Abstract
Hepatitis D virus (HDV) is a defective RNA virus that requires the surface antigens of hepatitis B virus (HBV) (HBsAg) for viral assembly and replication. Several commercial and in-house techniques have been described for HDV RNA quantification, but the methodologies differ widely, making a comparison of the results between studies difficult. In this study, a full-length genomic RNA standard was developed and used for HDV quantification by two different real-time PCR approaches (fluorescence resonance energy transfer [FRET] and TaqMan probes). Three experiments were performed. First, the stability of the standard was determined by analyzing the effect of thawing and freezing. Second, because of the strong internal base pairing of the HDV genome, which leads to a rod-like structure, the effect of intense thermal shock (95°C for 10 min and immediate cooling to −80°C) was tested to confirm the importance of this treatment in the reverse transcription step. Lastly, to investigate the differences between the DNA and RNA standards, the two types were quantified in parallel with the following results: the full-length genomic RNA standard was stable and reliably mimicked the behavior of HDV-RNA-positive samples, thermal shock enhanced the sensitivity of HDV RNA quantification, and the DNA standard underquantified the HDV RNA standard. These findings indicate the importance of using complete full-length genomic RNA and a strong thermal-shock step for optimal HDV RNA quantification.
INTRODUCTION
Hepatitis D virus (HDV) is a small defective RNA virus that requires hepatitis B virus surface antigens (HBsAg) for viral assembly and replication. The HDV genome is a circular, negative, and single-stranded RNA composed of 1,672 to 1,697 nucleotides. HDV virions present a nucleocapsid-like structure formed by genomic HDV-RNA, which is associated with the hepatitis delta antigen (HDAg) protein and surrounded by the three envelope proteins of the hepatitis B virus (1). The replication of HDV occurs intrahepatically through a double-rolling circle mechanism that leads to an antigenomic RNA intermediate that is the exact complement sequence of the genome. The genomic and antigenomic molecules both present 70% internal base pairing, enabling self-assembly of the molecule and leading to a rod-like structure. An additional RNA form, the 800-nucleotide mRNA responsible for expression of the hepatitis delta antigen (HDAg), is also known to be produced during replication (2).
HDV infection causes acute or chronic liver disease in patients with hepatitis B virus (HBV) infection. HDV infection is widespread, and the prevalence differs between areas, but it is estimated that among the 350 million individuals with chronic HBV infection, approximately 15 million are also exposed to HDV (2). Eight HDV genotypes with specific geographic patterns have been reported (3). Nonetheless, population migration has led to changes in the distributions of HDV infection and genotypes (4–6).
The diagnosis of HDV infection relies on the detection of specific antibodies (anti-HDV), but the presence of anti-HDV is not always associated with active infection. The marker that best indicates HDV replication is the detection of HDV RNA in the plasma or serum. Reliable HDV RNA quantification may improve monitoring and management of the infection and would be especially relevant for antiviral therapy follow-up.
A few commercial methods are now available to quantify HDV RNA; but, as was recently reported by Brichler et al. (7), they appear to underquantify non-genotype 1 sample. Several in-house methods based on quantitative reverse transcription-PCR (qRT-PCR) and targeting the HDAg or the ribozyme domain have been described and have proven effective for quantifying all HDV genotypes. These techniques involve the use of a one-step qRT-PCR procedure (8–13), the incorporation of heat shock treatment before the RT step (10, 14–16), or the addition of heterologous internal-control RNAs (11, 12). However, all these methods rely on the use of a specific in-house standard; therefore, a comparison between studies is impossible. Furthermore, most of the standards used are subgenomic RNA transcripts or DNA standards, which may not properly mimic the complex behavior of the HDV-RNA molecules present in clinical samples. A method based on the use of a complete full-length genomic RNA standard has been described recently (11), yet questions have been raised regarding its sensitivity (12).
The aim of the present study was to develop a reliable HDV standard that takes into consideration previous methodological improvements, and to test its applicability in the most commonly used qRT-PCR strategies for HDV-RNA quantification: TaqMan (11) and fluorescence resonance energy transfer (FRET) (10) probes.
MATERIALS AND METHODS
Development of the full-length genomic RNA standard.
