Abstract
An optimal antimicrobial drug regimen is the key to successful clinical outcomes of bacterial infections. To direct the choice of antibiotic, access to fast and precise antibiotic susceptibility profiling of the infecting bacteria is critical. We have developed a high-throughput nanowell antibiotic susceptibility testing (AST) device for direct, multiplexed analysis. By processing in real time the optical recordings of nanoscale cultures of reference and clinical uropathogenic Escherichia coli strains with a mathematical algorithm, the time point when growth shifts from lag phase to early logarithmic phase (Tlag) was identified for each of the several hundreds of cultures tested. Based on Tlag, the MIC could be defined within 4 h. Heatmap presentation of data from this high-throughput analysis allowed multiple resistance patterns to be differentiated at a glance. With a possibility to enhance multiplexing capacity, this device serves as a high-throughput diagnostic tool that rapidly aids clinicians in prescribing the optimal antibiotic therapy.
INTRODUCTION
Antibiotics have served as essential treatment of bacterial infections for several decades. Extensive use of antibiotics, however, has promoted the selection and emergence of resistant bacterial strains. The increasing prevalence of antibiotic-resistant infections accounts for at least 2 million illnesses and 23,000 deaths in the United States (1) and more than 25,000 deaths and €1.5 billion health care costs per year in Europe (2). Besides a general overuse of antibiotics, resistant pathogens emerge due to the frequent prescription of broad-spectrum antibiotics. Since such antibiotics kill a large proportion of different bacterial species apart from the disease-causing pathogen, the balance of the commensal flora is affected, thus favoring the emergence of previously outcompeted resistant bacteria (3). To confidently select a narrow-spectrum rather than broad-spectrum antibiotic therapy, the clinician requires the pathogen's antibiotic susceptibility profile. Currently, due to the relatively long time for diagnostic results to be finalized, doctors are often left with no alternative but to employ an empirically defined broad-spectrum antibiotic therapy to secure a patient's survival.
The general basis of antibiotic susceptibility testing (AST) methods for phenotypic resistance evaluation is to monitor the pathogen's ability to grow in the presence of defined concentrations of antibiotics as specified by the Clinical and Laboratory Standards Institute (CLSI) (4). Currently, the disk diffusion assay and the Etest (5, 6) are commonly used AST methods in clinical laboratories. Since both assays require a defined inoculum size, patient samples must be cultivated for at least 1 day prior to performing the AST assay. Antimicrobial susceptibility is then determined by visual examination of growth inhibition zones around the antibiotic-containing disk or Etest plastic strip after a minimum of 16 h of incubation of the agar plates. Despite being time-consuming, both methods are widely used in the clinic due to their low cost and ease of use. More recently, automated systems have been introduced. Following precultivation of a patient's sample to a defined inoculum size, turbidimetric monitoring of bacterial growth combined with colorimetric measurement of substrate metabolites enable the AST assay to be performed within 1 day for some, but far from all, bacterial species (7–11). Automated methods, however, often require costly equipment, and thus, their use can vary among different countries.
Molecular diagnostic techniques represent yet another category of AST that are based on the detection of genes or single-nucleotide polymorphisms (SNPs) associated with antibiotic resistance, using nucleic acid amplification tests, microarray, and sequencing technologies (12–15). Despite delivering results faster than the standard phenotypic ASTs, molecular methods are restricted to detection of currently known resistance-associated genetic loci. Moreover, phenotypic resistance often deviates from genotypic resistance, and therefore, molecular diagnostics are applied as a complement to currently used ASTs (16).
Positioned at the interface between nanotechnology and biomedical research, nanomedicine is evolving as a research area expected to provide tools for sensitive, high-throughput analysis of biological systems. Microfluidic droplet systems have been explored as AST devices, e.g., agarose microparticles and asynchronous magnetic bead rotation biosensors (17, 18). Though technically advanced, these systems are limited to testing only one antibiotic per assay, making them less suitable for high-throughput analysis. Miniaturization of growth chambers, such as the nanowell slide, has been demonstrated as a novel tool for single-cell analysis, including clonal expansion analysis of single embryonic stem cells (19) and leukemic primary cells (20), as well as PCR amplification and mutation analysis performed on cells directly in the nanowells (21).
In this study, we extended the use of the nanowell slide to prokaryotic organisms. Taking advantage of the multitude of miniaturized wells and real-time processing of optical data using a mathematical algorithm, a phenotypic multiplex AST device was developed, allowing heatmap representation of MIC determination within 4 h.
MATERIALS AND METHODS
Bacterial strains.
