Abstract
Clostridium difficile is a well-known nosocomial infectious pathogen. Research on C. difficile infection has primarily focused on strains such as the hypervirulent PCR ribotype 027 (sequence type 1 [ST1]) emerging in Europe and North America. However, other new emerging ribotypes in some countries have attracted attention, such as PCR ribotype 17 (ST37) in Asia and Latin America. We collected 70 strains and sequenced their toxin genes, tcdA and tcdB. Multilocus sequence typing (MLST) was used to study their population structure. In addition, tcdA and/or tcdB sequences of 25 other isolates were obtained from GenBank. Single nucleotide polymorphisms (SNPs) were identified and analyzed. Phylogenetic analyses were performed to study toxin gene evolution. All tcdA and tcdB sequences were divided into 1 of 16 types (denoted A01 to -16 and B01 to -16, respectively). Hypervirulent strain RT027 is A13B12, and RT078 is A14B10, whereas the newly epidemic strain RT017 is A15B13. SNP analysis suggests the possibility of recombination in tcdB, perhaps through horizontal gene transfer. SNPs were also found in the sequences corresponding to the PCR primers widely used for toxin detection. Our study shows that ST037 shares a few genotypic features in its tcdA and tcdB genes with some known hypervirulent strains, indicating that they fall into a unique clade. Our findings can be used to map the relationships among C. difficile strains more finely than can be done with less sensitive methods, such as toxinotyping or even MLST, to reveal their inherent epidemiological characteristics.
INTRODUCTION
Clostridium difficile is a nosocomial bacterial pathogen that causes antibiotic-associated diarrhea mediated by cellular exotoxins secreted into the intestine during bacterial growth (1, 2). In the past decade, the mortality from C. difficile infection (CDI) has increased from 6% to 13.5% overall among older patients (3). Research on CDI has primarily focused on strains such as the hypervirulent PCR ribotype 027, multilocus sequence type 1 (RT027, ST1), emerging in Europe and North America, which produces two major toxins, A and B, encoded by the genes tcdA and tcdB in the pathogenicity locus (PaLoc) (4, 5). In recent years, however, the epidemiology of CDI has changed dramatically, since other emerging PCR ribotypes have become prevalent (6–13), and several pathogenic A-negative B-positive (A− B+) strains (which produce toxin B, but not toxin A) have appeared in Asia and Latin America (8–12, 14, 15). Hence, it has become a matter of urgency to understand the new, complex pattern of A− B+ C. difficile strain variants and to reveal their relationships to other epidemic genotypes.
Recently, a particular variant strain of C. difficile, ST37, which has been ribotyped as RT017 and produces toxin B only, has attracted increasing attention (11, 12, 16). This type of strain shows increased resistance to clindamycin and erythromycin, with concomitant greater risk to inpatient health (13, 17, 18). Currently, the literature on CDI caused by A− B+ strains is limited, and there is some confusion over the correct identification of specific isolates. For example, the apparently high isolation rate of A− B+ strains in Asia and Latin America may reflect mismatching of PCR primers as a result of C. difficile polymorphisms (19). Nevertheless, it seems clear that C. difficile RT017 is prevalent in Asia and China, but the reasons for this distribution, as well as its relation to the highly virulent RT027 strain in Europe and North America, are still uncertain.
To address this situation, we collected C. difficile strains from five different geographical areas in China. The entire tcdA and tcdB genomic regions were sequenced and single nucleotide polymorphism (SNP) analysis was conducted (i) to provide information on genomic diversity and to aid PCR primer design for later epidemiological studies, (ii) to reveal the potential linkage between RT027 and RT017, and (iii) to throw new light on the widespread occurrence of RT017 in China. Our data also provide reference data for tcdA and tcdB sequences from the sampled regions, which will enable transmission patterns to be determined and allow forecasting of future distribution trends in these areas of China.
MATERIALS AND METHODS
Strain isolation and DNA extraction.
