Abstract
Recent advances in the molecular identification and serotyping of Streptococcus pneumoniae are useful for culture-negative samples; however, there are limitations associated with these methods. We aimed to assess the value of molecular assays for invasive pneumococcal disease (IPD) surveillance in South Africa from 2010 through 2012. Nonviable isolates and culture-negative clinical specimens were tested for the lytA gene and, if positive, were serotyped, using real-time PCRs. Multinomial regression analysis was used to determine the maximum lytA cycle threshold (CT) value useful for predicting the ability to detect a serotype for the sample. The χ2 test was used to compare the prevalence of serotypes between viable/nonviable isolates and culture-negative clinical specimens. Of 11,224 IPD cases reported, 1,091 (10%) were culture-negative samples and 981 (90%) of these were lytA positive. Samples with a lytA CT value of ≥35 were significantly less likely to be serotyped. A serotype/group was determined for 87% (737/844) of samples with a lytA CT value of <35, of which 60% (443/737) were identified as individual serotypes. The serotype prevalence did not differ significantly between isolates and culture-negative specimens. Although molecular serotyping added 7% (737/11,224) serotyping data, the inability to resolve 40% of samples to single serotypes remains a challenge for serotype-specific data analysis.
INTRODUCTION
Streptococcus pneumoniae is a commensal bacterium of the upper respiratory tract, as well as a significant human pathogen. Each year, pneumococcal diseases result in >1 million deaths globally in children <5 years of age (1). The high burden of pneumococcal disease is also observed in elderly individuals, causing substantial morbidity and mortality (2). The polysaccharide capsule of the pneumococcus is an important virulence determinant (3, 4) and is the target for a number of pneumococcal vaccine formulations. At least 94 serotypes have been identified (5); however, only approximately 15 to 20 serotypes are responsible for the majority of disease worldwide (6).
Culture remains the gold standard for diagnosis of pneumococcal disease due to its high specificity, but it has a low sensitivity and requires long incubation periods. Antibiotic therapy prior to specimen collection and suboptimal culturing conditions hinder the yield of cultures (7, 8). PCR-based methods targeting pneumococcus-specific genes, such as lytA, have resulted in improved and timely diagnosis of pneumococcal disease (9–11).
Determination of a pneumococcal serotype is important for surveillance and determining the effectiveness of polysaccharide-based vaccines. For decades, the Quellung reaction (12) has been the gold standard for serotyping; however, the method is culture dependent and therefore is not useful for culture-negative lytA-positive specimens. Sequencing of the capsular polysaccharide synthesis genes (13, 14) has enabled the development of conventional (15) and real-time PCR (16–18) assays. The increased sensitivity of real-time PCR assays compared to that of conventional PCR assays increases the potential to serotype specimens with low bacterial loads.
Molecular serotyping assays have a number of limitations. First, the high genotypic similarities between the capsular loci of certain serotypes make it difficult to develop a serotype-specific assay, and, therefore, certain serogroups will remain unresolved to specific serotypes (14). This is problematic for serotype-specific disease surveillance and specifically in cases where vaccine and nonvaccine serotypes cannot be differentiated. Second, limited serotypes are included in most assays to reduce the time and labor required, and, therefore, not all serotypes will be detected. Most assays target vaccine serotypes and potentially relevant nonvaccine serotypes. Despite the widespread use of PCR-based serotyping assays, the challenges of analyzing the results of these assays in a surveillance setting have not been fully assessed. In the South African national invasive pneumococcal disease (IPD) surveillance, real-time PCR-based serotyping was initiated in January 2010 and is performed routinely on all culture-negative samples in order to increase the proportion of serotyped samples. Acknowledging the above-mentioned limitations, we aimed to evaluate the utility of molecular pneumococcal serotyping over a 3-year period of surveillance by determining the following: (i) the lytA cycle threshold (CT) value cutoff for optimal serotype identification, (ii) the proportion of samples for which a single serotype could be identified, and (iii) the serotype distribution between culture-positive and culture-negative samples.
MATERIALS AND METHODS
Invasive pneumococcal disease surveillance.