The reference Escherichia coli strain ATCC 25922 and uropathogenic Escherichia coli (UPEC) clinical isolates ARD144, ARD145, and ARD146 from patients with urinary tract infections (Karolinska University Hospital, Sweden) were initially cultivated at 37°C on Luria-Bertani (LB) agar plates, and then single colonies from the plates were transferred to cation-adjusted Mueller-Hinton II broth (M-H II broth; Becton, Dickinson), according to CLSI instructions. After culturing overnight at 37°C, bacteria were diluted 1:100 in M-H II broth and cultivation was continued under shaking conditions to an optical density at 600 nm (OD600) of 0.4. From this exponential phase, dilutions in prewarmed M-H II broth equivalent to 104, 105 (equal to 0.5 McFarland unit as suggested by the CLSI), or 106 CFU/ml (5, 50, and 500 CFU/well, respectively) were immediately used as inocula for both the broth microdilution and the nanowell AST device assays. Technical replicates of the nanowell AST device assay were performed using bacterial inocula of the same strain, derived from liquid cultures prepared on different days. Biological replicates were performed by testing the different strains of Escherichia mentioned above. The number of bacteria in each inoculum was defined using viable-count assays.
Antibiotics.
Ampicillin sodium salt (A9518; Sigma-Aldrich), ciprofloxacin hydrochloride (PHR1044-1G; Sigma-Aldrich), and cefotaxime sodium salt (C7039; Sigma-Aldrich) were dissolved and diluted in sterile deionized water to 50 mg/ml (ampicillin), 0.5 mg/ml (ciprofloxacin), and 10 mg/ml (cefotaxime) and stored at −80°C until use.
Broth microdilution assay.
Broth microdilution assays were performed in 96-well plates with ampicillin used to precoat the wells. Precoating was achieved by adding ≤40 μl of ampicillin dissolved in sterile water to wells in triplicates, to generate final concentrations of 0.5, 1, 2, 4, 8, 16, and 32 μg/ml in the 150-μl cultures. Subsequently, ampicillin was dried by incubation at 35°C for 24 h. Exponential-phase bacteria cultivated in M-H II broth were serially diluted in order to generate inocula of 104, 105, and 106 CFU/ml. Positive (bacteria in M-H II medium with no antibiotic) and negative (M-H II broth with no bacteria) controls were included in each experiment. Plates were incubated at 37°C for 20 h under aerated conditions, and absorbance (600 nm) was recorded either at the end of the experiment or every 10 min using a SpectraMax M5 multimode microplate reader (Molecular Devices) with SoftMax Pro 6.1. The MIC was determined as the lowest antibiotic concentration at which no increase in OD600 was measured compared to the negative control. Testing the ATCC 25922 strain served also as the quality control of the broth microdilution assay, since its MIC breakpoints for ampicillin are defined by the CLSI. Average absorbance and standard deviations were calculated for the triplicates of each concentration.
Nanowell slide design.
The nanowell slide consists of a glass slide (75 by 0.175 by 25 mm) anodically bonded to a nanowell-etched silicon grid (75 by 0.5 by 25 mm) with tapered sides (19, 21). The glass slide-to-grid bonding creates a 14 by 48 matrix of 672 wells, each with a volume of 500 nl. The 650-μm by 650-μm surface area at the bottom of the well gradually increases to 1,360 by 1,360 μm at the top due to the outward-tilted walls. The transparent gas-permeable membrane (74 by 1.5 by 24 mm) was made with a Sylgard 184 silicone elastomer kit (DowCorning) according to the manufacturer's instructions. The membrane was autoclaved before being applied on the nanowell slide (20).
Functionalization of the nanowell slide.
