Abstract
This study involves longitudinal and point-in-time surveys of Salmonella carriage and environmental contamination on two commercial cage layer farms positive for Salmonella enterica subsp. enterica serovar Typhimurium (flock A age, 32 weeks; flock B age, 34 weeks). Salmonella-positive fecal, egg belt, and dust samples were all unconditionally associated with eggshells testing positive for Salmonella. The odds of an eggshell testing positive for Salmonella were 91.8, 61.5, and 18.2 times higher when fecal, egg belt, and dust samples, respectively, tested positive for Salmonella. The agreement between the culture-based methods and real-time PCR on preenriched broths for detecting Salmonella was almost perfect for eggshell (observed agreement, 99.19%; kappa coefficient, 0.94) and egg belt samples (observed agreement, 95%; kappa coefficient, 0.88), and it was substantial for fecal (observed agreement, 87.14%; kappa coefficient, 0.47) and floor dust samples (observed agreement, 80.61%; kappa coefficient, 0.58). A 1-log increase in the load of Salmonella detected in the fecal, egg belt, and floor dust samples resulted in 35%, 43%, and 45% increases, respectively (P < 0.001), in the odds of an eggshell testing positive for Salmonella. The multilocus variable-number tandem-repeat analysis (MLVA) patterns of the S. Typhimurium strains isolated from flock A were distinct from those of flock B. S. Typhimurium strains detected from human food poisoning cases exhibited an MLVA pattern similar to those of the strains isolated from flocks A and B.
INTRODUCTION
Eggs and their derived products are often linked to cases of Salmonella food poisoning. Salmonella outbreaks have been associated with uncooked products, like mayonnaise, ice cream, and cold desserts that contain raw egg (1). A very low dose of Salmonella, 10 to 20 CFU, is sufficient to cause human salmonellosis (2, 3). In Australia in 2010, the incidence risk of Salmonella infection was 53.7 cases per 100,000 people, almost 30% higher than the average risk of 41.8 cases per 100,000 people in the previous five years (1).
Salmonella enterica subsp. entericaserovar Enteritidis is a major concern for most egg industries around the world. Although S. Enteritidis is associated with the majority of egg-related outbreaks of human salmonellosis occurring in the European Union (77.2%) (4), it is not endemic to Australian layer flocks (5). Instead, S. enterica subsp. enterica serovar Typhimurium was the most frequently reported serovar in the 21 egg-related food poisoning outbreaks in Australia in 2010 (1). In Australia, a study showed that S. enterica subsp. enterica serovar Infantis was the most frequently reported serovar from the eggshell wash of eggs collected from 31 flocks (6). Furthermore, in Australia, S. Infantis has had the largest percentage increase in reported human infections, with 2.2 times more notifications reported nationally in 2010 than in the previous year (1).
Residual contamination of the environment with Salmonella is a major problem in commercial layer farms (7–9). Davies and Breslin (10) concluded that in cage systems, environmental samples, such as those from egg belts, dust near cages, and pooled accumulated feces should be tested while screening flocks for Salmonella. There is little information available in the literature about the risks of Salmonella contamination of eggs from infected birds and a contaminated shed environment. Chemaly et al. (11) investigated the prevalence of Salmonella on eggshells in infected layer flocks, whereas Wales et al. (12) correlated environmental contamination with fecal contamination by Salmonella. However, the rate at which an infected flock can produce Salmonella-contaminated eggs is unclear. The possible transmission of Salmonella from the environment to the egg might be explained with the help of longitudinal studies (12). However, cooperation from egg producers over a period of months or years and the requirement of resources are limiting factors to such studies (12) There are a few reports in which the levels of Salmonella contamination in laying houses and hens were examined over time during lay (12–14). However, these studies did not investigate the degree of internal or external egg contamination. Furthermore, the focus of these studies was mainly on S. Enteritidis. Although S. Typhimurium has an established ability to be transmitted to humans via contaminated shell eggs, there is little published data on field studies, natural infections, and long-term experiments on this topic (15).
In the present study, longitudinal and point-in-time surveys were conducted on two known S. Typhimurium-contaminated commercial layer farms, both with multiaged flocks housed in the same shed. The primary objectives of this study were to (i) evaluate the association between Salmonella load in the shed environment and the probability of eggshells being contaminated with Salmonella, (ii) investigate the dynamics of Salmonella shedding of various serovars over a prolonged period of time during longitudinal samplings, (iii) detect S. Typhimurium- and S. Infantis-positive samples using multiplex PCR, and (iv) investigate the relatedness of various S. Typhimurium strains using multilocus variable-number tandem-repeat analysis (MLVA).
MATERIALS AND METHODS
This study was conducted in two stages. In stage 1, with the help of a cross-sectional study, cages infected with various serovars of Salmonella spp. were identified. Based on the results of the cross-sectional study, in stage 2, Salmonella-positive and -negative cases were selected for longitudinal study, and the association between eggs and environmental Salmonella contamination was investigated.
Stage 1, cross-sectional survey to select cages for longitudinal study.
Two commercial layer cage sheds were selected from two different farms with a history of Salmonella infection. The study sheds, shed A (from farm A) and shed B (from farm B), included multiaged flocks, with each age class housed in separate rows, and only a single age class flock was selected for sampling in each shed. In shed A, the selected flock included 1,320 cages of 32-week-old birds (5 birds per cage, for an approximate total of 6,600 birds), while the selected flock in shed B included 1,300 cages of 34-week-old birds (5 birds per cage, for an approximate total of 6,500 birds). To ensure that at least several cages positive for Salmonella would be identified, a representative sample size of 78 cages per flock was targeted. Accounting for field constraints, two adjacent cages were selected at equal intervals along the three lowest tiers (tiers 1, 2, and 3) out of the five tiers.
For the isolation of Salmonella spp., the fecal samples were inoculated in buffered peptone water (BPW) (Oxoid, Australia) (1:4). The inoculated samples were incubated at 37°C overnight, and 100 μl of this sample was transferred into Rappaport-Vassiliadis soya peptone (RVS) broth (Oxoid), which was then incubated at 42°C for 24 h. A loopful of the incubated RVS broth was streaked onto Brilliance Salmonella agar (BSA) (Oxoid) and xylose lysine deoxycholate agar (XLD) (Oxoid) plates. Two to three presumptive Salmonella colonies from BSA and XLD agars were selected and used to stab inoculate triple sugar iron (TSI) agar slopes (Oxoid). After incubation at 37°C, the inoculated TSI slopes were examined at intervals of 24 h up to 72 h for typical Salmonella reactions. The presumptive Salmonella colonies were also tested for ortho-nitrophenyl-β-d-galactopyranoside (Oxoid), lysine decarboxylase (LDC), and urease (Oxoid) activity. Depending upon the results of the biochemical reactions, the presumptive Salmonella isolates were sent for serotyping to the Salmonella Reference Laboratory, Adelaide, Australia.