The main requirement of the study was to obtain an RNA molecule containing full-length genomic HDV. The complete HDV-RNA standard was obtained after in vitro transcription of an expression plasmid (pBluescript) obtained after subcloning from the HDV trimeric plasmid pSVL(D3) (kindly provided by John Taylor [17]). The pBluescript expression plasmid contained 1.0 copies of the HDV genome. The plasmid was linearized with HindIII (New England BioLabs, Hitchin, United Kingdom) and transcribed from the T3 promoter (MAXIscript, Ambion, United Kingdom). The product of in vitro transcription was purified with TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA). The control experiments showed the presence of residual plasmid HDV DNA, but DNase treatment for 30 min (MasterPure Complete DNA and RNA purification kit; Epicentre, Madison, WI, USA) completely removed the plasmid DNA. RNA was then quantified with the Qubit RNA assay kit (Invitrogen). HDV-RNA aliquots were diluted in RNase-free water containing 1 ng carrier RNA/ng HDV RNA.
The HDV-DNA plasmid and HDV-RNA standard (free of residual DNA) were diluted to obtain aliquots containing defined amounts (100 to 10E9 copies/μl).
HDV RNA quantification: thermal-shock and one-step qRT-PCR.
Due to the high rate of internal base pairing (≥70%) of the HDV RNA genome, it seemed essential to maximize the breaking of hydrogen bridges to facilitate priming at the reverse transcription (RT) step. For this reason, the application of strong thermal shock to the isolated HDV-RNA was considered. Thermal shock consisted of a denaturation step with 10 μl of HDV RNA at 95°C for 10 min, immediately followed by cooling to −80°C to minimize the possible recovery of natural internal base pairing (renaturation). HDV quantification was performed using two different real-time PCR approaches, FRET technology using the primers and probes reported by Schaper et al. (10) and TaqMan technology using the primers and probes reported by Ferns, Nastouli, and Garson (11). HDV RNA was determined via one-step reverse transcription- and quantitative RT-PCR (qRT-PCR) using the LightCycler RNA Master HybProbe (Roche, Mannheim, Germany) on a LightCycler 2.0 system (Roche) (10) for the FRET strategy, and the ABI fast virus 1-step master (Applied Biosystems, Carlsbad, CA, USA) on an ABI ViiA7 system (Applied Biosystems) (12) for the TaqMan strategy. RT-PCR was carried out according to the manufacturer's instructions for each system. Briefly, using the LightCycler, RT was performed at 61°C for 20 min, and amplification was performed in 45 cycles at 95°C for 2 s, 55°C for 12 s, and 72°C for 15 s. Using the ABI ViiA7 system, RT was done at 50°C for 15 min, and amplification was done in 40 cycles at 95°C for 3 s and 60°C for 30 s.
All experiments were performed in triplicate in independent runs. The mean and standard deviation (SD) of the crossing threshold (CT) values were calculated. Statistical analyses were performed using parametric tests with SPSS 20 (IBM SPSS, Inc., Chicago, IL, USA).
RESULTS
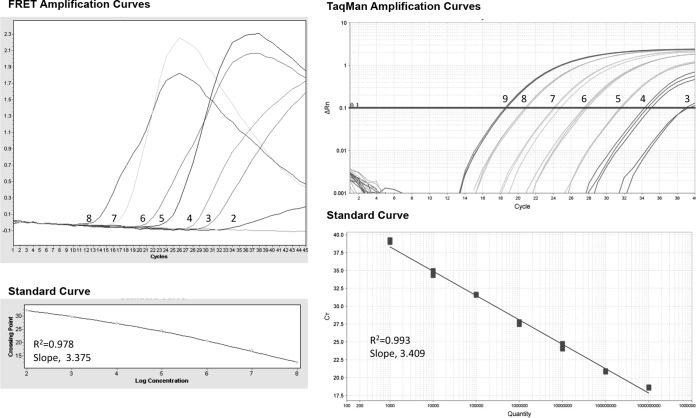
The amplification curves of the HDV-RNA standard (Fig. 1) show that the detection of the standard was feasible by both types of probes. However, each system presented a specific range of detection; experiments performed by FRET resulted in a linear range of 2- to 8-log HDV-RNA copies/μl, whereas TaqMan yielded a linear range of 3- to 9-log HDV-RNA copies/μl.