Laboratory-based surveillance for invasive pneumococcal disease (IPD) was initiated nationwide in South Africa in 1999 (19). By 2012, 215 laboratories routinely submitted isolates or clinical specimens in the event that an organism could not be cultured, to the reference laboratory at the National Institute for Communicable Diseases (NICD). The case definition for IPD is illness requiring hospitalization with the isolation of S. pneumoniae or detection of pneumococcal DNA (lytA) by a PCR or a positive latex antigen test with a matched Gram stain result from a normally sterile site specimen (e.g., blood, cerebrospinal fluid [CSF], or joint or pleural fluid). Upon receipt of specimens, the bacterial strains are subcultured from Dorset transport medium onto 5% horse blood Columbia base agar (Oxoid, Hampshire, UK). The culture identification is confirmed using standard microbiological procedures (20). Strains are serotyped by the Quellung reaction (12) using type-specific antisera (Statens Serum Institute, Copenhagen, Denmark). Nonviable isolates include (i) isolates transported on Dorset medium that failed to grow upon receipt at the NICD (nonviable transport medium [NVTM] samples) and (ii) autolyzed blood culture broths flagged by the BacT/Alert system as positive signals for microbial detection at the sending laboratories and confirmed to be pneumococcus by antigen detection (21). Culture-negative samples include clinical specimens (e.g., CSF, blood, and pleural fluid) yielding no growth. Confirmed IPD cases identified through laboratory audits are also included.
For surveillance purposes, all nonviable isolates are considered IPD cases, whereas culture-negative clinical specimens received for diagnostic purposes are only considered IPD cases if identified as positive for the lytA gene.
Molecular identification and serotyping of culture-negative samples. (i) DNA extraction.
DNA extraction was performed using the MagNA Pure Compact or MagNA Pure LC 2.0 instrument (Roche, Mannheim, Germany) with a DNA isolation kit I or DNA isolation kit III (Roche), respectively. DNA extraction was performed according to the manufacturer's instructions from 200 μl of sample and eluted into 100 μl of elution buffer.
(ii) Molecular identification.
The lytA gene was detected using either a singleplex real-time PCR assay (10) or a multiplex real-time PCR assay (22) as previously described. Briefly, for the singleplex assay, each 25-μl reaction consisted of 1× TaqMan gene expression master mix (Applied Biosystems, Foster City, CA), forward and reverse primers (200 nM), a FAM-labeled TaqMan minor groove-binding (MGB) probe (200 nM) (Applied Biosystems), and 2.5 μl of DNA. For the multiplex assay, each 25-μl reaction consisted of 12.5 μl of Platinum PCR SuperMix-UDG (Invitrogen, Carlsbad, CA), primers and probes as previously described (22), and 2 μl of DNA. The singleplex reaction was performed to confirm suspected pneumococcus-positive nonviable samples, including NVTM samples and autolyzed blood culture broths, while the multiplex reaction, which simultaneously detects S. pneumoniae, Haemophilus influenzae, and Neisseria meningitidis, was performed as a diagnostic test for all culture-negative clinical specimens. Samples were considered positive if the lytA CT value was <40. Additionally, a PCR inhibition control assay targeting the human RNase P (RNaseP) gene was performed on all clinical specimens and blood culture broths to exclude the negative results due to PCR inhibition, sample degradation, or DNA extraction failure (10).
(iii) Molecular serotyping.
Molecular serotyping was performed on the lytA-positive nonviable isolates and culture-negative clinical specimens (Table 1) as described by Azzari et al. (16), with an additional primer/probe set for serotype 6C/D (17). The assay enables the identification of some serotypes as single serotypes (1, 3, 4, 5, 8, 14, 20, 19A, 23F, and 35B) and others as mixed serotypes or serogroups that cannot be identified individually (6A/B, 6C/D, 7A/F, 9A/V/N/L, 10A/B, 12A/B/F, 15A/B/C/F, 18A/B/C, 19B/F, 22A/F, 33A/F/37, and 38/25A/F). The PCR was interpreted as being positive for a serotype/group if the CT value was <40. Altogether, taking into account the mixed serotypes or serogroups, the assay detected 42 serotypes. Samples negative for all reactions were recorded as negative for the 42 serotypes detected by the assay (NEG42).
TABLE 1.