Stage 2, longitudinal study to investigate the association between eggs and environmental Salmonella contamination.
Based on the Salmonella typing results, five Salmonella-positive cages each from farm A (3 cages of S. Typhimurium phage type 9, 1 cage each of S. Infantis and Salmonella enterica subsp. enterica serovar Orion) and farm B (2 cages of S. Typhimurium phage type [PT] 9, 1 cage each of S. Infantis, Salmonella enterica subsp. enterica serovar Agona, and Salmonella enterica subsp. enterica serovar Oranienburg), as well as two Salmonella-negative cages per farm, were selected for the longitudinal study. The reason for selecting cages positive with different Salmonella serovars was to investigate the dynamics of Salmonella shedding of various serovars over a prolonged period of time during the longitudinal samplings. The selected cages were sampled at 4-week intervals. Both farms were sampled with a gap of 1 week. For each flock, 10 longitudinal samplings were performed over the period of 40 weeks (i.e., 4-week intervals).
Environmental and egg sampling.
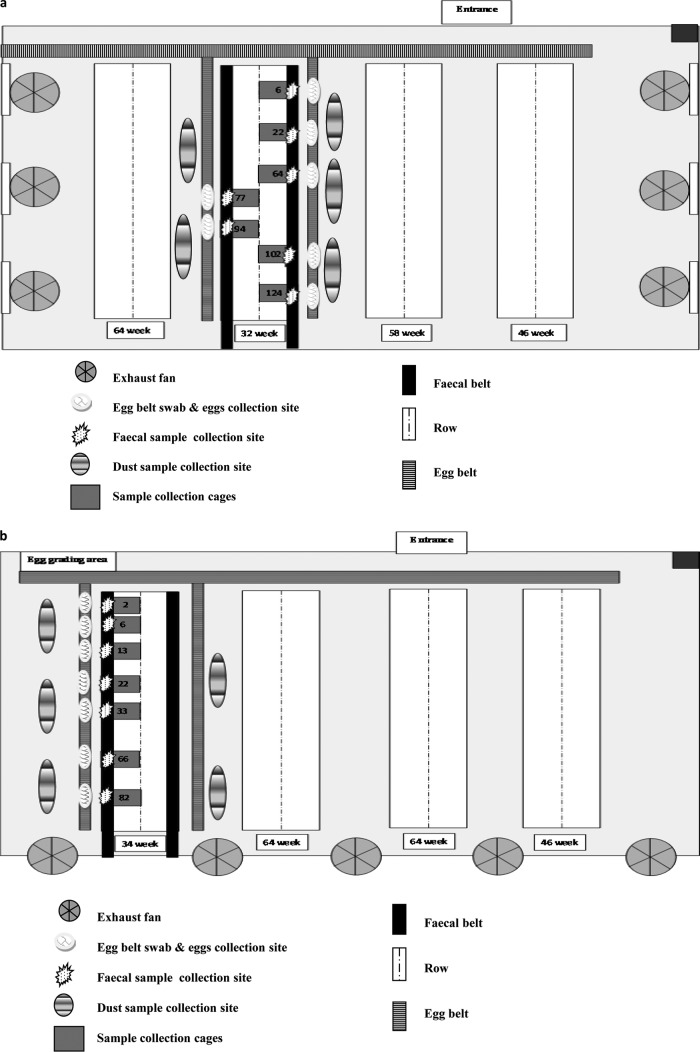
A composite fecal sample (one per study cage, n = 7) from each flock was collected in a sterile Whirl-Pak plastic bag (150 by 230 mm; Thermo Fisher Scientific, Australia) from underneath the individual selected cages. The full length of the manure belt under each cage was covered while collecting fecal samples. To avoid cross-contamination, disposable gloves were changed between each fecal sample collection. Egg belt samples (one per study cage, n = 7) were collected from the front of the cage. Whirl-Pak Speci-Sponge bags (115 by 239 mm; Thermo Fisher Scientific) were used for sample collection. The swabs were premoistened using 25 ml of BPW and dragged to cover the whole area in front of the individual cages. During each sampling period, five dust samples were collected from different parts of the poultry shed and from the floor near the selected cages for longitudinal study. Dust (n = 5) was collected in gamma-irradiated sterile containers (Pacific Laboratory Products, Australia). Figure 1a and b show the layout of the layer sheds, along with sample collection areas.
FIG 1.
Layouts of the shed with flock A (A) and flock B (B) showing the areas of sample collection (with number of samples in the collection area).
All the eggs at the front of the seven selected cages were collected. Each egg was collected in a separate sterile Whirl-Pak plastic bag to avoid cross-contamination. Flock A molted at the 67th week of lay; therefore, no eggs were obtained in the ninth week of sampling.
Sample processing for Salmonella isolation.
For feces and dust, 2 g of sample was added to 8 ml of BPW. Fecal, egg belt swab, and dust samples were processed as mentioned above to isolate Salmonella spp. Eggshell and egg internal content samples were individually processed. Individual eggs were placed in 10 ml of sterile BPW in Whirl-Pak bags and rinsed by massaging for 2 min. Before rinsing, BPW was prewarmed to 37°C to facilitate bacterial recovery. After a rinse sample was obtained, each egg was removed and transferred to a new sterile bag. The BPW samples were incubated at 37°C overnight, and 100 μl of this sample was inoculated into RV broth (Oxoid, Australia), which was then incubated at 42°C for 24 h. The incubated RV broths were further processed for Salmonella isolation, as mentioned above. The egg internal contents, collected in sterile containers, were thoroughly mixed, and 2 ml of egg internal content was inoculated into 8 ml of BPW. The inoculated BPW samples were further processed for Salmonella isolation, as mentioned above.
Multilocus variable-number tandem-repeat analysis of S. Typhimurium isolates.
After serotyping, all Salmonella strains that were identified as S. Typhimurium were further analyzed by MLVA, as described by Ross et al. (16), at the Salmonella Reference Laboratory, Adelaide, Australia.
Quantitative PCR detection of S. enterica.
