently, multiplex PCR assays up to 9-plex with a sensitivity of 89 to 100% and specificity of 93 to 100% for all targets have been developed (26, 27). In this study, the PCR assay is flexible, in that additional target genes can be added to detect additional enteric pathogens. Also, if some species are rare, as in case with Vibrio and Yersinia spp., these reagents can be withheld. Our PCR assay revealed a lower LoD, high sensitivity and specificity, and a reduced detection time. Therefore, this assay offers clinical laboratories a new rapid, simple, feasible, and accurate tool. Its application allows early detection of hemorrhagic bacteria from stool samples, which makes this assay an alternative to conventional methods for the future.
This study has some limitations. First, our assay detects only a limited number of clinically significant pathogens and does not detect protozoa or viral targets. Second, the assay did not detect enterotoxigenic E. coli strains, the main bacterial agents implicated in diarrhea. However, we evaluated stool samples for the most common hemorrhagic bacteria.
In conclusion, the multiplex PCR assay developed in this study can obtain results within a short time with only a few processing steps, which minimizes processing errors. The approach presented here was able to identify four genes that are essential for the detection of most common hemorrhagic bacteria at the genus level simultaneously in a single test. The multiplex PCR assay was highly sensitive and could provide results within a few hours, thus allowing clinicians to start early antibiotic treatment in critically ill patients. The built-in internal control in this assay prevented false-negative results. Hence, this test can be used as an effective diagnostic tool for detection of hemorrhagic bacteria. Further studies are needed to evaluate the cost-effectiveness of the present test and, in the long run, the impact of this assay on the clinical outcomes of patients.
ACKNOWLEDGMENTS
This study was supported by a Research Intensive Faculty grant entitled Development of a Heptaplex PCR Assay for the Rapid Detection of Diarrheal Pathogens among Children (600-RMI/DANA 5/3/RIF; 685/2012).
We gratefully acknowledge the Institute for Medical Research, Malaysia, for providing isolates and the Institute of Medical Molecular Biotechnology for providing facilities.
Footnotes
Published ahead of print 25 June 2014
REFERENCES
- 1.Lim VK. 2007. Infectious diarrhoea. Med. J. Malaysia 62:187–188. [PubMed] [Google Scholar]
- 2.Baron EJ, Miller JM, Weinstein MP, Richter SS, Gilligan PH, Thomson RB, Jr, Bourbeau P, Carroll KC, Kehl SC, Dunne WM, Robinson-Dunn B, Schwartzman JD, Chapin KC, Snyder JW, Forbes BA, Patel R, Rosenblatt JE, Pritt BS. 2013. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. 57:e22−e121. 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Talib H, Yean CY, Al-Khateeb A, Hassan H, Singh KK, Al-Jashamy K, Ravichandran M. 2009. A pentaplex PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine leucocidin. BMC Microbiol. 9:113. 10.1186/1471-2180-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancini N, Carletti S, Ghidoli N, Cichero P, Burioni R, Clementi M. 2010. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin. Microbiol. Rev. 23:235–251. 10.1128/CMR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doern GV, Vautour R, Gaudet M, Levy B. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. 2010. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J. Clin. Microbiol. 48:2929–2933. 10.1128/JCM.00339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer RF, Ott A, Kesztyus B, Kooistra-Smid AM. 2010. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J. Clin. Microbiol. 48:4140–4146. 10.1128/JCM.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Talib H, Yean CY, Al-Khateeb A, Hasan H, Ravichandran M. 2013. Rapid detection of methicillin-resistant Staphylococcus aureus by a newly developed dry reagent-based polymerase chain reaction assay. J. Microbiol. Immunol. Infect., in press. 10.1016/j.jmii.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Haarman M, Knol J. 2006. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72:2359–2365. 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McOrist AL, Jackson M, Bird AR. 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50:131–139. 10.1016/S0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2006. Molecular diagnostic methods for infectious diseases; approved guideline—second edition. CLSI document MM3-A2 Clinical and Laboratory Standards, Wayne, PA. [Google Scholar]
- 12.Thapar N, Sanderson IR. 2004. Diarrhoea in children: an interface between developing and developed countries. Lancet 363:641–653. 10.1016/S0140-6736(04)15599-2. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra N, Upadhyay RP. 2013. Why are there delays in seeking treatment for childhood diarrhoea in India? Acta Paediatr. 102:e413–e418. 10.1111/apa.12304. [DOI] [PubMed] [Google Scholar]
- 14.Quetz Jda S, Lima IF, Havt A, Prata MM, Cavalcante PA, Medeiros PH, Cid DA, Moraes ML, Rey LC, Soares AM, Mota RM, Weigl BH, Guerrant RL, Lima AA. 2012. Campylobacter jejuni infection and virulence-associated genes in children with moderate to severe diarrhoea admitted to emergency rooms in northeastern Brazil. J. Med. Microbiol. 61:507–513. 10.1099/jmm.0.040600-0. [DOI] [PubMed] [Google Scholar]
- 15.El Metwally HR, Ibrahim HH, El-Athamna MN, Amer MA. 2007. Multiplex PCR for detection of diarrheagenic Escherichia coli in Egyptian children. J. Med. Sci. 7:255–262. 10.3923/jms.2007.255.262. [DOI] [Google Scholar]
- 16.Hegde A, Ballal M, Shenoy S. 2012. Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J. Med. Microbiol. 30:279–284. 10.4103/0255-0857.99485. [DOI] [PubMed] [Google Scholar]
- 17.Wiemer D, Loderstaedt U, von Wulffen H, Priesnitz S, Fischer M, Tannich E, Hagen RM. 2011. Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in fecal samples. Int. J. Med. Microbiol. 301:577–584. 10.1016/j.ijmm.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Rossen L, Norskov P, Holmstrom K, Rasmussen OF. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37–45. 10.1016/0168-1605(92)90017-W. [DOI] [PubMed] [Google Scholar]
- 19.Wilson IG. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson EJ, Harris JB, Morris JG, Jr, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7:693–702. 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theron J, Morar D, Du Preez M, Brozel VS, Venter SN. 2001. A sensitive seminested PCR method for the detection of Shigella in spiked environmental water samples. Water Res. 35:869–874. 10.1016/S0043-1354(00)00348-1. [DOI] [PubMed] [Google Scholar]
- 22.Thong KL, Hoe SL, Puthucheary SD, Yasin R. 2005. Detection of virulence genes in Malaysian Shigella species by multiplex PCR assay. BMC Infect. Dis. 5:8. 10.1186/1471-2334-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, Molina M, Amemiya I, Verastegui H, Hall ER, Cleary TG, Lanata CF. 2009. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from periurban areas in Lima, Peru. Clin. Infect. Dis. 49:1694–1702. 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatta M, Smits HL. 2007. Detection of Salmonella typhi by nested polymerase chain reaction in blood, urine, and stool samples. Am. J. Trop. Med. Hyg. 76:139–143. [PubMed] [Google Scholar]
- 25.Linton D, Lawson AJ, Owen RJ, Stanley J. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, Howlader AM, Sobuz SU, Haque R, Talukder KA, Qureshi S, Zaidi A, Haverstick DM, Houpt ER. 2012. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-Luminex assay. J. Clin. Microbiol. 50:98–103. 10.1128/JCM.05416-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yean CY, Yin LS, Lalitha P, Ravichandran M. 2007. A nanoplex PCR assay for the rapid detection of vancomycin and bifunctional aminoglycoside resistance genes in Enterococcus species. BMC Microbiol. 7:112. 10.1186/1471-2180-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]