Nanowells were functionalized by precoating the bottoms and walls of wells with defined concentrations of antibiotics. Antibiotic-containing aqueous solutions were pipetted manually into defined areas of the slide until a flat mirror image appeared on the surface. The ampicillin-containing AST slide was coated with 0.5, 1, 2, 4, 8, 16, and 32 μg/ml. One area for positive control of bacterial growth in the absence of antibiotic and one for negative control to ensure contamination-free M-H II medium were left uncoated. Additionally, a negative-control area, to check for contamination in antibiotic solutions, was included by coating wells with 32 μg/ml of ampicillin. To generate a multiplex AST slide, areas were functionalized with 0.002 μg/ml and 0.03 μg/ml of ciprofloxacin, 0.015 μg/ml and 0.3 μg/ml of cefotaxime, and 2 μg/ml and 16 μg/ml of ampicillin (see scheme in Fig. S3 in the supplemental material), which correspond to values lower and higher than the individual MICs against strain ATCC 25922 (CLSI). Uncoated areas were used for positive control (ATCC 25922 in M-H II broth), whereas 4 negative-control areas were included as follows: (i) M-H II broth, (ii) M-H II broth plus 16 μg/ml of ampicillin, (iii) M-H II broth plus 0.03 μg/ml of ciprofloxacin, and (iv) M-H II broth plus 0.03 μg/ml of cefotaxime. When the UPEC clinical isolates were tested, the multiplex nanowell AST slide was coated with 1, 2, 4, 8, 16, 32, and 64 μg/ml of cefotaxime and 0.25, 0.5, 1, 2, and 4 μg/ml of ciprofloxacin. A positive-control area (no antibiotic) and 3 negative-control areas (M-H II broth, M-H II broth plus 64 μg/ml of cefotaxime, and M-H II broth plus 4 μg/ml of ciprofloxacin) were included. After addition of antibiotics, slides were dried at 37°C overnight and then stored in the dark for ≤4 days at 4°C before use.
MIC analysis in the nanowell AST device.
The membrane, attached to the slide with a clip, was lifted at one end, and approximately 400 μl of inoculum was added underneath. The inoculum was evenly spread into wells by lowering the membrane onto the device. An adaptor the size of a standard 96-well plate (designed with AutoCAD 2013 for Mac and three-dimensionally [3D] printed at Shapeways) was manufactured to place the nanowell device in the SpectraMax M5 multimode microplate reader (Molecular Devices) (Fig. 1E). In addition to a position for the device, the adaptor included a well aimed for holding 6 ml of water to prevent evaporation from the nanowells. The slide was secured on the adaptor using flat-top screws. Because of an inherent limitation of the available instrument to read maximally 384 points, the microplate reader was reprogrammed (using SoftMax Pro 6.1 software) to spectrophotometrically record each row of wells in every second column of the nanowell device. The distance between wells was set to 3,000 μm in x direction and 1,500 μm in y direction and the bottom area set to 650 μm2. Recordings at 600 nm were performed every 5 or 10 min for 10 or 12 h at 37°C. The shorter length of the light path in the nanowell device compared to standard 1-cm cuvettes generated lower OD values in our device. Data from each growth curve were internally normalized by subtracting the average of 5 baseline recordings from all values of each growth curve. The MIC was determined as the lowest antibiotic concentration inhibiting bacterial growth.
FIG 1.
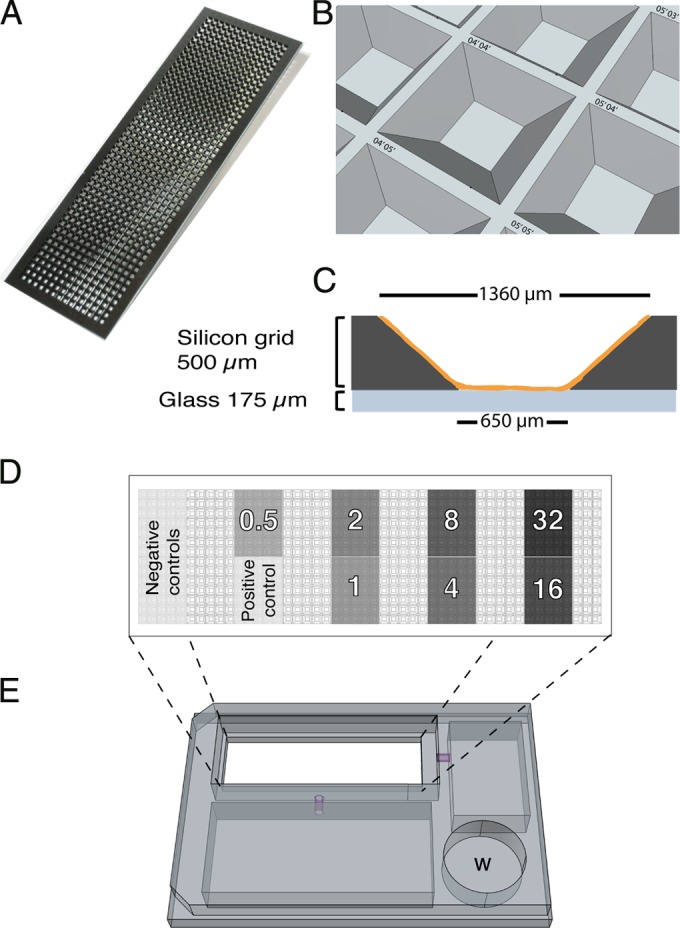
Nanowell AST device. (A) Illustration of the nanowell slide with 672 wells (500 nl/well). (B) Illustration of the tilted walls of the wells providing large surface-to-volume ratio, fast diffusion, and gas exchange. Each well is labeled with a unique x-y position indicator. (C) Side view illustrating a well precoated with an antibiotic (orange). (D) Top-view illustration of a slide with defined areas for bacterial growth in the absence or presence of an antibiotic concentration. Each number represents a different antibiotic concentration in μg/ml. (E) Custom-designed adaptor for placing the nanowell AST device in a microplate reader. W, water bath.