FIG 1.
Amplification curves obtained from the standard points, using the LightCycler (FRET technology) and the ABI ViiA7 (TaqMan technology). The number in each amplification curve corresponds to the logarithmic expression of HDV-RNA level, illustrating the detection ranges (2- to 8-log HDV-RNA copies/μl for FRET and 3 to 9 for TaqMan). The slope and R2 are indicated.
Stability of the HDV-RNA standard.
The considerable stability of HDV-RNA in the serum samples results from the highly self-complementary nature of the genome (rod-like structure). The HDV-RNA standard developed here contains the complete HDV genome sequence, and therefore, it might also include the highly self-complementary stabilizing structure. However, in contrast to natural HDV-RNA, the standard is linear, not a covalently closed molecule, and this fact questions its stability, reproducibility, and traceability for routine quantitative analysis. Therefore, we tested the stability of the standard by applying cycles of thawing and freezing in three independent experiments (Table 1). No differences were found in the CT values obtained following thawing and freezing (P = 0.6). These results indicate that despite the lack of a covalently closed structure, the HDV-RNA standard is stable and reliably mimics the behavior of natural HDV-RNA from patient samples.
TABLE 1.
Mean CT and SD of the three experiments testing the effect of thawing and freezing (1 to 5 cycles) in different aliquots of the same standard point
| Value | Expt 1 | Expt 2 | Expt 3 |
|---|---|---|---|
| Mean CT | 20.41 | 21.23 | 20.81 |
| SD | 0.32 | 0.58 | 0.39 |
Effect of thermal shock.
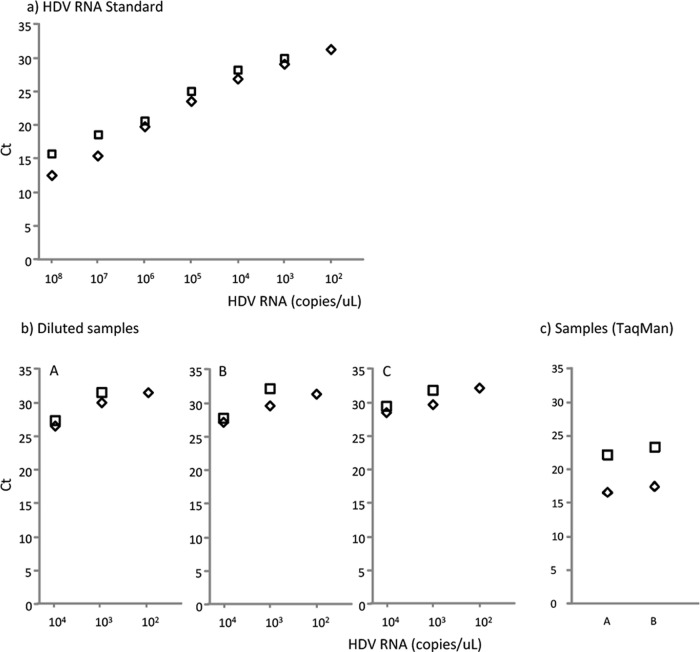
The relevance of the thermal-shock step for optimal reverse transcription in HDV RNA quantification was assessed in triplicate. Dilutions of HDV-RNA standard were quantified with and without thermal shock (Fig. 2a). On average, the CT values were 1.79 lower with this treatment than without, which strongly suggested greater sensitivity with thermal shock. In fact, at the lowest dilution (102 copies/μl), the standard was only detected when thermal shock had been performed.
FIG 2.
Effect of thermal shock (denaturation) on the standard and on samples with (◇) and without (□) thermal shock. (a) HDV RNA standard; (b) diluted samples; (c) TaqMan quantification. The CT values were consistently higher in the absence of thermal shock. Points with 102 copies/μl of standard and samples without thermal-shock treatment were not detected.
In addition, the effect of thermal shock was assessed in serially diluted clinical samples (Fig. 2b) using TaqMan probes (Fig. 2c). In these samples, lower CT values were observed following denaturing by thermal shock than without denaturation, confirming the importance of this step in the technique. Keeping in mind that the higher the CT, the lower the quantification, these findings indicate that thermal shock increased the efficiency of the RT step.