Summary of invasive pneumococcal disease surveillance cases, South Africa, 2010-2012
| Cases reported to NICD as | No. of cases in: |
Total no. (%) of cases | ||
|---|---|---|---|---|
| 2010 | 2011 | 2012 | ||
| Viable isolates | 2,873 | 2,409 | 2,160 | 7,442 (66) |
| Nonviable isolatesa | ||||
| Nonviable transport medium | 262 | 228 | 143 | 633 (6) |
| Blood culture broths | 136 | 84 | 79 | 299 (3) |
| lytA-positive, culture-negative clinical specimensb | 40 | 69 | 50 | 159 (1) |
| Isolate/specimen not received | 886 | 1,014 | 791 | 2,691 (24) |
| Total | 4,197 | 3,804 | 3,223 | 11,224 (100) |
All nonviable isolates received are considered invasive pneumococcal disease (IPD) cases.
Culture-negative clinical specimens are only considered IPD cases if identified as positive for the lytA gene.
Data analysis.
We assessed the correlation between the CT values obtained from the serotyping and lytA assays using linear regression analysis. To assess the performance of the serotyping assay over different lytA CT values, we used multinomial regression analysis. Multinomial regression allows modeling of outcome variables with >2 categories and relates the probability of being in category j to the probability of being in a baseline category. A complete set of coefficients are estimated for each of the j levels being compared with the baseline, and the effect of each predictor in the model is measured as the relative risk ratio (RRR). For this analysis, we used the proportion of serotypable samples with lytA CT values of ≤30 as the baseline category and compared it with the proportion of serotypable samples with individual lytA CT values from 31 to 38. Only 2 samples with lytA CT values of 39 were available in the data set and were excluded from the analysis because of the small sample size in this group. The proportions of serotypes included in the molecular serotyping assay that were detected among the viable isolates and culture-negative samples were compared using the χ2 test for categorical variables. In order to determine whether there was a difference in the proportion of a specific serotype detected between viable/nonviable isolates and culture-negative clinical specimens, we compared the proportions of serotypes between different sample types using the χ2 test for categorical variables. Statistical significance was assessed at a value of P <0.05 for all models. The statistical analysis was implemented using STATA version 12 (StataCorp., TX).
Ethics.
Ethics approvals for the national surveillance (protocol no. M08117) and this project (protocol no. M10364) were obtained from the University of the Witwatersrand, Johannesburg.
RESULTS
National surveillance.
During the 3-year period, 11,224 IPD cases were reported: 4,197, 3,804, and 3,223 for 2010, 2011, and 2012, respectively (Table 1). The majority of cases (66%, 7,442) had a viable isolate. Nonviable isolates (NVTM and blood culture broths) and lytA-positive, culture-negative clinical specimens accounted for 10% (1,091) of cases. For 24% (2,691) of cases, an isolate or specimen was never received at the NICD.
Identification of the lytA CT value cutoff for optimal serotype identification.
Of the 1,091 culture-negative samples received, 981 (90%) were lytA positive with a median lytA CT value of 25. A total of 941 (96%) samples were available for serotyping, and a serotype/group was determined for 797 (85%). For two of these samples, two serotypes/groups were detected, indicative of mixed infections, and these samples were excluded from further analysis. In addition, two samples with lytA CT values of 39 were excluded from the analysis because of the small sample size in this group. A positive linear correlation was observed between the lytA CT values and serotyping CT values (linear regression coefficient 0.9, P < 0.001) (see Fig. S1 in the supplemental material).
We observed a decrease in the proportion of samples for which a serotype/group was determined for samples with a lytA CT value of ≥34 (Table 2). Compared to the proportion of samples with a lytA CT value of ≤30, the decrease in the proportion of serotypable samples was statistically significant for samples with a lytA CT value of ≥35 (P = 0.003). The proportion of serotypable samples declined from 89% for samples with a lytA CT value of ≤30 to 78% for those with a CT value of 34, although the decline was not statistically significant. From this analysis, a lytA CT value cutoff of <35 was used. Samples with a lytA CT value of <35 which could not be assigned a serotype/group were regarded as true negatives for the serotypes included in the assay.
TABLE 2.