Total nucleic acid was extracted from samples using a modification (17) of a South Australian Research and Development Institute (SARDI) (Adelaide, Australia) proprietary method (18). All samples had been subjected to preenrichment in BPW, with 2 g of feces or dust samples in 8 ml BPW, egg belt swabs in 25 ml BPW, and six pooled individual eggshell washes in 10 ml BPW each. Following incubation at 37°C overnight, the incubated broths were frozen and freeze-dried. Ten milliliters of extraction buffer (17) was added to the freeze-dried samples and incubated at 70°C for an hour before proceeding with the SARDI proprietary extraction method. The quantitative PCR (qPCR) detection of Salmonella was done using the TaqMan S. enterica detection kit system (Applied Biosystems, Australia) in a total reaction volume of 20 μl containing an 8-μl sample, 10 μl of 2× environmental master mix, and 2 μl of 10× target assay mix. All real-time PCR assays were run in a 384-well format, with master mix and template being dispensed using a Biomek 3000 laboratory automation workstation (Beckman Coulter, USA). All reactions were run on a 7900HT sequence detection system (Applied Biosystems), with the following conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 60 s. All data were analyzed using the 7900HT v2.3 SDS software (Applied Biosystems). The raw data were analyzed for target-specific S. enterica and internal positive control (IPC) using a cycle threshold (CT) of 0.8 and baseline of 3 to 10. Salmonella copies were calculated using a standard curve prepared by a serial 10-fold dilution of cultured S. enterica serovar Infantis. A cutoff CT of 34 was used to exclude the detection of false positives. A CT of 34 corresponded to 200 CFU of Salmonella.
Multiplex PCR to identify S. Typhimurium- and S. Infantis-positive samples.
Two multiplex PCRs, one for the detection of S. Typhimurium and the other for S. Infantis, were performed using the primers published by Akiba et al. (19). The primer sequences and the expected amplification product sizes are shown in Table S1 in the supplemental material. The multiplex PCR assays were used on various Salmonella serovars isolated from the Australian layer industry to confirm the specificities of the assays (see Table S2 in the supplemental material). For the S. Typhimurium multiplex PCR, each reaction mixture contained 1× PCR buffer II (Applied Biosystems, Australia), 2.5 mM MgCl2, 1.6 mM deoxynucleoside triphosphates (dNTPs) (Invitrogen, Australia), 0.5 μM (each) InvAF, InvAR, TMP2F, and TMP2R primers, 0.3 μM (each) TMP1F, TMP1R, TMP3F, and TMP3R primers, 1 U of AmpliTaq DNA polymerase LD (Applied Biosystems), and 5 μl DNA template in a reaction volume of 20 μl. For the S. Infantis multiplex PCR, each reaction mixture contained 1× PCR buffer II (Applied Biosystems), 2.5 mM MgCl2, 1.6 mM dNTPs, 0.5 μM (each) InvAF, InvAR, IMP3F, and IMP3R primers, 0.3 μM (each) IMP1F, IMP1R, IMP2F, and IMP2R primers, 1 U of AmpliTaq DNA polymerase LD (Applied Biosystems), and 5 μl DNA template in a reaction volume of 20 μl. The samples were amplified in a MJ Research PTC-225 Peltier thermal cycler (GeneWorks, Adelaide, Australia), with an initial denaturation step at 95°C for 2 min, followed by 35 cycles of amplification (denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s), and a final extension step at 72°C for 10 min. The PCR products were separated by 2% agarose gel electrophoresis in Tris-acetate-EDTA (TAE) buffer, stained with GelRed (Jomar Diagnostics, Australia), and visualized under ultraviolet light.
In order to determine the limit of detection for each multiplex PCR assay, fecal samples were spiked with various concentrations (108 CFU/ml to 102 CFU/ml) of the corresponding Salmonella serovar (S. Typhimurium or S. Infantis). DNA was extracted from Salmonella-spiked fecal samples, and the multiplex PCRs were performed as mentioned above. The limit of detection was determined by running the PCR products on a 2% agarose gel.
Statistical analysis.
Binomial exact confidence intervals were computed for the prevalence of Salmonella-positive cages estimated in each flock. Multilevel logistic regression was used to estimate the association between an eggshell being Salmonella positive and Salmonella-positive feces from the cage that held the egg, a Salmonella-positive egg belt at the front of a cage, and Salmonella-positive floor dust at the front of a cage. Random effects for “flock” and for “cage within flock” were added to the model to account for the fact that eggs were clustered within the cage and within the flock. Multilevel logistic regression was also used to evaluate the association among the Salmonella test outcomes of the corresponding cage fecal, egg belts, and floor dust samples (i.e., only included “flock” as random effect). Kappa statistics were computed to assess the agreement between culture isolation and real-time PCR. The association between the Salmonella burden (using log-transformed CT values from qPCR) in the fecal, egg belt, and floor dust samples and the odds of an eggshell testing positive for Salmonella was investigated using the same structure for multilevel logistic regressions. The parameters of all models (odds ratio) were interpreted at a 5% significance level. The models assumptions were assessed using standard diagnostic plots. Statistical analyses were performed using the statistical package Stata v12.1 (20).
RESULTS
Selection of cages for longitudinal study.
The culture isolation results indicated that in flock A, 21 cages (26.9%; confidence interval [CI], 17.5 to 38.2) were positive for Salmonella spp. at 32 weeks. Flock B had a higher prevalence of Salmonella-positive cages, with 31 cages (39.7%; CI, 28.8 to 51.5) reported to be positive for Salmonella at 34 weeks. Based on the Salmonella typing results, five Salmonella-positive cages each from farm A (3 cages for S. Typhimurium phage type 9 and 1 cage each for S. Infantis and S. Orion) and farm B (2 cages for S. Typhimurium PT 9, and 1 cage each for S. Infantis, S. Agona, and S. Oranienburg), as well as two Salmonella-negative cages per farm were selected for the longitudinal study.
Salmonella prevalence in flocks A and B in longitudinal study.
The details of the number of samples that were reported as Salmonella positive, along with types of serovars identified over the period of 40 weeks from flocks A and B, are described in Tables 1 and 2, respectively. Figure 2 shows the prevalence of Salmonella in different types of samples. In both flocks, the Salmonella prevalence was higher in dust samples than in egg belt, fecal, and eggshell samples. In flock B, from the 6th sampling (58 week onwards), there was an increase in the prevalence of Salmonella in all types of samples, with the highest prevalence in dust samples. It was observed that there was a higher fluctuation in Salmonella contamination in fecal samples than in the dust and egg belt samples. Out of all eggs tested, in flock B, 7.17% (19/265) of the eggshells were Salmonella positive; however, in flock A, only one eggshell out of 256 (0.39%) was reported as Salmonella positive. All of the egg internal contents from flocks A and B were Salmonella negative.
TABLE 1.