Sixty-six of the 70 C. difficile strains analyzed derived from five provinces and municipalities of China, including Beijing (21), Shanghai (15), Shandong (11), Henan (7), and Guangdong (12). The strains in each province were collected from one general hospital and randomly selected regardless of the years of isolation to include as many strains as possible. The remaining four strains were isolated from the United States, the United Kingdom, Japan, and France (see Table S1 in the supplemental material). Stool specimens from diarrhea patients were collected using Transwabs (MW&E Ltd., Wiltshire, England), and then cultured on cycloserine-cefoxitin-fructose agar (CCFA) with 5% egg yolk. Colonies that demonstrated a typical morphology (flat, yellow, ground-glass appearance) and odor on the CCFA and in Gram staining were Gram-positive bacilli with subterminal spores, and those that yielded positive results in response to the commercially available latex agglutination test (Oxoid, Ltd., Basingstoke, United Kingdom) were identified as C. difficile. Isolates that were not confirmed by these methods were further identified via API 20A (Bio Mérieux, France) and 16S rRNA gene sequencing (with the primer set 5′-GGAGGCAGCAGTGGGGAATA-3′ [forward] and 5′-TGACGGGCGGTGTGTACAAG-3′ [reverse]) and glucose dehydrogenase (GDH) gene amplification and sequencing (6) (with the primer set 5′-TTCCTAATTTAGCAGCAGCTTC-3′ [forward] and 5′-GTCTTGGATGGTTGATGAGTAC-3′ [reverse]).
Characterization of toxin genes by PCR and multilocus sequence typing.
The toxigenic property of each C. difficile isolate was determined by characterization of the tcdA and tcdB genes. A PCR assay for tcdB was performed using primers NK104 and NK105, which resulted in a 203-bp amplicon for a tcdB-positive strain (9). The tcdA gene was detected using primers tcdA-F and tcdA-R, which yielded a 369-bp amplicon for tcdA-positive strains and a 110-bp amplicon for tcdA-negative strains (20). All 70 strains were characterized by the MLST method with seven housekeeping genes (21), and sequences were submitted to the Clostridium difficile multilocus sequence typing (MLST) database (http://pubmlst.org/cdifficile) to acquire a sequence type (ST). Thirty-five strains were part of the 104 strains used in our previous MLST study (16). A minimum spanning tree was also constructed to exhibit the population structure of Chinese strains using the categorical data for MLST via BioNumerics v4.0 software (Applied Maths BVBA, Belgium).
Sequencing of tcdA and tcdB.
Using a primer-walking method, nine pairs of primers for tcdA and eight pairs for tcdB were designed via Primer Premier 5.0 software to cover the whole length of each gene (8,133 bp and 7,101 bp, respectively) in overlapping segments. The sequence of C. difficile 630 (downloaded from GenBank) was used as a reference (see Table S2 in the supplemental material). Amplicon sizes were from 700 to 1,800 bp. PCR assays were performed in a total volume of 50 μl, containing 3 μl of chromosomal DNA (approximately 100 ng), 25 μl of premixed Hot Start Taq (TaKaRa, Japan), 20 μl of molecular biology-grade water, and 1 μl of a 25-μM concentration of each primer. The PCR program comprised a 5-min predenaturation at 94°C, followed by 34 cycles of 30 s of denaturation at 94°C, 30 s of annealing at an individually specified temperature (depending on the primers used), and 120 s of extension at 72°C, with a 5-min final extension at 72°C. The crude PCR products were purified and then sequenced with PCR forward and reverse primers using an ABI-PRISM BigDye Terminator sequencing kit V3.1 (PE Biosystems, USA) on a 3730 XL DNA analyzer (Applied Biosystems, USA).
Analysis of sequence data.
Sequences for each gene were assembled into complete sequences via DNASTAR Lasergene v7.1 software. The initiation and stop codon positions were recognized by aligning to the tcdA and tcdB sequences of C. difficile 630. In addition, 15 tcdA and 25 tcdB sequences were obtained from GenBank. Multiple sequence alignments were performed using MUSCLE v3.6, and phylogenic trees were constructed using the neighbor-joining (NJ) method via MEGA v5 software. The cytotoxin gene from Clostridium sordellii (GenBank accession number X82638) was used as an outgroup. Using PERL scripts, SNPs were obtained from the multiple sequence alignments by comparison with the corresponding sequences of C. difficile strain VPI 10463. The numbers of SNPs in different functional domains were calculated based on the ABCD models of both TcdA and TcdB proteins (22, 23).
RESULTS
Overview of representative sequenced strains.