Heatmaps.
Heatmaps were generated in RStudio (RStudio, Inc.) using the “Heatmap” package. Each line of the heatmap illustrates the change in OD600 over time for one well where the shift from yellow to red represents 0 to 0.500 or 0 to 0.550 absorbance unit as indicated below. Individual time points when growth shifts from lag phase to early logarithmic phase (Tlag), indicated by black dots, were added manually to the heatmaps in Adobe Illustrator CS4. The heatmaps were generally produced by including wells 1 to 4 from each experiment for all conditions tested. At 4 μg/ml of ampicillin, however, the variable growth patterns among wells prompted us to select 4 specific wells out of the 12 to ensure that the full range of growth was represented.
RESULTS
Nanowell AST device development.
The nanowell slide features 672 wells holding 500 nl each (Fig. 1A) within the dimensions of a 25- by 75-mm standard microscope slide. The slide consists of an etched silicon grid bonded to a glass slide, enabling spectrophotometric analysis and high-resolution imaging of individual wells. The silicon walls of each well are tilted outwards to facilitate dispensing of bacterium-containing samples or other liquids by manual spreading of the appropriate volume across the slide (Fig. 1B). Moreover, each well is designated with defined x and y coordinates to enable sample identification. To prepare the slide for AST analysis, 35 wells in each defined region were functionalized with 0 to 32 μg/ml of ampicillin by drying an antibiotic-containing solution in the wells, leaving a coating on the bottom and walls (Fig. 1C and D). Together with regions for positive and negative control of bacterial growth, the slide represents a high-throughput AST device with each antibiotic concentration tested in multiple replicates.
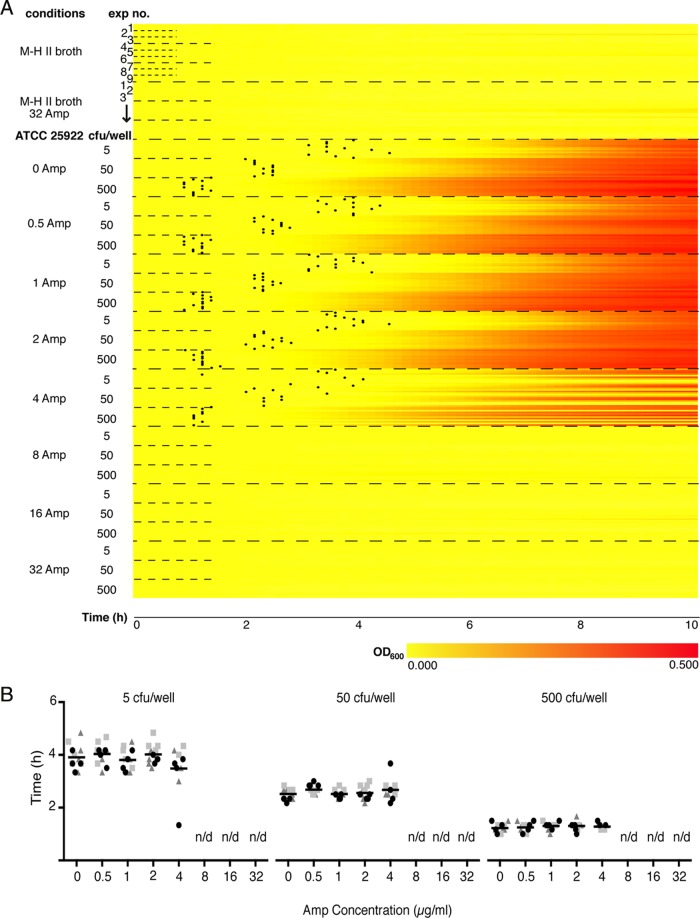
To validate the nanowell AST device, individual slides were inoculated with either of three different inoculum sizes of the reference Escherichia coli strain, ATCC 25922, in M-H II broth. Evaporation from the wells was limited by positioning a transparent gas-permeable membrane on top of the slide. Using a custom-designed adaptor (Fig. 1E), the nanowell device was positioned in a temperature-controlled microplate reader, allowing absorbance of each bacterial culture to be monitored at 600 nm every 10 min for 10 h. Traditional growth curve visualization is far from optimal when presenting absorbance data from the hundreds of nanowells. Inspired by the field of transcriptomics, we introduced heatmaps for this purpose. Each line in the heatmap shows the color-coded OD600 values collected every 10 min in individual wells for 10 h (Fig. 2A). In wells with bacterial growth, the increase over time is visualized as a gradual shift from yellow to red. In cases of no growth, OD600 remains low, shown by a continuous yellow line.