Proportion of serotypeable culture-negative Streptococcus pneumoniae samples (n = 793) by lytA CT value, South Africa, 2010-2012
| lytA CT value | No. of specimens with serotype detected/total no. of specimens (%) | Relative risk ratio (95% CIa) | P |
|---|---|---|---|
| ≤30 | 643/720 (89) | Base outcome | |
| 31 | 32/37 (86) | 0.7 (0.3–2.0) | 0.592 |
| 32 | 33/38 (86) | 0.8 (0.3–2.1) | 0.634 |
| 33 | 17/19 (89) | 1.0 (0.2–4.5) | 0.981 |
| 34 | 22/28 (78) | 0.4 (0.2–1.1) | 0.084 |
| 35 | 18/26 (69) | 0.3 (0.1–0.6) | 0.003 |
| 36 | 24/33 (72) | 0.3 (0.1–0.7) | 0.005 |
| 37 | 11/23 (48) | 0.10 (0.05–0.20) | <0.001 |
| 38 | 5/13 (38) | 0.07 (0.02–0.23 | <0.001 |
CI, confidence interval.
Molecular detection and serotyping from culture-negative samples.
From 2010 to 2012, 1,091 IPD culture-negative samples were received, and lytA was detected in 981 (90%) (Table 3). Of the 844 samples that had a lytA CT value of <35 and had specimen/DNA for serotyping, a serotype/group was detected for 737 (87%), of which 443 (60%) were single serotypes, 292 (40%) were mixed serotypes/serogroups, and 2 tested positive for two serotypes, indicative of mixed infections. For 13% (107/844) of the samples, a serotype/group was not detected (NEG42).
TABLE 3.
Summary of lytA and molecular serotyping results for culture-negative samples received as part of invasive pneumococcal disease surveillance in South Africa, 2010-2012
| Sample data | Nonviable transport media |
Blood culture broths |
Clinical specimens |
Total for samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | Total | 2010 | 2011 | 2012 | Total | 2010 | 2011 | 2012 | Total | ||
| No. of samples | 262 | 228 | 143 | 633 | 136 | 84 | 79 | 299 | 40a | 69a | 50a | 159a | 1,091 |
| lytA positive | |||||||||||||
| No. | 222 | 194 | 112 | 528b | 136 | 84 | 74c | 294c | 40 | 69 | 50 | 159 | 981 |
| %d | 85 | 85 | 78 | 83 | 100 | 100 | 94 | 98 | 100 | 100 | 100 | 100 | 90 |
| lytA CT value | |||||||||||||
| Range | 13–37 | 13–39 | 12–38 | 12–39 | 11–36 | 12–34 | 11–37 | 11–37 | 17–38 | 14–37 | 16–38 | 14–38 | 11–39 |
| Median | 26 | 26 | 26 | 26 | 19 | 19 | 19 | 19 | 26 | 27 | 25 | 26 | 25 |
| lytA CT values of <35 | |||||||||||||
| No. | 191 | 157 | 94 | 442 | 135 | 84 | 73 | 292 | 36 | 63 | 47 | 146 | 880 |
| %e | 86 | 81 | 84 | 84 | 99 | 100 | 99 | 99 | 90 | 91 | 94 | 92 | 90 |
| Available for serotyping | |||||||||||||
| No. | 185 | 152 | 94 | 431 | 120 | 82 | 73 | 275 | 32 | 60 | 46 | 138 | 844 |
| %f | 97 | 97 | 100 | 98 | 89 | 98 | 100 | 92 | 89 | 95 | 98 | 95 | 96 |
| Serotype/group detected | |||||||||||||
| No. | 158c | 140 | 76 | 374g | 112 | 77 | 57 | 246 | 28 | 51 | 38 | 117 | 737g |
| %h | 85 | 92 | 81 | 87 | 93 | 94 | 78 | 89 | 88 | 85 | 83 | 85 | 87 |
| Single serotype | |||||||||||||
| No. | 107 | 74 | 46 | 227 | 76 | 44 | 33 | 153 | 16 | 23 | 24 | 63 | 443 |
| %i | 69 | 53 | 61 | 61 | 68 | 57 | 58 | 62 | 57 | 45 | 63 | 54 | 60 |
| Mixed serotypes/groups | |||||||||||||
| No. | 49 | 66 | 30 | 145 | 36 | 33 | 24 | 93 | 12 | 28 55 | 14 | 54 | 292 |
| %j | 31 | 47 | 39 | 39 | 32 | 43 | 42 | 38 | 43 | 37 | 46 | 40 | |
Only lytA-positive specimens were included; lytA-negative clinical specimens were not considered IPD cases.
A total of 92 samples were lytA negative, and 13 were not tested.
Five samples were lytA negative.
Percentage of lytA-positive samples of the samples received.
Percentage of samples with a lytA CT value of <35 of lytA-positive samples.