Sixty-six Chinese strains used in this study were isolated from the 1980s to 2012, and 42 of them (63.6%) were from inpatients. All 70 individual C. difficile isolates include the sequence types 1 (1 strain) 2 (11 strains), 3 (12 strains), 8 (2 strains), 35 (13 strains), 37 (18 strains), 46 (1 strain), 53 (1 strain), 54 (4 strains), 55 (2 strains), 92 (1 strain), 99 (1 strain), 102 (1 strain), 129 (1 strain) and 221 (1 strain). Their tcdA and tcdB genes were successfully amplified and sequenced. In addition, the tcdA and tcdB sequences of 25 further C. difficile isolates were distilled from GenBank (see Table S1 in the supplemental material), for 10 of which only one of the two genes was available and in other cases the ST types were uncertain. In total, 25 A− B+, 63 A+ B+, and 7 strains of unknown toxin status were analyzed. All strains were divided into 16 STs by MLST analysis, of which ST11 was the only type not found in China in this study (see Table S1 in the supplemental material). Unlike in Europe and the United States, in China the predominant type in our sampled strains is ST37 (which is quite different from other STs) followed by ST35. All A− B+ strains obtained in our study are ST37, and tcdA from these strains contains partially truncated sequences.
tcdA sequence analysis.
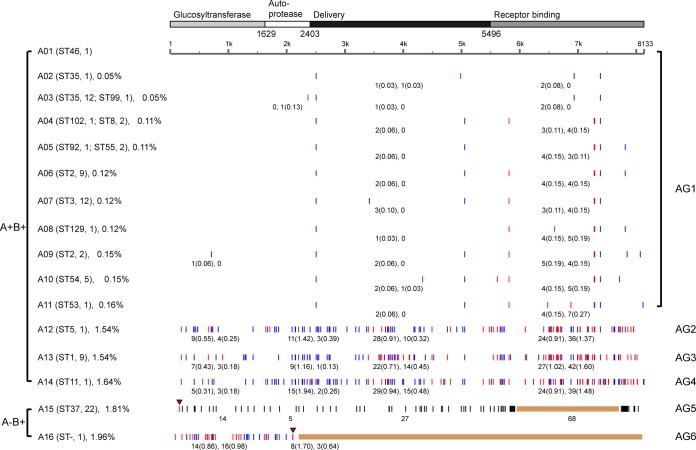
In total, 16 tcdA variants were obtained from 85 sequences, of which 10 sequence types (A02, A03, A04, A05, A06, A07, A08, A09, A11, and A12) from Chinese strains were new compared to those in GenBank; multiple SNPs were found only in A12 (Fig. 1; also see Table S3 in the supplemental material). There were 14 different sequence types (A01 to A14) for tcdA from A+ B+ strains, which clustered into four groups (AG1 to AG4). AG1 is the largest group. It contained 11 sequence types in which the average number of SNPs was 9.3, with mutation rates all below 0.2% (Fig. 1). In the other three A+ B+ sequence groups (AG2 to AG4), a large number of SNPs distinguished the respective strains, which had mutation rates of about 1.5%. The C. difficile isolate containing sequence type A12 (AG2) is a Chinese A+ B+ strain with numerous tcdA SNPs distinguishing it from other Chinese A+ B+ strains. Strains with A13 (AG3) and A14 (AG4) existed only in Europe and North America. A detailed analysis of functional domains encoded by tcdA revealed that mutations mainly occur in the receptor-binding domain, and largely consist of nonsynonymous SNPs. The numbers and rates of nonsynonymous SNPs in the receptor-binding domain are significantly higher than in the other three domains, indicating that this region is undergoing rapid evolution.
FIG 1.
Identification of single nucleotide polymorphisms (SNPs) in tcdA sequences using VPI 10463 as reference. The coding regions of the gene are divided into four domains, glucosyltransferase, autoprotease, delivery domain, and receptor-binding domain, according to the molecular structure of the toxin. STs and numbers of strains are given in brackets following the toxin gene sequence types, and ST- indicates unknown ST. The rates of SNPs in each tcdA type are given in brackets after the type name on the left; the names of groups are marked on the right. Nonsynonymous SNPs (red lines) and synonymous SNPs (blue lines) are identified. The number of SNPs in each domain is marked under the lines, with the SNP rates shown in the brackets (nonsynonymous rate and synonymous rate). In A− B+ strains, the mutant stop codons of tcdA are marked by red triangles and deletions are shown as brown bars. In A15, the SNPs downstream of the stop codon are represented by black lines.