FIG 2.
Heatmap representation and Tlag determinations for MIC analysis. (A) Heatmap representation of OD600 recordings from ATCC 25922 in M-H II broth with 0 to 32 μg/ml of ampicillin (Amp) at different inoculum sizes (5, 50, and 500 CFU/well). The vertical axis indicates experimental conditions and inoculum sizes (5, 50, 500 CFU/well). “Exp no.” refers to the 3 experiments for each inoculum, each including 4 rows corresponding to OD600s recorded from wells 1 to 4 every 10 min over 10 h. Increased absorbance is depicted as a change from yellow to red along the horizontal axis. Black dots indicate Tlag in each culture positive for growth. (B) Dot plots showing Tlag calculated from the 4 wells per experiment at 0 to 32 μg/ml of Amp for 5 CFU/well (left), 50 CFU/well (middle), and 500 CFU/well (right). Shapes and colors of dots represent data collected from wells derived from the same experiment. The grand mean for each condition is depicted as a black horizontal line.
The heatmap shown in Fig. 2A compiles 360 growth curves originating from 9 independent experiments (3 experiments for each of the inoculum sizes of 5, 50, and 500 CFU/well) performed on separate nanowell AST devices. Data were recorded from 120 wells of each experiment (12 wells from each of the 10 conditions), but for clarity, data from only 4 wells/experiment are presented. The different colors in the heatmap visualize distinctly the overall effect of ampicillin on bacterial growth, while the 360 individual lines at the same time allow inspection of growth in each individual well (Fig. 2A). Conversely, growth curves cannot fully exploit the multiplexity of the device, and only the average absorbance from a group of wells for each condition can be efficiently shown (see Fig. S1 in the supplemental material). A quick glance at the color-coding is sufficient to identify a MIC of 8 μg/ml regardless of inoculum size. This is identical to information given by the CLSI (4), as well as to results from parallel experiments using broth microdilution assays in 96-well plates (see Fig. S2) and Etest (see Table S1). At the intermediate concentration of 4 μg/ml, bacterial growth was gradually inhibited as the MIC of 8 μg/ml was approached. The growth of strain ATCC 25922 in all wells under drug-free conditions (positive control) revealed that the manual spreading technique resulted in an even distribution of the sample.
Algorithm-based determination of Tlag for rapid MIC testing.
Bacterial growth curves include distinct growth phases termed lag, exponential, and stationary phases. The lag phase, representing the delay before the start of exponential growth, allows bacterial cells to adapt to and exploit new environmental conditions (22). Translated to the AST situation, this means that the point of transition from lag to the exponential phase represents the earliest time when an antibiotic-resistant strain can be differentiated from a susceptible one. This point, representing the duration of the lag phase, is designated Tlag. Since accurate determination of Tlag is difficult to achieve by visual inspection of heatmaps and growth curves, we modified an algorithm previously used for definition of Tlag (23). To calculate the change in average OD600 (ΔMOD), a mathematical formula was used:
| (1) |
ΔMOD is identified by subtracting the average OD600 of 7 measurements preceding and including the time point (t) in question from the average OD600 of 7 measurements including and following this time point. Then, the difference between two ΔMOD values of subsequent time points is calculated:
| (2) |
If ΔΔMOD is ≥0.001 for six subsequent time points (tn … tn + 5), then tn is defined as Tlag. If no Tlag was obtained within 12 h, bacteria were considered susceptible to that antibiotic.