Percentage of samples available for serotyping of lytA-positive samples with a CT value of <35.
Two samples had two serotypes detected and were excluded from further analysis.
Percentage of samples for which a serotype/group was detected of samples available for serotyping.
Percentage of samples for which a single serotype was detected of samples for which a serotype/group was detected.
Percentage of samples for which a mixed serotype/group was detected of samples for which a serotype/group was detected.
Nonviable transport medium samples.
Of the 633 NVTM samples received, 83% (n = 528) were lytA positive with a median CT value of 26. A serotype/group was detected for 87% (374/431) of samples that had a lytA CT value of <35 and had specimen/DNA for serotyping. Two samples had mixed infections with two serotypes detected and were excluded from further analysis. For the remaining 372 samples, a single serotype was identified for 227 (61%) and mixed serotypes/groups were detected for 145 (39%).
Blood culture broths.
Of the 299 blood culture broths received, 294 (98%) were lytA positive. A total of 275 samples had a lytA CT value of <35 and had specimen/DNA for serotyping, and a serotype/group was detected for 246 (89%), including 153 (62%) single serotypes and 93 (38%) mixed serotypes/groups.
Clinical specimens.
A total of 468 culture-negative clinical specimens were received. Of these, 34% (159; 157 from CSF and 2 from pleural fluid) were positive for S. pneumoniae, 24% (112; 100 from CSF and 12 from blood) were positive for N. meningitidis, 3% (15; 13 from CSF and 2 from blood) were positive for H. influenzae, and 39% (182) were negative for all three pathogens. A serotype/group was detected for 85% (117/138) of specimens that had a lytA CT value of <35 and specimen/DNA for serotyping. Of these, 54% (63) were identified as single serotypes, and 46% (54) were identified as mixed serotypes/groups.
Comparison of serotype distributions between sample types.
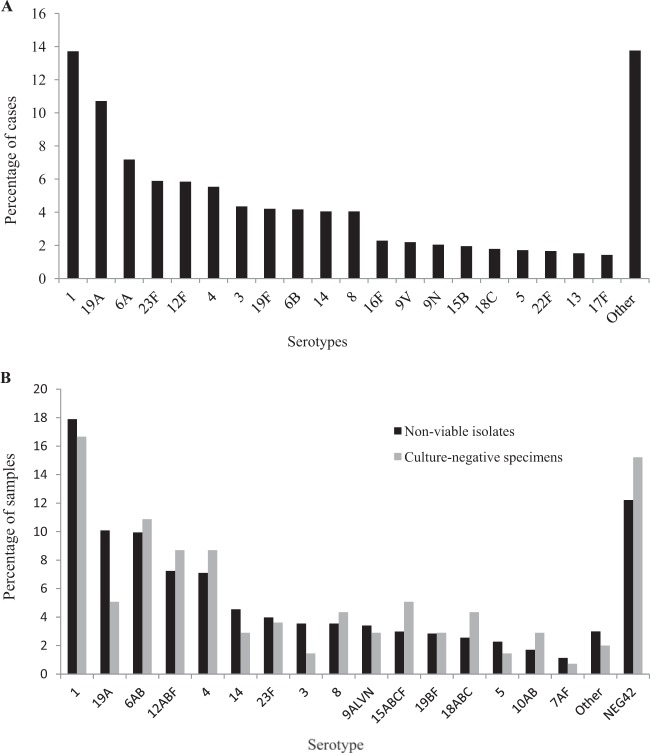
For the viable isolates, 49% (3,636/7,442) belonged to one of six serotypes, including serotype 1 (1,020/7,442, 14%), 19A (797/7,442, 11%), 6A (534/7,442, 7%), 23F (438/7,442, 6%), 12F (435/7,442, 6%), and 4 (412/7,442, 6%) (Fig. 1A). Among the nonviable isolates (including nonviable transport medium and blood culture broths), serotypes 1 (126/704, 18%), 19A (71/704, 10%) 6A/B (70/704, 10%), 12A/B/F (51/704, 7%), 4 (50/704, 7%), and 14 (32/704, 5%) were the most prevalent serotypes/serogroups detected, accounting for 57% (400/704) of nonviable isolates (Fig. 1B). Among culture-negative clinical specimens, serotypes 1 (23/138, 17%), 6A/B (15/138, 11%), 12A/B/F (12/138, 9%), 4 (12/138, 9%), 15A/B/C/F (7/138, 5%), and 19A (7/138, 5%) were the predominant serotypes, accounting for 55% (76/138) of serotypes in this group (Fig. 1B). The 42 serotypes detected by the molecular serotyping assay accounted for 89% (6,598/7,442) of the viable isolates and 87% (737/844) of the culture-negative samples with a lytA CT value of <35 (P = 0.27). In contrast, 62% (60/97) of the culture-negative samples with a lytA CT value of ≥35 were serotypes included in the assay (6,598/7,442 versus 60/97, P < 0.001).