Two variant gene types (A15 and A16), with truncated or deleted sequences and mutation rates near 2.0% in tcdA, were found in A− B+ strains and assigned to groups AG5 and AG6, respectively. A16 was an unusual sequence from strain 8864 that was only 2,091 bp long. Nonsense mutations were found in both groups, but compared to A16, the premature stop codon in A15 appeared much earlier, along with an approximately 6-kb nonfunctional region containing a large number of mutations. All 22 ST37 strains contained A15-type tcdA genes.
tcdB sequence analysis.
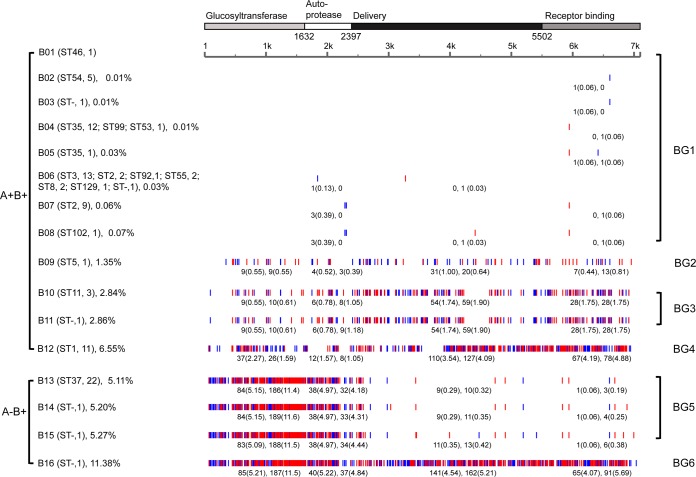
We obtained 16 tcdB variants from 95 sequences, of which five sequence types (B04, B05, B07, B08, and B09) were new compared to those in GenBank. B09 was derived from the same Chinese A+ B+ strain that contained the highly mutated tcdA type A12 and also contained multiple SNPs in tcdB (Fig. 2; also see Table S4 in the supplemental material). There were 12 different tcdB sequence types (B01 to B12) in the A+ B+ strains, which are clustered into four groups (BG1 to BG4). BG1 was the largest group. It contained eight sequence types (B01 to B08), in which there were on average 2.4 mutations per sequence; all mutation rates were below 0.1%. The remaining four sequence types (B09 to B12) were divided into the other three A+ B+ groups (BG2 to BG4) and showed mutation rates of about 1.35%, 2.85%, and 6.55%, respectively (Fig. 2). While AG2 to AG4 shared a number of mutations with each other, BG2 to BG4, which comprised many of the same strains found in AG2 to AG4, had far more diversity in their SNPs. BG2 possessed fewer mutations than BG3, which in turn had far fewer SNPs than BG4. Most SNPs, the majority of which in these groups were nonsynonymous, were concentrated in the delivery domain and receptor-binding domain.
FIG 2.
Identification of single nucleotide polymorphisms (SNPs) in tcdB sequences using VPI 10463 as reference. The coding regions of the gene are shown as four domains, glucosyltransferase, autoprotease, delivery domain, and receptor-binding domain, according to the molecular structure of the toxin. SNP rates and nonsynonymous and synonymous SNPs are indicated as in Fig. 2.
Four tcdB sequence types (B13 to B16) found in the A− B+ strains were 7,104 bp in length, which differed from the 7,101 bp seen in most A+ B+ strains. B13, B14, and B15 clustered into the same group (BG5), with an average mutation rate above 5.0% and with SNPs mainly distributed in the first two domains, which have glucosyltransferase and autoprotease functions. All 22 ST37 strains contained the B13 tcdB sequence type. B14 and B15 differed only slightly from B13.
Interestingly, BG6, comprising sequence type B16 only, contained SNPs throughout its length and had the highest mutation rate (11.38%). In its first two domains, BG6 possessed mutations that were similar to those in BG5. However, BG6 mutations in the third and fourth domains, which have delivery and receptor-banding functions, resembled mutations found in BG4, particularly in the fourth domain. Although the first two domains in BG5 (A− B+ [RT017]) were very similar to those of BG6 (A− B+ [8864]), its last two domains resembled those of BG1 (A+ B+ [common]). In another example of this phenomenon, the last 3 kb of the tcdB variant defining BG4 (A13B12 [RT027]) also showed a high degree of identity to the corresponding region of BG6 (A− B+ [8864]), but the first 4 kb of BG4 was more similar to that of BG3 (A+ B+ [RT078]), which is 203 SNPs (95% similarity) with BG3 versus 420 SNPs (90% similarity) with BG6. Taken together, this analysis suggests the possibility of recombination in tcdB, perhaps through horizontal gene transfer (HGT) of the toxin genes or the whole PaLoc. This may be significant in the context of the evolution of hypervirulent strains.