As an alternative to the visual inspection of MIC, Tlag was exploited as a means to phenotypically discern the antibiotic susceptibility profile. Tlag values at different inocula of strain ATCC 25922 were determined by applying the algorithm to the optical recordings in the presence of ampicillin. Tlag values represented by black dots plotted on the corresponding well of the heatmap were obtained for all cultures grown at sub-MICs (0.5 to 4 μg/ml), whereas no Tlag was obtained in cultures exposed to ≥8 μg/ml of ampicillin due to lack of growth (Fig. 2A). Regardless of inoculum size, Tlag calculations accurately defined a MIC of 8 μg/ml. Moreover, sub-MICs of ampicillin had no effect on bacterial growth, since Tlag values were approximately the same as in the absence of ampicillin among bacteria from the same inoculum (Fig. 2B). The inoculum did, however, affect the length of the lag phase in all cultures exposed to sub-MICs (0 to 4 μg/ml) of ampicillin. The Tlag for 5 CFU/well ranged around 4 h (Fig. 2B, left), whereas the Tlag values for 50 and 500 CFU/well ranged between 2 to 3 h and 1 to 2 h, respectively (Fig. 2B, middle and right). Collectively, this illustrates the applicability of Tlag to define the MIC within 4 h.
When recording the absorbance of bacterial cultures, slight fluctuations are common during lag phase before a steady increase is observed. Theoretically, the algorithm is designed to cope with such fluctuations, since 13 data points are included for Tlag determination. To empirically test the accuracy of Tlag, individual growth curves were plotted for all wells in the heatmap and examined for growth and assignment of Tlag. False positives (cultures assigned a Tlag although no growth occurred) were observed in 1.4%, whereas false negatives (growth although no Tlag was assigned) were observed in as few as 0.3% of the cultures. In the remaining 98.3%, Tlag was accurately determined.
Multiplexed AST in nanowell devices.
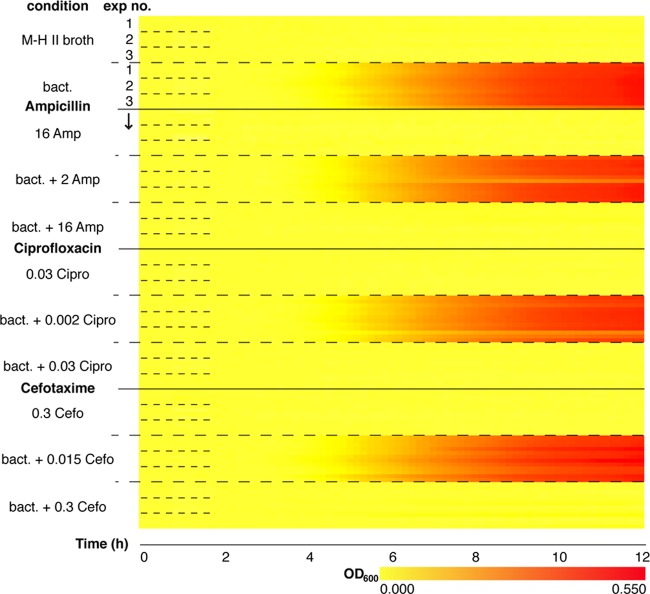
To test the use of the nanowell AST device for concurrent differentiation between multiple antibiotics, separate regions were coated with two concentrations each of ciprofloxacin, cefotaxime, and ampicillin, representing sub- and supra-MICs as previously reported for strain ATCC 25922 (see Fig. S3 in the supplemental material). We performed 3 independent assays (50 CFU/well) and monitored OD600 every 10 min for 12 h. Recordings from 4 wells of each of the 11 conditions per assay were used to compile the 132 growth curves into one heatmap (Fig. 3). The shifts from yellow to red reveal bacterial growth in the absence of antibiotics (positive control) and in the presence of sub-MICs. In contrast, solid yellow lines at concentrations exceeding the individual MICs demonstrate lack of growth and, accordingly, antibiotic susceptibility. Since the results for all antibiotics are in accordance to their respective CLSI MIC breakpoints, and no cross-contamination occurred between the defined regions, we conclude that the nanowell device appears highly suitable for multiplexed MIC determinations. The minimum time for susceptibility testing is 4 h 10 min. This includes time for cultures to reach Tlag (2 h 20 min under all growth-permitting conditions) and collection of the six data points required for the ΔMOD algorithm (equations 1 and 2).
FIG 3.
Multiplex nanowell AST for MIC determination. Shown is a heatmap representation of growth curves (ATCC 25922) in the multiplex nanowell AST. The vertical axis indicates the 11 conditions tested. Positive (ATCC 25922 in M-H II broth) and negative (MH-II broth and M-H II broth plus antibiotics: 16 μg/ml of ampicillin [Amp], 0.03 μg/ml of ciprofloxacin [Cipro], and 0.03 μg/ml of cefotaxime [Cefo]) controls were included. “Exp no.” refers to the 3 independent experiments, each including 4 rows corresponding to OD600s from wells 1 to 4 recorded every 5 min over 12 h. Increased absorbance is depicted as a change from yellow to red along the horizontal axis.
Clinical isolates tested in the multiplexed AST.