FIG 1.
(A) Serotype distribution of the viable Streptococcus pneumoniae isolates causing invasive disease in patients of all ages, South Africa, 2010-2012 (n = 7,442). Other refers to the remaining serotypes detected by the Quellung reaction. (B) Serotype distribution of the nonviable isolates (n = 704) and culture-negative clinical specimens (n = 138) of Streptococcus pneumoniae (lytA CT value of <35) causing invasive disease in patients of all ages, South Africa, 2010-2012. Other refers to the remaining serotypes included in the PCR assay, and NEG42 refers to the samples negative for the 42 serotypes detected by the assay.
Overall there were no significant differences determined in the proportions of specific serotype/group between viable/nonviable isolates and culture-negative clinical specimens (P = 0.17) (Table 4). The proportions of serotype 19A differed significantly (P = 0.048) between viable/nonviable isolates (11%, 868/8,146) and culture-negative clinical specimens (5%, 7/138).
TABLE 4.
Comparison of the proportion of Streptococcus pneumoniae cases by serotype between viable/nonviable isolates and culture-negative clinical specimens received as part of invasive pneumococcal disease surveillance, South Africa, 2010-2012
| Serotype | No. (%) of samples that were: |
P | |
|---|---|---|---|
| Viablea/nonviableb isolates (n = 8,146) | Culture-negative clinical specimensb (n = 138) | ||
| 1 | 1,146 (14) | 23 (17) | 0.46 |
| 4 | 462 (6) | 12 (9) | 0.18 |
| 6A/B | 914 (11) | 15 (11) | 0.90 |
| 8 | 326 (4) | 6 (4) | 0.84 |
| 12A/B/F | 486 (6) | 12 (9) | 0.25 |
| 14 | 333 (4) | 4 (3) | 0.63 |
| 15A/B/C/F | 296 (4) | 7 (5) | 0.51 |
| 18A/B/C | 177 (2) | 6 (4) | 0.15 |
| 19A | 868 (11) | 7 (5) | 0.05 |
| 23F | 466 (6) | 5 (4) | 0.38 |
| Other | 2,672 (33) | 41 (30) | 0.50 |
Serotype determined by the Quellung reaction. Individual serotypes were grouped according to the PCR assay for comparative purposes.
Serotype determined by the real-time PCR assay.
DISCUSSION
We analyzed 3 years of IPD surveillance data during which culture-negative samples accounted for 10% (1,091/11,224) of cases. Overall, an additional 7% (737/11,224) of serotyping data were added to our surveillance as a result of molecular serotyping, highlighting the added value of these methods in a surveillance setting. This proportion may increase significantly in parts of the developing world where culturing practices are suboptimal and/or specimens are transported over long distances, compromising specimen quality and resulting in a greater number of culture-negative specimens.
There are, however, a number of important factors that should be considered in the decision to implement molecular serotyping and analysis of the data. A sample should only be considered negative for the serotypes included in the assay if the sample had a sufficient pneumococcal load. In this study, samples with a lytA CT value of ≥35 had a significantly reduced probability of the serotype being detected, and inclusion of these data would therefore result in an overestimation of the serotypes not included in the assay. For accurate analysis and interpretation of molecular serotyping data, a lytA CT value cutoff should be determined for the individual setting, as this value may differ depending on the assay used and only samples with lytA CT values lower than this cutoff should be included. This may be less problematic for certain sample types, such as blood culture broths, which inherently have high bacterial loads. In our study, 99% of blood culture broths had a CT value of <35 in comparison with 84% of the nonviable transport media and 92% of clinical specimens. High blood pneumococcal loads are associated with severe disease and outcome (23), and, therefore, patients who are sampled as part of a systematic pneumonia surveillance are likely to have lower blood pneumococcal loads than patients from whom a specimen is collected for etiological diagnosis of severe disease.