Molecular characteristics of tcdA and tcdB sequence types in epidemic strains.
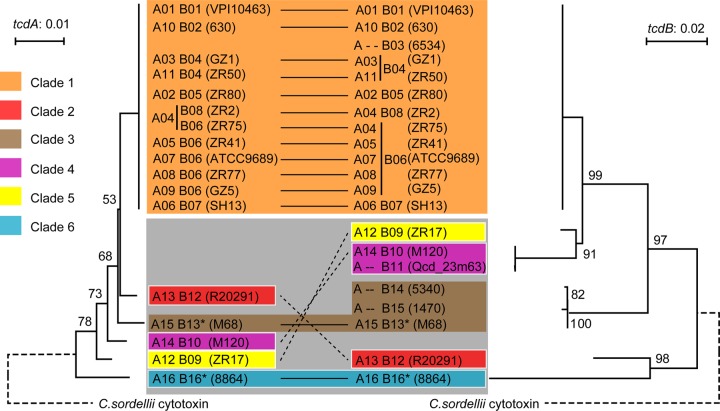
Based on sequence similarity and phylogenetic analysis, the 16 tcdA types and 16 tcdB types were clustered into six clades, in keeping with the six groups of both tcdA and tcdB variants (Fig. 3). The epidemic strains can be analyzed with respect to their toxin gene sequence type. Thus, the hypervirulent strain RT027 (ST1), which presents as A+ B+ and is binary toxin positive, is included in clade 2, with toxin gene sequence type A13B12. Epidemic strain RT078 (ST11), which is also A+ B+ and binary toxin positive, is a member of clade 4, with toxin gene sequence type A14B10. An examination of the phylogenetic relationship between RT027 and RT078, based on their tcdA and tcdB sequences, showed that the toxin genes in these strains are evolving separately.
FIG 3.
Phylogenetic analyses of tcdA and tcdB. The Clostridium sordellii cytotoxin gene (GenBank accession number X82638) was used to root the phylogenetic trees of tcdA (left) and tcdB (right). Clades are represented by different colors, and A− B+ clades are marked with asterisks. Clade 1, as a new lineage, is distinguished from the older cluster, comprising clade 2 to clade 6, which is marked by a gray square.
Epidemic strain RT017 (ST37), which presents as A− B+ and has a high recent isolation rate in Asia and Latin America, forms part of clade 3, with toxin gene sequence type A15B13. Multiple nonsynonymous mutations downstream of the glucosyltransferase domain were found in its tcdB gene, together with a nonsynonymous mutation at nucleotide position 139 (i.e., C139T) in its tcdA gene, generating a premature stop codon in the TcdA amino acid sequence (Q47X). Currently, the major strains in China are ST37, but ST37 isolates have also been recorded in other countries, including Ireland, Belgium, and the United States. Furthermore, the sequences of both tcdA and tcdB from all 22 ST37 strains isolated worldwide since the 1980s are identical, suggesting that ST37 may be a rapidly spreading sequence type. Whole-genome analysis also showed a high degree of similarity between ST37 strains (24, 25).
Mutations in the genomic sequences used for toxin detection.
In a number of tcdA and tcdB sequence types, SNPs were found in the sequences corresponding to the PCR primers widely used for toxin detection. For example, at the loci corresponding to primers NK104 and NK105 (19), which are commonly used for tcdB detection, there are between one and three SNPs in B09 to B15 and six in B16. At the loci of a frequently used primer set for tcdA detection, i.e., NK2 and NK3 (19), there are one or two SNPs in A02, A09, A12, A13, A14, and A16. Such a high level of polymorphisms at genome loci that are important for tcdA and tcdB identification might cause false-negative results in toxin gene detection and typing by PCR.