Prevalence of Salmonella in flock A during longitudinal sampling
| Collection no. | Week of lay | No. of positive isolates/total no. of indicated sample type |
Sample type(s) and Salmonella serovar(s) (no. of isolates)a | |||
|---|---|---|---|---|---|---|
| Eggshells | Feces | Egg belt | Dust | |||
| 1 | 36 | 0/33 | 0/7 | 0/7 | 1/5 | Dust: S. Typhimurium PT 9 |
| 2 | 40 | 0/33 | 0/7 | 1/7 | 1/5 | Egg belt and dust: S. Worthington |
| 3 | 44 | 0/30 | 1/7 | 0/7 | 3/5 | Feces and dust: S. Typhimurium PT 9 |
| 4 | 48 | 0/30 | 0/7 | 0/7 | 0/5 | |
| 5 | 52 | 0/30 | 0/7 | 0/7 | 1/5 | Dust: S. Typhimurium PT 9 |
| 6 | 56 | 0/30 | 0/7 | 0/7 | 1/5 | Dust: S. Typhimurium PT 9 |
| 7 | 60 | 0/32 | 1/7 | 1/7 | 1/5 | Feces: S. Worthington; egg belt: Salmonella subsp. 1 serotype 4,5,12; dust: S. Typhimurium PT 9 |
| 8 | 64 | 0/28 | 1/7 | 1/7 | 0/5 | Feces: S. Worthington; egg belt: Salmonella subsp. 1 serotype 4,5,12:-:- |
| 9 | 68 | 2/7 | 1/7 | 2/5 | Feces and egg belt: S. Worthington; dust: S. Worthington (1), S. Typhimurium PT 9 (1) | |
| 10 | 72 | 1/10 | 0/7 | 1/7 | 3/5 | Egg: S. Worthington; egg belt: S. Typhimurium PT 9; dust: S. Typhimurium PT 9 (2), S. Worthington |
PT, phage type.
TABLE 2.
Prevalence of Salmonella in flock B during longitudinal sampling
| Collection no. | Week of lay | No. of positive isolates/total no. of indicated sample type: |
Sample type(s) and Salmonella serovar(s) (no. of isolates)a | |||
|---|---|---|---|---|---|---|
| Eggshells | Feces | Egg belt | Dust | |||
| 1 | 38 | 0/31 | 1/7 | 0/7 | 1/5 | Feces and dust: S. Oranienburg |
| 2 | 42 | 0/36 | 0/7 | 0/7 | 1/5 | Dust: S. Oranienburg |
| 3 | 46 | 0/39 | 0/7 | 0/7 | 1/5 | Dust: S. Agona |
| 4 | 50 | 0/27 | 0/7 | 2/7 | 2/5 | Egg belt: S. Oranienburg (1), S. Agona (1); dust: S. Oranienburg (1), S. Agona (1) |
| 5 | 54 | 0/27 | 2/7 | 3/7 | 3/5 | Feces, dust, and egg belt: S. Oranienburg |
| 6 | 58 | 1/30 | 5/7 | 5/7 | 1/5 | Feces, dust, egg belt, and eggshells: S. Oranienburg |
| 7 | 62 | 10/34 | 5/7 | 6/7 | 5/5 | Feces, dust, and egg belt: S. Oranienburg; eggshells: S. Oranienburg (10), S. Typhimurium PT 9 (1) |
| 8 | 66 | 8/33 | 6/7 | 7/7 | 5/5 | Eggshells, feces, and egg belt: S. Oranienburg; dust: S. Oranienburg (5), S. Agona (1) |
| 9 | 70 | 0/1 | 2/7 | 6/7 | 5/5 | Feces, dust, and egg belt: S. Oranienburg |
| 10 | 74 | 0/4 | 2/7 | 6/7 | 5/5 | Feces: S. Oranienburg (1), S. Typhimurium PT 9 (1); egg belt: S. Oranienburg (5), S. Typhimurium PT 9 (2); dust: S. Oranienburg (5), Salmonella subsp. 1 serovar rough: g,s,t:- |
PT, phage type.
FIG 2.
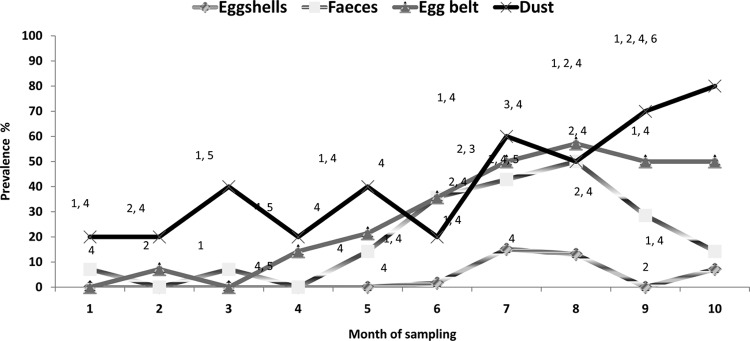
Percent prevalence of Salmonella in different types of samples over a period of 10 months. 1, Salmonella Typhimurium PT 9; 2, Salmonella Worthington; 3, Salmonella subsp. 1 serotype 4,5,12:-:-; 4, Salmonella Oranienburg; 5, Salmonella Agona; 6, Salmonella subsp. 1 serotype rough g,s,t:-.
Serotyping results confirmed that S. Oranienburg was the most frequently (76.92%) reported S. enterica subsp. enterica serovar, followed by S. Typhimurium PT 9 (11.54%), S. Worthington (8.46%), S. Agona (3.08%), Salmonella subsp. 1 serotype 4,5,12:-:- (1.54%), and Salmonella subsp. 1 serotype rough g,s,t:- (0.77%). Table 3 provides the percentages of various Salmonella serovars isolated from different types of samples. The results of MLVA indicated that S. Typhimurium strains isolated from flocks A and B were genetically distinct. In flock B, all the S. Typhimurium isolates possessed the same MLVA pattern (03 15 07 11 550). On other hand, the S. Typhimurium strains isolated from flock A exhibited three different MLVA patterns (03 24 11 10 523, 03 24 11 11 523, and 03 24 11 12 523).
TABLE 3.
Serovars detected in various Salmonella-positive sample types from layer farms
| Salmonella serovara | Frequency (% [no. positive/total no.]) for indicated sample type |
||||
|---|---|---|---|---|---|
| Feces | Egg belt | Dust | Eggshells | Total | |
| S. Typhimurium PT 9 | 7.14 (2/28) | 7.50 (3/40) | 21.43 (9/42) | 5 (1/20) | 11.54 (15/130) |
| S. Oranienburg | 78.57 (22/28) | 82.5 (33/40) | 64.28 (27/42) | 90 (18/20) | 76.92 (100/130) |
| S. Worthington | 14.28 (4/28) | 5 (2/40) | 9.52 (4/42) | 5 (1/20) | 8.46 (11/130) |
| S. Agona | 0 (0/28) | 2.5 (1/40) | 7.14 (3/42) | 0 (0/20) | 3.08 (4/130) |
| Salmonella subsp. 1 Serotype 4,5,12:-:- | 0 (0/28) | 5 (2/40) | 0 (0/42) | 0 (0/20) | 1.54 (2/130) |
| Salmonella subsp. 1 serotype rough: g,s,t:- | 0 (0/28) | 0 (0/40) | 2.38 (1/42) | 0 (0/20) | 0.77 (1/130) |
PT, phage type.
Relationship between the environmental contamination of Salmonella with Salmonella-positive eggshells.