Comparison between DNA and RNA standards.
Some reported methods have used HDV-cDNA as the standard for HDV-RNA quantification (9, 10, 14–16, 18), which does not allow an evaluation of the RT step. In the present research, we used the HDV-DNA standard to evaluate the HDV-RNA standards as if they were samples. The results showed underquantification of HDV RNAs (Table 2). These findings indicate that the efficiency of the RT step was low and that DNA standards do not take this limiting step into account. Therefore, proper quantification of HDV RNA in clinical samples requires an RNA standard.
TABLE 2.
Quantification of RNA standard using the DNA standard as reference to investigate differences in quantification between the two types of standardsa
| Std HDV RNA log10 (c/μl)b | Log10 (c/μl) |
|---|---|
| 8 | 6.57 |
| 7 | 5.75 |
| 6 | 4.84 |
| 5 | 4.04 |
| 4 | 2.48 |
| 3 | 1.37 |
| 2 | 1.06 |
DISCUSSION
The complete full-length genomic HDV RNA standard obtained in the present study, free of residual DNA, was found to be useful for HDV-RNA quantification. The experiments demonstrated that the molecule obtained is stable and reliably mimics the behavior of clinical samples.
It is thought that because of its strong internal base pairing, the HDV genome may be able to reassemble its rod-like structure. Hence, it was possible that the complete genomic linear RNA standard evaluated in this study might also have the capability to adopt this secondary structure, a fact that would explain its high stability after thawing and freezing. The application of thermal shock before the qRT-PCR likely decreased the probability of reassembling the rod-like structure, facilitated primer hybridization in the RT step, and improved HDV RNA quantification. This hypothesis seems to be confirmed by the lower CT values obtained with the use of thermal shock than without this treatment, and as a consequence, the higher HDV RNA levels.
The HDV genome is characterized by considerable heterogeneity, a fact that may affect quantification. In this study, we used primers and probes that are useful for the quantification of all HDV genotypes (10, 11). We found that the TaqMan and FRET probes were feasible for HDV RNA standard quantification, but the range of detection was different for each technology, at 2- to 8-log HDV-RNA copies/μl for FRET and 3- to 9-log HDV-RNA copies/μl for TaqMan. These results suggest that HDV quantification can undergo further optimization, especially in the RT step, which seems to be the critical point of the process.
In conclusion, the complete genomic HDV-RNA standard obtained in this study is highly stable and exhibits behavior similar to that of HDV-RNA in clinical samples. The control experiments demonstrated an improvement in HDV quantification by applying thermal shock before the RT step, using both FRET and TaqMan technology. In the absence of an international standard for HDV quantification, the full-length genomic HDV-RNA standard developed in this study is clearly better than DNA plasmids for clinical HDV RNA quantification. Interlaboratory studies should be performed to confirm the reliability of this standard as a prelude to its widespread use, which would enable comparability between studies. Lastly, the differing linear ranges of the FRET and TaqMan procedures indicate that users should select the optimal strategy for their purposes, while always including a complete genomic HDV-RNA standard.