DISCUSSION
Toxin A and B mediate the main clinical symptoms of CDI, consistent with their coding genes within the same genomic region, and are expressed together (26, 27). However, the newly prevalent cases caused by A− B+ strains appear to challenge the requirement for both toxins in the etiology of this disease. According to the size of deletions at the 3′ end of tcdA, A− B+ strains are divided into deletion and truncated forms (9). Yet, our previous study demonstrated significant genetic diversity in tcdA and tcdB, consistent with the results of toxinotyping based on the restriction fragment length polymorphism (RFLP)-PCR method (19, 28). However, the simple classification of RFLP-PCR failed to explain more details about the epidemiological relevance and phylogenetic relationships between strains with different types of toxin genes, while the recently increased ST37 (RT017 [A− B+]) contribution (48%) for CDI cases and its epidemic potentiality in Asia (11, 16) make it urgent to build a more finely described pattern to reveal the intrinsic epidemic characteristics. In the present study, we introduce a novel approach that is more discerning and accurate; for example, 13 ST35 and 11 ST2 strains were divided into 2 groups (A03B04 and A02B05 and A06B07 and A09B06, respectively).
We also uncovered details of the genetic variation of the toxin genes. For example, ST37 was characterized as A15B13. Multiple nonsynonymous mutations were found downstream of the glucosyltransferase domain in B13, in line with previous findings (29), and a nonsynonymous mutation at nucleotide 139 in A15 introduces a premature stop codon, which is presumably responsible for the negative toxin A-specific immunoassay in these strains. For two A+ B+ strains, ZR75 was described as A04B06, and ZR2 as A04B08. Their sequence types are ST8 and ST102, respectively. Their combined toxin gene types are different, which is in line with MLST, but more details were described. They are of the same tcdA type (A04) and their tcdB types (B06 and B08) are both in BG1, which shows the sequence similarity of their toxin genes and the connection of their virulence.
Clade 2 was representative of RT027 strains, including the “historic” nonepidemic RT027 (CD196) and the hypervirulent RT027 (R20291). These strains have characteristic polymorphisms that distinguish them from other types. Their tcdB genes both belong to BG4. In the receptor-binding domain, BG4 has a high similarity with BG6, which is from an A− B+ strain 8864 with exceptional pathogenicity (30, 31). And BG5, including the tcdB gene from ST37, has a high similarity with BG6 in the glucosyltransferase and autoprotease domains. These sequence similarities with the same confirmed highly pathogenic type indicate that BG6 may be the common ancestor of BG4 and BG5 and might be associated with the virulence characteristics of RT027 and ST37 strains, although other factors must also be involved (32, 33). For example, CD196 was not of clinical significance when it was isolated in 1985, despite having the same tcdA and tcdB sequences as R20291. Clades 2 to 6 are regarded as an “old” cluster, with clade 1 representing a relatively new cluster. So, we hypothesize that A− B+ strains evolved from an ancestral A+ B+ strain after deletion of the 3′ region of the tcdA gene, consistent with that of RT017 (ST37) strains shown by whole-genome analysis to occupy a distinct lineage (24).
All of the ST37 isolates from different regions in our study contained the same sequences of tcdA and tcdB, consistent with its low genetic diversity and the possible international spread. As an old, virulent A− B+ strain (30, 31), 8864 (B16) has a variant 5′ end similar to those of common A− B+ strains (B13 to B15) and a variant 3′ end similar to that of the hypervirulent A+ B+ RT027 strain (B12). This unusual arrangement may reflect a connection between RT027 and A− B+ strains and highlights the potential epidemic threat of an A− B+ strain in China. Another possibility is that the increased pathogenicity of toxin B enhances the virulence and spread of A− B+ strains. Some of the ST37 strains derive from the 1980s. The fact that their toxin genes have not changed in 30 years is quite remarkable and may be due to the inherent success of this strain.
This is the first systematic analysis of tcdA and tcdB in C. difficile from different regions of China, and it provides us a complete map of these two major toxin genes. This accurate map provides us not only the possible typing sites for nucleotide detection, but also the potential connection of prevalent strains among the world, such as the ST37 threat in China. Together with other genetic typing tools, such as MLST, the result could provide more accurate toxinic orientation, relationship maps, and interpretive classifications, which will facilitate epidemic risk evaluation beyond different period, population, and geographical spans to specified strains and improve CDI prevention and control.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 81101218, 81201322, and 81301402).
We thank Yuanyuan Zhang, Haiyin Wang, and Fengjuan Li from the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, for the technical support, and we thank Haihui Huang from Huashan Hospital, Shanghai, China, for generously providing strains.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03487-13.