Salmonella-positive fecal, egg belt, and dust samples were all unconditionally (analysis did not account for other factors) associated with eggshells testing positive for Salmonella. The odds of an eggshell testing positive for Salmonella were 91.8 times higher when the fecal sample from the cage tested positive for Salmonella (odds ratio, 91.8; P < 0.001; CI, 11.2 to 749.7). The odds of an eggshell testing positive for Salmonella were 61.5 times higher when the corresponding section of the egg belt tested positive for Salmonella (odds ratio, 61.5; P < 0.001; CI, 7.65 to 494.8). The odds of an eggshell testing positive for Salmonella were 18.2 (odds ratio, 18.2; P < 0.001; CI, 3.93 to 84.2) times higher when the corresponding floor dust sample tested positive for Salmonella. In the final multifactorial model (designed to study the possible environment/bird/egg transmission of Salmonella), the fecal and dust sample results were conditionally (analysis accounted for other factors) associated with an eggshell testing positive for Salmonella. The odds of an eggshell testing positive for Salmonella were 58.9 times higher when the fecal sample from the cage tested positive for Salmonella (odds ratio, 58.9; P < 0.001; CI, 6.9 to 501.0) and 9.2 times higher when the corresponding floor dust tested positive for Salmonella (odds ratio, 9.2; P = 0.007; CI, 1.8 to 45.8).
Quantification of Salmonella load in environmental samples using qPCR and its relationship with Salmonella eggshell contamination.
The TaqMan S. enterica detection system does not provide quantification of positive samples. Therefore, to determine the limit of detection of the assay, a standard curve prepared from a known concentration of S. Infantis (2 × 106 to 2 × 100 CFU Salmonella organisms per qPCR) was used. The standard curve produced a slope of −3.2, a y intercept of 41, and R2 of 0.99. Despite the good PCR assay efficiency (105%), confident detection was not possible at <200 CFU per qPCR or 25 CFU/μl extracted nucleic acid template. When a cutoff CT of 34 was used (CFU, >200 per PCR), qPCR identified 87 Salmonella-positive samples, of which 7 were not detected by the culture-based method. qPCR failed to detect Salmonella in 38 samples from which Salmonella was cultured. The qPCR resulted in 68% (80/118) of the samples being identified as containing Salmonella by microbiological culturing also testing positive by qPCR (see Table S3 in the supplemental material).
Table 4 provides the details of the Salmonella-positive and -negative samples detected by culture-based analysis and qPCR. Agreement between culture-based methodology and qPCR in detecting Salmonella was almost perfect for eggshell (observed agreement, 99.19%; kappa coefficient, 0.94) and egg belt samples (observed agreement, 95%; kappa coefficient, 0.88), and it was substantial for fecal (observed agreement, 87.14%; kappa coefficient, 0.47) and floor dust samples (observed agreement, 80.61%; kappa coefficient, 0.58). The overall (in all samples) agreement between the culture-based and qPCR detections of Salmonella was good (observed agreement, 91.02%; kappa coefficient, 0.73).
TABLE 4.
Agreement between culture method and real-time PCR to detect Salmonella-positive and -negative samples
| Sample type | Sample identification by culture | No. of samples identified by real-time PCR to be: |
Total no. of samples | Observed agreement (%) | Kappa coefficient | |
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| Eggshells | Positive | 8 | 1 | 9 | 99.14 | 0.94 |
| Negative | 0 | 114 | 114 | |||
| Total | 8 | 115 | 123 | |||
| Feces | Positive | 10 | 18 | 28 | 87.14 | 0.47 |
| Negative | 0 | 112 | 112 | |||
| Total | 10 | 130 | 140 | |||
| Egg belt | Positive | 37 | 3 | 40 | 95 | 0.87 |
| Negative | 4 | 96 | 100 | |||
| Total | 41 | 99 | 140 | |||
| Dust | Positive | 25 | 16 | 41 | 80.61 | 0.58 |
| Negative | 3 | 54 | 57 | |||
| Total | 28 | 70 | 98 | |||
| All types | Positive | 80 | 38 | 118 | 91.02 | 0.73 |
| Negative | 7 | 376 | 383 | |||
| Total | 87 | 414 | 501 | |||
Using the qPCR standard curve, the load of Salmonella in fecal, egg belt, eggshell, and dust samples was determined. Figure 3 shows the load of Salmonella (average log CFU per PCR) in feces, egg belt, dust, and eggshells. The results indicated that the levels of Salmonella detected in the fecal, egg belt, and floor dust samples were unconditionally associated with an eggshell testing positive for Salmonella. A 1-log increase in the load of Salmonella detected in fecal samples resulted in a 35% increase (odds ratio, 1.35; P < 0.001) in the odds of an eggshell testing positive for Salmonella. Similarly, a 1-log increase in the load of Salmonella detected on the egg belt and floor dust samples resulted in 43% (odds ratio, 1.43; P < 0.001) and 45% (odds ratio, 1.45; P < 0.001) increases, respectively, in the odds of an eggshell testing positive for Salmonella. When averaging Salmonella environmental burden across the fecal, egg belt, and dust samples, a 1-log increase in environmental Salmonella burden resulted in a 51% (odds ratio, 1.51; P < 0.001) increase in the odds of an eggshell testing positive for Salmonella. In the final multifactorial model (not considering combined environment burden), only the Salmonella burden detected in the egg belt appeared to be conditionally associated with an eggshell testing positive for Salmonella (odds ratio, 1.43, P < 0.001).
FIG 3.
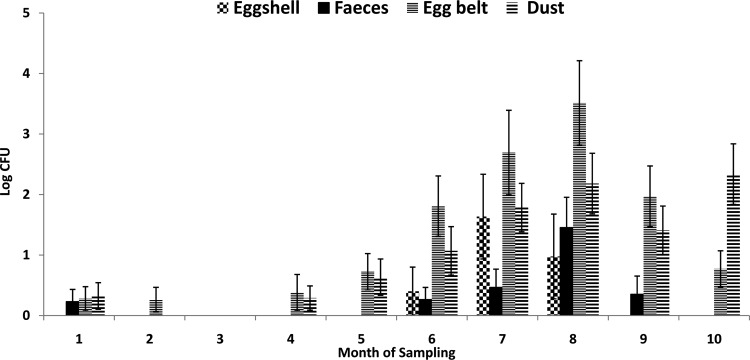
The load of Salmonella (average log CFU/qPCR) in feces, egg belt, dust, and eggshell samples over a period of 10 months. Error bars indicate standard errors.
Multiplex PCR to detect S. Typhimurium- and S. Infantis-positive samples.