Footnotes
Published ahead of print 2 July 2014
REFERENCES
- 1.Pascarella S, Negro F. 2011. Hepatitis D virus: an update. Liver Int. 31:7–21. 10.1111/j.1478-3231.2010.02320.x. [DOI] [PubMed] [Google Scholar]
- 2.Alvarado-Mora MV, Locarnini S, Rizzetto M, Pinho JRR. 2013. An update on HDV: virology, pathogenesis and treatment. Antivir. Ther. 18:541–548. 10.3851/IMP2598. [DOI] [PubMed] [Google Scholar]
- 3.Radjef N, Gordien E, Ivaniushina V, Gault E, Anaïs P, Drugan T, Trinchet JC, Roulot D, Tamby M, Milinkovitch MC, Dény P. 2004. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J. Virol. 78:2537–2544. 10.1128/JVI.78.5.2537-2544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buti M, Homs M, Rodriguez-Frias F, Funalleras G, Jardí R, Sauleda S, Tabernero D, Schaper M, Esteban R. 2011. Clinical outcome of acute and chronic hepatitis delta over time: a long-term follow-up study. J. Viral Hepat. 18:434–442. 10.1111/j.1365-2893.2010.01324.x. [DOI] [PubMed] [Google Scholar]
- 5.Wedemeyer H, Heidrich B, Manns MP. 2007. Hepatitis D virus infection–not a vanishing disease in Europe! Hepatology 45:1331–1332, author reply, 1332–1333. 10.1002/hep.21590. [DOI] [PubMed] [Google Scholar]
- 6.Wedemeyer H. 2010. Re-emerging interest in hepatitis delta: new insights into the dynamic interplay between HBV and HDV. J. Hepatol. 52:627–629. 10.1016/j.jhep.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Brichler S, Le Gal F, Butt A, Chevret S, Gordien E. 2013. Commercial real-time reverse transcriptase PCR assays can underestimate or fail to quantify hepatitis delta virus viremia. Clin. Gastroenterol. Hepatol. 11:734–740. 10.1016/j.cgh.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 8.Yamashiro T, Nagayama K, Enomoto N, Watanabe H, Miyagi T, Nakasone H, Sakugawa H, Watanabe M. 2004. Quantitation of the level of hepatitis delta virus RNA in serum, by real-time polymerase chain reaction–and its possible correlation with the clinical stage of liver disease. J. Infect. Dis. 189:1151–1157. 10.1086/382133. [DOI] [PubMed] [Google Scholar]
- 9.Mederacke I, Bremer B, Heidrich B, Kirschner J, Deterding K, Bock T, Wursthorn K, Manns MP, Wedemeyer H. 2010. Establishment of a novel quantitative hepatitis D virus (HDV) RNA assay using the Cobas TaqMan platform to study HDV RNA kinetics. J. Clin. Microbiol. 48:2022–2029. 10.1128/JCM.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaper M, Rodriguez-Frias F, Jardi R, Tabernero D, Homs M, Ruiz G, Quer J, Esteban R, Buti M. 2010. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J. Hepatol. 52:658–664. 10.1016/j.jhep.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Ferns RB, Nastouli E, Garson JA. 2012. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J. Virol. Methods 179:189–194. 10.1016/j.jviromet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Scholtes C, Icard V, Amiri M, Chevallier-Queyron P, Trabaud MA, Ramière C, Zoulim F, André P, Dény P. 2012. Standardized one-step real-time reverse transcription-PCR assay for universal detection and quantification of hepatitis delta virus from clinical samples in the presence of a heterologous internal-control RNA. J. Clin. Microbiol. 50:2126–2128. 10.1128/JCM.06829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodani M, Martin A, Mixson-Hayden T, Drobeniuc J, Gish RR, Kamili S. 2013. One-step real-time PCR assay for detection and quantitation of hepatitis D virus RNA. J. Virol. Methods 193:531–535. 10.1016/j.jviromet.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann J, Frenzel K, Minh BQ, von Haeseler A, Edelmann A, Ross SR, Berg T, Krüger DH, Meisel H. 2010. Quantitative detection and typing of hepatitis D virus in human serum by real-time polymerase chain reaction and melting curve analysis. Diagn. Microbiol. Infect. Dis. 67:172–179. 10.1016/j.diagmicrobio.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Shang D, Hughes SA, Horner M, Bruce MJ, Dong Y, Carey I, Suddle AR, Agarwal K, Harrison PM, Atkins M. 2012. Development and validation of an efficient in-house real-time reverse transcription polymerase chain reaction assay for the quantitative detection of serum hepatitis delta virus RNA in a diverse South London population. J. Virol. Methods 184:55–62. 10.1016/j.jviromet.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Katsoulidou A, Manesis E, Rokka C, Issaris C, Pagoni A, Sypsa V, Hatzakis A. 2013. Development and assessment of a novel real-time PCR assay for quantitation of hepatitis D virus RNA to study viral kinetics in chronic hepatitis D. J. Viral Hepat. 20:256–262. 10.1111/jvh.12000. [DOI] [PubMed] [Google Scholar]
- 17.Kuo MY, Chao M, Taylor J. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Gal F, Gordien E, Affolabi D, Hanslik T, Alloui C, Dény P, Gault E. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J. Clin. Microbiol. 43:2363–2369. 10.1128/JCM.43.5.2363-2369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]