In our study, approximately 60% of samples were identified as single serotypes with the remainder identified as mixed serotypes/groups. This is problematic for serotype-specific analyses such as capsular-based vaccine effectiveness studies (24) and serotype association studies (4, 25), where the separation of individual serotypes is essential, and is particularly relevant for countries that have introduced the pneumococcal conjugate vaccines and are evaluating the effect of the vaccine in their setting. Methods such as the Quellung reaction, which are able to separate individual serotypes, specifically vaccine serotypes, remain the preferred methodology. In South Africa, the 7-valent pneumococcal conjugate vaccine was introduced into the routine child immunization program in April 2009 and was replaced by the 13-valent vaccine in July 2011. Our data show decreasing numbers of IPD samples from 2010 to 2012, with an increasing proportion of samples with serotypes not detected by the real-time PCR assay, which may reflect decreases in disease rates and changing serotypes in our population; however, attribution of these changes to the vaccine would be strengthened by analysis of individual serotypes.
There were no significant differences observed in the predominant serotypes detected among the viable isolates and culture-negative samples in our study. Serotypes 1, 4, 6A/B, 12A/B/F, and 19A were among the most prevalent serotypes/groups detected for the viable isolates, nonviable isolates, and culture-negative samples. This indicates that serotype is not associated with the initial culture of the diagnostic specimen and that it is more likely other factors such as host, specimen culture, and/or transport practices that influence the viability of pneumococci.
While molecular serotyping methods enable serotyping from culture-negative samples and add data to surveillance systems, caution should be taken when a molecular assay is used as a stand-alone serotyping method. Samples should have sufficient pneumococcal loads, and serotype-specific analysis of the data will be limited. With ongoing advances in molecular methods, including increased sensitivity and the ability to detect more individual serotypes (17), many of these limitations will likely be overcome. However, the large number of pneumococcal serotypes and changing serotype trends due to vaccine use will, for now, continue to challenge the pneumococcal research community.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of GERMS-SA (Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa) for contributing to the national surveillance and making this work possible.
This work was supported by the Medical Research Council of South Africa and the National Institute for Communicable Diseases (NICD) of the National Health Laboratory Service (NHLS), South Africa.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01061-14.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Fedson DS, Scott JA. 1999. The burden of pneumococcal disease among adults in developed and developing countries: what is and is not known. Vaccine 17(Suppl 1):S11–S18. 10.1016/S0264-410X(99)00122-X. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger DM, Trzcinski K, Lu YJ, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 5:e1000476. 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberger DM, Harboe ZB, Sanders EA, Ndiritu M, Klugman KP, Ruckinger S, Dagan R, Adegbola R, Cutts F, Johnson HL, O'Brien KL, Scott JA, Lipsitch M. 2010. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin. Infect. Dis. 51:692–699. 10.1086/655828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, Abeygunwardana C, Nahm MH. 2012. Biochemical, genetic, and serological characterization of two capsule subtypes among Streptococcus pneumoniae serotype 20 strains: discovery of a new pneumococcal serotype. J. Biol. Chem. 287:27885–27894. 10.1074/jbc.M112.380451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson HL, Deloria-Knoll M, Levine OS, Stoszek SK, Freimanis Hance L, Reithinger R, Muenz LR, O'Brien KL. 2010. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 7:e1000348. 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigrovic LE, Malley R, Macias CG, Kanegaye JT, Moro-Sutherland DM, Schremmer RD, Schwab SH, Agrawal D, Mansour KM, Bennett JE, Katsogridakis YL, Mohseni MM, Bulloch B, Steele DW, Kaplan RL, Herman MI, Bandyopadhyay S, Dayan P, Truong UT, Wang VJ, Bonsu BK, Chapman JL, Kuppermann N. 2008. Effect of antibiotic pretreatment on cerebrospinal fluid profiles of children with bacterial meningitis. Pediatrics 122:726–730. 10.1542/peds.2007-3275. [DOI] [PubMed] [Google Scholar]