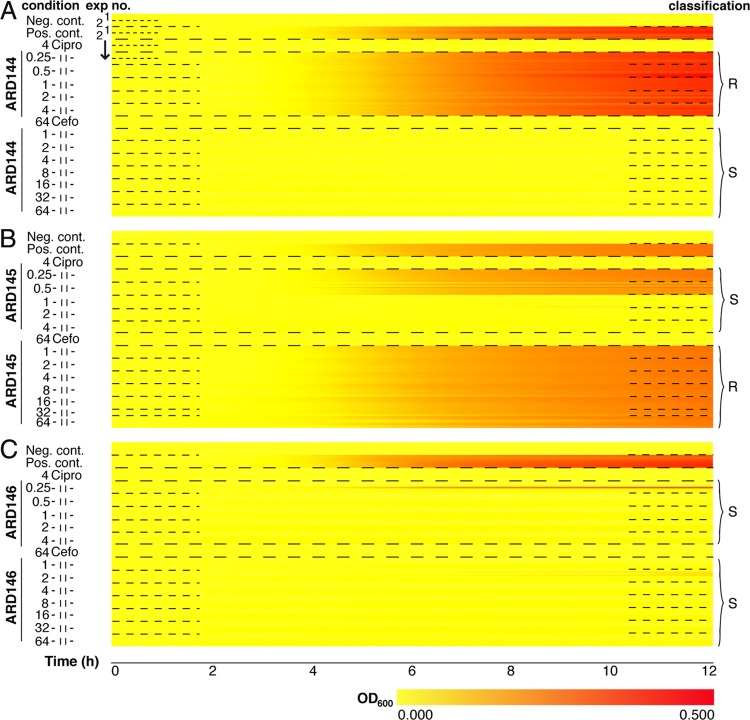
To evaluate the nanowell AST device's clinical performance, a test was conducted blind with three unknown uropathogenic E. coli (UPEC) isolates, ARD144, ARD145, and ARD146, from patients who presented with symptoms of urinary tract infections. We functionalized 6 nanowell slides with 0.25 to 4 μg/ml of ciprofloxacin and 1 to 64 μg/ml of cefotaxime, as well as regions for positive (bacteria seeded under drug-free conditions) and negative (medium only or medium with antibiotic) controls. We performed two independent assays for every clinical isolate (50 CFU/well as the inoculum), and data were recorded every 5 min for 12 h. Sampling the data every 5 min allowed Tlag to be calculated with only a 30-min delay (the time required for collecting the 6 data points for the ΔMOD algorithm). This shortening required, however, that the cutoff for statistical significance for ΔΔMOD be set to 0.0005. Data recorded from 128 wells (4 wells per 16 conditions per assay) was used to compile one heatmap per strain (Fig. 4).
FIG 4.
Clinical strains analyzed in the multiplexed nanowell AST. Shown is a heatmap representation of antibiotic susceptibility of clinical strains ARD144 (A), ARD145 (B), and ARD146 (C) against ciprofloxacin (Cipro) and cefotaxime (Cefo) at the indicated concentrations (μg/ml). Positive (strains in M-H II broth) and negative (M-H II broth and M-H II broth plus antibiotics [4 μg/ml of Cipro or 64 μg/ml of Cefo]) controls were included. “Exp no.” refers to the 2 independent experiments, each including 4 rows corresponding to OD600s from wells 1 to 4 recorded every 5 min over 12 h. Increased absorbance is depicted as a change from yellow to red along the horizontal axis. Classification of strains at each condition is indicated as susceptible (S) and resistant (R) based on CLSI guidelines.
Strain ARD144 showed a clear-cut response (Fig. 4A). The shift from yellow to intense red reveals bacterial growth despite exposure to the full range of ciprofloxacin concentrations. With a MIC of ≥4 μg/ml, the nanowell AST assay classifies this strain as resistant, which corresponds to values indicated for this antibiotic against Enterobacteriaceae in the CLSI guide (4). Being resistant to ciprofloxacin, strain ARD144 showed an average Tlag of 2 h 23 min when cultivated in the presence of this antibiotic, which correlates well to the Tlag of the strain under the drug-free condition. In contrast, the solid yellow lines for the full range of cefotaxime concentrations represent lack of growth. With a MIC of <1 μg/ml, well below the breakpoint of ≤8 μg/ml given by the CLSI, ARD144 is characterized as susceptible to cefotaxime. No Tlag was detected in the presence of cefotaxime, since the strain is sensitive to all concentrations used.