The multiplex PCR, specific for S. Typhimurium and S. Infantis, was used to test various Salmonella serovars isolated from Australian layer farms (see Table S2 in the supplemental material). All tested Salmonella serovars were amplified by the InvA primers, which identify Salmonella spp. All three primer pairs specific to either S. Typhimurium or S. Infantis were able to correctly detect the respective serovar. The primers specific for S. Typhimurium or S. Infantis did not produce specific PCR amplification with other Salmonella serovars. However, there was an exception for Salmonella subsp. 1 serotype 4,5,12:-:-, which produced PCR amplification patterns similar to those of S. Typhimurium. In addition to the Salmonella-specific InvA amplicon, the following S. enterica subsp. enterica serovars also produced an additional single amplicon with one of the TSR or ISR primer pairs: S. Agona, S. Adelaide, S. Havana, S. Kiambu, S. Livingstone, S. Mbandaka, and S. Ohio (see Table S2). To determine the limit of detection of multiplex PCR, the fecal samples were spiked with a known concentration of Salmonella. The results indicated that the limit of detection by multiplex PCR was either 2,000 CFU/PCR or 400 CFU/μl extracted nucleic acid template.
The samples that were Salmonella positive by qPCR (n = 87) were all analyzed by S. Typhimurium and S. Infantis multiplex PCR. Multiplex PCR identified six potential S. Typhimurium- and no S. Infantis-positive samples. The latter result is in agreement with the serotyping results. Of the six samples identified as potentially S. Typhimurium positive by multiplex PCR, only one sample had S. Typhimurium (in addition to S. Oranienburg) isolated by microbial culturing. The other five samples identified as S. Typhimurium by multiplex PCR had either S. Worthington (n = 1) or S. Oranienburg (n = 3) isolated by culturing. These multiplex-positive samples all contained Salmonella at levels of >10,000 CFU/qPCR or 1,250 CFU/μl nucleic acid template. None of the four qPCR-positive samples that had S. Typhimurium isolated by culturing tested positive by the S. Typhimurium multiplex PCR assay.
DISCUSSION
The current study involved longitudinal and point-in-time surveys of Salmonella carriage and environmental contamination on two commercial cage layer farms. The initial prevalences of Salmonella (based on fecal sampling, n = 78) in flocks A and B were 26.9% and 39.7%, respectively.
In the longitudinal study of 40 weeks, the highest prevalence of Salmonella was detected in dust samples (42%), followed by in egg belt (28%), fecal (20%), and eggshell (4%) samples. The high prevalence of Salmonella in dust may result in an airborne spread of infection in the layer flock within the shed. It has been observed that S. Typhimurium is capable of surviving in aerosol form for long periods of time (21). A low dose of S. Typhimurium DT 104 infection (2 × 102 or 2 × 104 CFU per bird) resulted in increased Salmonella contamination of eggs (22). In the present study, the qPCR results indicated that the level of Salmonella in dust samples peaked up to 5 log CFU per qPCR, which may have resulted in the lateral spread of Salmonella in the flock. Hence, the presence of Salmonella in dust is a risk factor for the spread of infection in layer flocks.
Of the 140 fecal samples tested, 20% were reported as Salmonella positive. It was observed that there was higher variation in Salmonella contamination of feces than in that of the dust samples. This may be due to the increased frequency of the removal of feces from the systems compared to that of the dust (12). Fecal samples are believed to be better indicators of the infection status of flocks, whereas dust samples are more likely to indicate previous infection status (23).
The prevalence of Salmonella in both flocks A and B increased during the later stages of lay. There is no information available in the literature that clearly indicates the relationship between the stage of lay and degree of Salmonella shedding. In flock B, following 58 weeks of age, there was a substantial increase in the prevalence of Salmonella in all types of samples. During this period, a new flock was introduced into the same shed. This new flock was housed adjacent to the flock that was sampled in the current study. There is a possibility that the introduction of a new batch of birds into the same shed may have stressed the birds under investigation, resulting in increased shedding of Salmonella. However, further studies are essential to confirm these observational findings.
Flock A molted at the 67th week of lay. In the following week (week 68), it was observed that the shedding of Salmonella in feces increased and, subsequently, dust contamination also increased. As a result of molting, no eggs were obtained for Salmonella isolation in the 68th week. However, in the 72nd week, one eggshell was reported as Salmonella positive. Molting along with immunosuppression can alter gut microbiota and physiology, and these changes may influence the host-pathogen relationship (24). Holt (25) reported that induced molting resulted in higher shedding of Salmonella Enteritidis in feces and increased colonization of internal organs. The higher numbers of S. Enteritidis-positive eggs were produced within the first 5 weeks after molting (25).
Out of all eggs tested, 4% (20/521) of the eggshells were reported to be Salmonella positive. The serovars that were detected on the eggshells were the same as those detected from the farm environmental samples. However, all the egg internal contents were Salmonella negative. These findings are in agreement with our previous survey (6), in which all egg internal contents were Salmonella negative. The egg penetration experiment indicated that S. Worthington has a capacity to penetrate the eggshell but lacks the ability to survive in the egg internal contents (V. C. Gole, unpublished data), whereas S. Typhimurium has a capacity to penetrate and survive in egg internal contents at 20°C (26). In the present experiment, even though chickens were positive for S. Typhimurium, the egg internal contents were Salmonella negative. There is a lack of reliable information regarding the ability of S. Typhimurium to transmit vertically.
In the current study, of 20 Salmonella-positive eggshell samples, 18 were positive for S. Oranienburg, whereas one sample was positive for S. Worthington and another positive for S. Typhimurium. In Australia, egg-associated S. Oranienburg outbreaks have not been reported so far. However, in Germany, a large chocolate-related outbreak of this serovar was reported in 2005 (27).
Salmonella-positive fecal, egg belt, and dust samples were all unconditionally associated with eggshells testing positive for Salmonella. The odds of an eggshell testing positive for Salmonella were 91.8, 61.5, and 18.2 times higher when fecal, egg belt, and dust samples tested Salmonella positive. This clearly suggests that fecal contamination with Salmonella is the most important factor for the production of Salmonella-positive eggshells. On other hand, the qPCR results suggest that not only the qualitative score (positive or negative) but also the quantitative level of Salmonella in environmental samples is very important in order to have Salmonella-positive eggshells. A 1-log increase in the load of Salmonella detected in fecal, egg belt, and dust samples resulted in 35%, 43%, and 45% increases, respectively, in the odds of an eggshell testing positive for Salmonella. These findings are very important for developing management strategies for reducing the incidences of Salmonella-positive eggshells by decreasing the level of Salmonella in the environment of layer shed.
The prevalence with which Salmonella was detected in samples using qPCR was lower than traditional microbiological culturing, with 68% of known Salmonella positives being identified. Furthermore, the sample type also influenced the variation in the agreement between the culture- and qPCR-based detection methods. An almost-perfect agreement was reported between the two methods for identifying Salmonella-positive eggshell and egg belt samples, with only a moderate agreement observed when fecal and dust samples were investigated. The microbiological culture-based method involved the preenrichment of samples in BPW, followed by selective enrichment in RVS, while qPCR analysis was done on the preenriched samples only. This might explain the lower probability of detection by qPCR, especially if samples were contaminated with only low levels of Salmonella, as has been observed by others (28). Despite the traditional microbiological culture-based methods being more sensitive due to selection, the qPCR results did indicate that the probability of eggshell contamination was significantly increased with as little as a 10-fold increase in Salmonella levels within the shed environment. Therefore, qPCR does have potential as an initial rapid and high-throughput screening tool for identifying Salmonella in the environment.