- 8.Resti M, Micheli A, Moriondo M, Becciolini L, Cortimiglia M, Canessa C, Indolfi G, Bartolini E, de Martino M, Azzari C. 2009. Comparison of the effect of antibiotic treatment on the possibility of diagnosing invasive pneumococcal disease by culture or molecular methods: a prospective, observational study of children and adolescents with proven pneumococcal infection. Clin. Ther. 31:1266–1273. 10.1016/j.clinthera.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Azzari C, Moriondo M, Indolfi G, Massai C, Becciolini L, de Martino M, Resti M. 2008. Molecular detection methods and serotyping performed directly on clinical samples improve diagnostic sensitivity and reveal increased incidence of invasive disease by Streptococcus pneumoniae in Italian children. J. Med. Microbiol. 57:1205–1212. 10.1099/jmm.0.2008/000935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho M, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J. Clin. Microbiol. 45:2460–2466. 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacchi CT, Fukasawa LO, Goncalves MG, Salgado MM, Shutt KA, Carvalhanas TR, Ribeiro AF, Kemp B, Gorla MC, Albernaz RK, Marques EG, Cruciano A, Waldman EA, Brandileone MC, Harrison LH. 2011. Incorporation of real-time PCR into routine public health surveillance of culture negative bacterial meningitis in Sao Paulo, Brazil. PLoS One 6:e20675. 10.1371/journal.pone.0020675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Austrian R. 1976. The Quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. 43:699–709. [PubMed] [Google Scholar]
- 13.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, Reeves PR, Bentley SD, Spratt BG. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7841–7855. 10.1128/JB.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124–131. 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azzari C, Moriondo M, Indolfi G, Cortimiglia M, Canessa C, Becciolini L, Lippi F, de Martino M, Resti M. 2010. Real-time PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS One 5:e9282. 10.1371/journal.pone.0009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, von Gottberg A, McGee L, Carvalho M, Beall B. 2013. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J. Clin. Microbiol. 51:647–652. 10.1128/JCM.02927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarragó D, Fenoll A, Sanchez-Tatay D, Arroyo LA, Munoz-Almagro C, Esteva C, Hausdorff WP, Casal J, Obando I. 2008. Identification of pneumococcal serotypes from culture-negative clinical specimens by novel real-time PCR. Clin. Microbiol. Infect. 14:828–834. 10.1111/j.1469-0691.2008.02028.x. [DOI] [PubMed] [Google Scholar]
- 19.Huebner RE, Klugman KP, Matai U, Eggers R, Hussey G. 1999. Laboratory surveillance for Haemophilus influenzae type B meningococcal, and pneumococcal disease. Haemophilus Surveillance Working Group. S. Afr. Med. J. 89:924–925. [PubMed] [Google Scholar]
- 20.Winn WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Woods GL. 2006. Gram-positive cocci. Part II: streptococci, enterococci, and the “streptococcus-like” bacteria, p 672–764 In Konemans's color atlas and textbook of diagnostic microbiology, 6th ed. Lippincott Williams & Wilkins, Baltimore. [Google Scholar]
- 21.Baggett HC, Rhodes J, Dejsirilert S, Salika P, Wansom T, Jorakate P, Kaewpan A, Olsen SJ, Maloney SA, Peruski LF. 2012. Pneumococcal antigen testing of blood culture broth to enhance the detection of Streptococcus pneumoniae bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 31:753–756. 10.1007/s10096-011-1370-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Theodore MJ, Mair R, Trujillo-Lopez E, du Plessis M, Wolter N, Baughman AL, Hatcher C, Vuong J, Lott L, von Gottberg A, Sacchi C, McDonald JM, Messonnier NE, Mayer LW. 2012. Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J. Clin. Microbiol. 50:702–708. 10.1128/JCM.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolter N, Cohen C, Tempia S, Madhi SA, Venter M, Moyes J, Walaza S, Malope-Kgokong B, Groome M, du Plessis M, Pretorius M, Dawood H, Kahn K, Variava E, Klugman KP, von Gottberg A. 2014. HIV and influenza virus infections are associated with increased blood pneumococcal load: a prospective, hospital-based observational study in South Africa, 2009–2011. J. Infect. Dis. 209:56–65. 10.1093/infdis/jit427. [DOI] [PubMed] [Google Scholar]
- 24.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201:32–41. 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 25.Weinberger DM, Harboe ZB, Viboud C, Krause TG, Miller M, Molbak K, Konradsen HB. 2013. Serotype-specific effect of influenza on adult invasive pneumococcal pneumonia. J. Infect. Dis. 208:1274–1280. 10.1093/infdis/jit375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.