REFERENCES
- 1.Heinlen L, Ballard JD. 2010. Clostridium difficile infection. Am. J. Med. Sci. 340:247–252. 10.1097/MAJ.0b013e3181e939d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536. 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 3.Karas JA, Enoch DA, Aliyu SH. 2010. A review of mortality due to Clostridium difficile infection. J. Infect. 61:1–8. 10.1016/j.jinf.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449. 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 5.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 6.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, Group ES. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 7.Clements AC, Magalhaes RJ, Tatem AJ, Paterson DL, Riley TV. 2010. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect. Dis. 10:395–404. 10.1016/S1473-3099(10)70080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549. 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y, Wasito EB. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbut F, Lalande V, Burghoffer B, Thien HV, Grimprel E, Petit JC. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079–2083. 10.1128/JCM.40.6.2079-2083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkey PM, Marriott C, Liu WE, Jian ZJ, Gao Q, Ling TK, Chow V, So E, Chan R, Hardy K, Xu L, Manzoor S. 2013. Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an underrecognized problem in Asia? J. Clin. Microbiol. 51:3308–3313. 10.1128/JCM.00587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins DA, Hawkey PM, Riley TV. 2013. Epidemiology of Clostridium difficile infection in Asia. Antimicrob. Resist. Infect. Control. 2:21. 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CH, Brazier JS, Lumelsky G, Kuijper EJ. 2009. Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin. Microbiol. Infect. 15:1080–1086. 10.1111/j.1469-0691.2009.02759.x. [DOI] [PubMed] [Google Scholar]
- 14.Rupnik M, Kato N, Grabnar M, Kato H. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 41:1118–1125. 10.1128/JCM.41.3.1118-1125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerding DN. 2010. Global epidemiology of Clostridium difficile infection in 2010. Infect. Control. Hosp. Epidemiol. 31(Suppl 1):S32–S34. 10.1086/655998. [DOI] [PubMed] [Google Scholar]
- 16.Yan Q, Zhang J, Chen C, Zhou H, Du P, Cui Z, Cen R, Liu L, Li W, Cao B, Lu J, Cheng Y. 2013. Multilocus sequence typing (MLST) analysis of 104 Clostridium difficile strains isolated from China. Epidemiol. Infect. 141:195–199. 10.1017/S0950268812000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, Weintraub A, Fang H, Nord CE. 2009. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 34:516–522. 10.1016/j.ijantimicag.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 18.van den Berg RJ, Claas EC, Oyib DH, Klaassen CH, Dijkshoorn L, Brazier JS, Kuijper EJ. 2004. Characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping. J. Clin. Microbiol. 42:1035–1041. 10.1128/JCM.42.3.1035-1041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Du P, Chen C, Yan S, Jia H, Wang J, Yan Q, Feng H, Lu J. 2011. Toxin A-negative, toxin B-positive Clostridium difficile infection diagnosed by polymerase chain reaction. Infect. Control. Hosp. Epidemiol. 32:520–522. 10.1086/659959. [DOI] [PubMed] [Google Scholar]
- 20.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, Pons JL. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 42:5710–5714. 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48:770–778. 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jank T, Aktories K. 2008. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 16:222–229. 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. 2010. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc. Natl. Acad. Sci. U. S. A. 107:13467–13472. 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107:7527–7532. 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297–7305. 10.1128/JB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263. 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 28.Rupnik M. 2010. Clostridium difficile toxinotyping. Methods Mol. Biol. 646:67–76. 10.1007/978-1-60327-365-7_5. [DOI] [PubMed] [Google Scholar]
- 29.Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int. J. Infect. Dis. 11:5–10. 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Soehn F, Wagenknecht-Wiesner A, Leukel P, Kohl M, Weidmann M, von Eichel-Streiber C, Braun V. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864—implications for transcription, expression and enzymatic activity of toxins A and B. Mol. Gen. Genet. 258:222–232. 10.1007/s004380050726. [DOI] [PubMed] [Google Scholar]
- 31.Haslam SC, Ketley JM, Mitchell TJ, Stephen J, Burdon DW, Candy DC. 1986. Growth of Clostridium difficile and production of toxins A and B in complex and defined media. J. Med. Microbiol. 21:293–297. 10.1099/00222615-21-4-293. [DOI] [PubMed] [Google Scholar]
- 32.Popoff MR, Rubin EJ, Gill DM, Boquet P. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.