In the present study, MLVA was used to investigate the relatedness of the different S. Typhimurium strains isolated from the two study flocks. As per the Australian coding system, strains isolated from flock A were distinct from and unrelated to the strains isolated from flock B. All the S. Typhimurium strains from flock B exhibited no allelic variation. In contrast to this, there was allelic variation in the strains isolated from flock A. However, as per the Australian MLVA coding system (29), this variation was not significant to consider them unrelated or distinct S. Typhimurium isolates. A quarterly report released from Institute of Medical and Veterinary Science (IMVS), Adelaide, Australia, indicated that S. Typhimurium strains responsible for human food poisoning cases exhibited similar MLVA patterns to those of the strains isolated from flocks A and B (30). Serotyping confirmed that S. Oranienburg was the most frequently reported serovar, followed by S. Typhimurium PT 9, S. Worthington, S. Agona, Salmonella subsp. 1 serotype 4, 5, 12:-:-, and Salmonella subsp. 1 serotype rough g,s,t:-.
Multiplex PCR assays were used to identify S. Typhimurium and S. Infantis from samples that had been identified as S. enterica positive by qPCR. All the samples were negative for S. Infantis, which is in agreement with the microbiological culture-based results. Of the qPCR-positive samples further analyzed by S. Typhimurium multiplex PCR, six potential S. Typhimurium positives were identified. Of these, only one sample had a positive S. Typhimurium strain isolated by culturing, while the others were positive for S. Oranienburg or S. Worthington. Four samples known to be positive for S. Typhimurium by culturing did not test positive by multiplex PCR. These results may be an indication of a mixed Salmonella infection within the sample and microbiological characterization based on a limited number of presumptive Salmonella-positive colonies. The limit of detection for multiplex PCR was 2,000 CFU/reaction, which may account for why no positive samples identified by culturing tested positive by multiplex PCR. This clearly suggests that culture-based methods are more sensitive and specific than multiplex PCR assays in characterizing S. Typhimurium. These findings are in agreement with previous experiment of Soumet et al. (31). They reported a poor sensitivity (107 CFU/ml) of multiplex PCR for the samples (obtained from poultry houses) preenriched in BPW and directly tested by multiplex PCR. Furthermore, when testing pure isolates of a range of Salmonella organisms, it was noted that Salmonella subsp. 1 serotype 4,5,12:-:- was amplified by all three S. Typhimurium primer pairs in multiplex PCR and hence was indistinguishable from S. Typhimurium. There is no previous information on the expected results for Salmonella subsp. 1 serotype 4,5,12:-:- using the multiplex PCR system described by Akiba et al. (19). In France, genomic analysis revealed that a nonmotile strain of S. enterica subsp. enterica with the antigenic formula 4,5,12:-:- is a nonmotile variant of S. Typhimurium and is responsible for egg-related food poisoning outbreaks (32). Within Australia, the present study is the first report of a nonmotile variant of S. Typhimurium in laying flocks. However, further genomic analysis is essential to reveal the similarity of this nonmotile variant with S. Typhimurium. In the future, such atypical Salmonella variants may emerge as a new challenge for the Australian layer industry.
In conclusion, the Salmonella-positive samples from feces, egg belt, and dust were significant predictors of eggshell contamination. A 1-log CFU increase in the level of Salmonella within the layer shed environment significantly increased the incidence of eggshell contamination. The flocks sampled during this study showed variation in Salmonella shedding over time. Stress induced by molting or the introduction of a new batch of birds within the shed may have resulted in higher shedding of Salmonella in the environment; however, further controlled studies are required to prove these observational findings. The results of this study might be helpful for determining the risks of having Salmonella-contaminated eggshells and also for developing strategies for risk management programs to control Salmonella.
Supplementary Material
ACKNOWLEDGMENTS
This research was conducted within the Poultry CRC, established and supported under the Australian Government's Cooperative Research Centres program.
Vaibhav Gole is an International Postgraduate Research Scholarship recipient.
We thank Amanda Kidsley, Geraldine Laven-Law, and Nigel Percy for their valuable technical help during this study.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00816-14.
REFERENCES
- 1.The OzFoodNet Working Group. 2012. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet Network, 2010. Commun. Dis. Intell. Q Rep. 36:E213–E241. [PubMed] [Google Scholar]
- 2.Vought KJ, Tatini SR. 1998. Salmonella enteritidis contamination of ice cream associated with a 1994 multistate outbreak. J. Food Prot. 61:5–10. [DOI] [PubMed] [Google Scholar]
- 3.Kapperud G, Gustavsen S, Hellesnes I, Hansen AH, Lassen J, Hirn J, Jahkola M, Montenegro MA, Helmuth R. 1990. Outbreak of Salmonella Typhimurium infection traced to contaminated chocolate and caused by a strain lacking the 60-megadalton virulence plasmid. J. Clin. Microbiol. 28:2597–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EFSA. 2010. Scientific opinion on a quantitative estimation of the public health impact of setting a new target for the reduction of Salmonella in laying hens. EFSA J. 8:1546. 10.2903/j.efsa.2010.1546. [DOI] [Google Scholar]
- 5.Sergeant E, Grimes TM, Jackson C, Baldock FC, Whan IF. 2003. Salmonella Enteritidis surveillance and response options: for the Australian egg industry: a report for the Rural Industries Research and Development Corporation. Rural Industries Research and Development Corporation, Barton, Australian Capital Territory, Australia. [Google Scholar]
- 6.Gole VC, Chousalkar KK, Roberts JR. 2013. Survey of Enterobacteriaceae contamination of table eggs collected from layer flocks in Australia. Int. J. Food Microbiol. 164:161–165. 10.1016/j.ijfoodmicro.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 7.van de Giessen AW, Ament AJ, Notermans SH. 1994. Intervention strategies for Salmonella Enteritidis in poultry flocks: a basic approach. Int. J. Food Microbiol. 21:145–154. 10.1016/0168-1605(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 8.Davies R, Breslin M. 2003. Observations on Salmonella contamination of commercial laying farms before and after cleaning and disinfection. Vet. Rec. 152:283–287. 10.1136/vr.152.10.283. [DOI] [PubMed] [Google Scholar]
- 9.Gradel K, Sayers A, Davies R. 2004. Surface disinfection tests with Salmonella and a putative indicator bacterium, mimicking worst-case scenarios in poultry houses. Poult. Sci. 83:1636–1643. 10.1093/ps/83.10.1636. [DOI] [PubMed] [Google Scholar]
- 10.Davies R, Breslin M. 2001. Environmental contamination and detection of Salmonella enterica serovar Enteritidis in laying flocks. Vet. Rec. 149:699–704. [PubMed] [Google Scholar]
- 11.Chemaly M, Huneau-Salaün A, Labbe A, Houdayer C, Petetin I, Fravalo P. 2009. Isolation of Salmonella enterica in laying-hen flocks and assessment of eggshell contamination in France. J. Food Prot. 72:2071–2077. [DOI] [PubMed] [Google Scholar]
- 12.Wales A, Breslin M, Carter B, Sayers R, Davies R. 2007. A longitudinal study of environmental Salmonella contamination in caged and free-range layer flocks. Avian Pathol. 36:187–197. 10.1080/03079450701338755. [DOI] [PubMed] [Google Scholar]
- 13.Davison S, Benson CE, Henzler DJ, Eckroade RJ. 1999. Field observations with Salmonella Enteritidis bacterins. Avian Dis. 43:664–669. 10.2307/1592735. [DOI] [PubMed] [Google Scholar]
- 14.Kinde H, Castellan DM, Kerr D, Campbell J, Breitmeyer R, Ardans A. 2005. Longitudinal monitoring of two commercial layer flocks and their environments for Salmonella enterica serovar Enteritidis and other salmonellae. Avian Dis. 49:189–194. 10.1637/7228-062704R. [DOI] [PubMed] [Google Scholar]
- 15.Wales AD, Davies RH. 2011. A critical review of Salmonella Typhimurium infection in laying hens. Avian Pathol. 40:429–436. 10.1080/03079457.2011.606799. [DOI] [PubMed] [Google Scholar]
- 16.Ross IL, Davos DE, Mwanri L, Raupach J, Heuzenrouder MW. 2011. MLVA and phage typing as complementary tools in the epidemiological investigation of Salmonella enterica serovar Typhimurium clusters. Curr. Microbiol. 62:1034–1038. 10.1007/s00284-010-9820-1. [DOI] [PubMed] [Google Scholar]
- 17.Torok VA, Ophel-Keller K, Loo M, Hughes RJ. 2008. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 74:783–791. 10.1128/AEM.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stirling GR, Griffin D, Ophel-Keller K, McKay A, Hartley D, Currar J, Stirling AM, Monsour C, Winch J, Hardie B. 2004. Combining an initial risk assessment process with DNA assays to improve prediction of soilborne diseases caused by root-knot nematode (Meloidogyne spp.) and Fusarium oxysporum f. sp. lycopersici in the Queensland tomato industry. Australas. Plant Pathol. 33:285–293. 10.1071/AP04004. [DOI] [Google Scholar]
- 19.Akiba M, Kusumoto M, Iwata T. 2011. Rapid identification of Salmonella enterica serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by multiplex PCR. J. Microbiol. Methods 85:9–15. 10.1016/j.mimet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 20.StataCorp. 2011. Stata Statistical Software: release 12.1. StataCorp LP, College Station, TX. [Google Scholar]
- 21.McDermid AS, Lever MS. 1996. Survival of Salmonella Enteritidis PT4 and Salm. Typhimurium Swindon in aerosols. Lett. Appl. Microbiol. 23:107–109. 10.1111/j.1472-765X.1996.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 22.Leach SA, Williams A, Davies AC, Wilson J, Marsh PD, Humphrey TJ. 1999. Aerosol route enhances the contamination of intact eggs and muscle of experimentally infected laying hens by Salmonella Typhimurium DT104. FEMS Microbiol. Lett. 171:203–207. 10.1111/j.1574-6968.1999.tb13433.x. [DOI] [PubMed] [Google Scholar]
- 23.Carrique-Mas JJ, Davies RH. 2008. Salmonella Enteritidis in commercial layer flocks in Europe: legislative background, on-farm sampling and main challenges. Rev. Bras. Cienc. Avic. 10:1–9. 10.1590/S1516-635X2008000100001. [DOI] [Google Scholar]
- 24.Golden NJ, Marks HH, Coleman ME, Schroeder CM, Bauer NE, Jr, Schlosser WD. 2008. Review of induced molting by feed removal and contamination of eggs with Salmonella enterica serovar Enteritidis. Vet. Microbiol. 131:215–228. 10.1016/j.vetmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Holt P. 2003. Molting and Salmonella enterica serovar Enteritidis infection: the problem and some solutions. Poult. Sci. 82:1008–1010. 10.1093/ps/82.6.1008. [DOI] [PubMed] [Google Scholar]
- 26.Gole VC, Chousalkar KK, Roberts JR, Sexton M, May D, Tan J, Kiermeier A. 2014. Effect of egg washing and correlation between eggshell characteristics and egg penetration by Various Salmonella Typhimurium strains. PLoS One 9:e90987. 10.1371/journal.pone.0090987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werber D, Dreesman J, Feil F, van Treeck U, Fell G, Ethelberg S, Hauri AM, Roggentin P, Prager R, Fisher I, Behnke SC, Bartelt E, Weise E, Ellis A, Siitonen A, Andersson Y, Tschäpe H, Kramer MH, Ammon A. 2005. International outbreak of Salmonella Oranienburg due to German chocolate. BMC Infect. Dis. 5:7. 10.1186/1471-2334-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen AN, Nielsen LR, Baggesen DL. 2013. Use of real-time PCR on faecal samples for detection of sub-clinical Salmonella infection in cattle did not improve the detection sensitivity compared to conventional bacteriology. Vet. Microbiol. 163:373–377. 10.1016/j.vetmic.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Heuzenroeder MW, Ross IL, Hocking H, Davos D, Young CC, Morgan G. 2013. An integrated typing service for the surveillance of Salmonella in chickens. Rural Industries Research and Development Corporation, Barton, Australian Capital Territory, Australia. [Google Scholar]
- 30.Australian Salmonella Reference Centre. 2013. A quarterly report for January–March 2013, p 1–16 Institute of Medical and Veterinary Sciences, SA Pathology, Adelaide, Australia. [Google Scholar]
- 31.Soumet C, Ermel G, Rose N, Rose V, Drouin P, Salvat G, Colin P. 1999. Evaluation of a Multiplex PCR assay for simultaneous identification of Salmonella sp., Salmonella Enteritidis and Salmonella Typhimurium from environmental swabs of poultry houses. Lett. Appl. Microbiol. 28:113–117. 10.1046/j.1365-2672.1999.00488.x. [DOI] [PubMed] [Google Scholar]
- 32.Le Hello S, Brisabois A, Accou-Demartin M, Josse A, Marault M, Francart S, Da Silva NJ, Weill FX. 2012. Foodborne outbreak and nonmotile Salmonella enterica variant, France. Emerg. Infect. Dis. 18:132–134. 10.3201/eid1801.110450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.