When testing strain ARD145 against ciprofloxacin, no growth occurred above 0.5 μg/ml (Fig. 4B). This characterized the strain as susceptible, which is in accordance with the CLSI MIC breakpoint (MIC ≤ 1 μg/ml). The Tlag was 2 h 43 min in the presence of sub-MICs, whereas no Tlag was generated at higher concentrations due to lack of growth. In contrast, ARD145 was resistant to cefotaxime, and an estimated MIC of >64 μg/ml is in agreement with the CLSI classification. Due to the resistant phenotype, a Tlag of 2 h 43 min was obtained in the presence of the full range of concentrations of this antibiotic, which is the same as under drug-free conditions.
Yet another pattern was observed for strain ARD146, which showed susceptibility to both ciprofloxacin (MIC ≤ 0.25 μg/ml) and cefotaxime (MIC ≤ 1 μg/ml) (Fig. 4C). Growth did, however, occur in one of eight wells at the lowest concentration (0.25 μg/ml), due to possible heterogeneity of this clinical isolate. Interestingly, MIC determination for all strains tested on the multiplexed nanowell AST correlated precisely with results from parallel Vitek 2 and Etests of the same strains performed in the clinical laboratory (see Table S1 in the supplemental material). Likewise, the susceptibility of strain ARD146 to both ciprofloxacin and cefotaxime was revealed by the absence of Tlag in antibiotic-containing cultures, whereas drug-free conditions revealed an average Tlag of 2 h 1 min. Collectively, the times for determination of antibiotic susceptibility, including the time required for collection of the six data points required for the ΔMOD algorithm (equations 1 and 2), are as short as 3 h 18 min (ARD144), 3 h 38 min (ARD145), and 2 h 56 min (ARD146).
DISCUSSION
Obtaining a fast antibiotic susceptibility profile of infecting bacteria is of utmost importance to aid clinicians in deescalating from broad- to narrow-spectrum antibiotic therapy, thus avoiding selection for resistant bacteria. Here, we demonstrate the use of the nanowell AST device as a high-throughput tool enabling rapid determination of antimicrobial susceptibility phenotypes in bacteria. The nanowell AST device delivers precise MIC determinations for clinical UPEC strains against multiple antibiotics in as short a time as 3 h. This is 6 times faster than traditional broth microdilution assays in 96-well plates. Key to the short time to result is the method's ability to define Tlag as the point in time when a bacterial culture shifts from lag to logarithmic phase. We demonstrate how Tlag could be applied as a very early indicator differentiating whether bacterial cultures are resistant or susceptible to an antibiotic. Sensitivity of the method is obtained by the application of a mathematical algorithm when data from frequent spectrophotometric recordings of multiple nanowell cultures are processed. Detecting initiation of growth at this very early stage is not possible by methods based on naked-eye inspections of agar plates. The commonly used agar disk diffusion tests and Etest require, for instance, >16 h of incubation for bacteria to reach a density high enough for results to be accurately interpreted.
Time to result in the AST device can be modulated by performing the spectrophotometric recordings of cultures at different intervals. By collecting data every 10 min, the first ΔMOD can theoretically be calculated at 120 min, since the algorithm is based on 13 sequential recordings. The minimum time for diagnostic results requires an additional 60 min, since Tlag is determined as the point when 6 consecutive ΔMODs give a value of ≥0.001 each. By reducing the interval between spectrophotometric recordings to 5 min, diagnostic results can be obtained in as little as 90 min. This requires, however, an adjustment of the cutoff for significant ΔΔMOD from 0.001 to 0.0005. Whereas a change in optical density as low as 0.0005 might be considered insignificant, specificity of the assay is left uncompromised since ΔMOD for each time point is based on 13 consecutive recordings. Indeed, bacterial growth was clearly differentiated from random fluctuations, since the algorithm accurately detected antibiotic susceptibility and resistance in 98.3% of the total number of nanowells analyzed in the current study.
In current methods for MIC breakpoint determination, a trade-off must be considered between the inclusion of several types of antibiotics, their concentration ranges, and the number of experimental replicas. These limitations do not exist in the AST device owing to the large number of wells, which were shown to generously accommodate an extensive concentration range of each of several antibiotics, thereby enabling precise definition of MIC breakpoints. Such precise information is essential when characterizing new antimicrobial compounds against different bacteria or when designing the optimal antimicrobial therapy, especially in severe, complicated infections (24). Whether the pattern of antimicrobial activity is concentration or time dependent, optimal dosing of an antibiotic regimen can only be estimated by considering the exact MIC and the pharmacodynamic and pharmacokinetic parameters of a given antibiotic. Additionally, since the number of nanowells can be increased to 3,243 on one device (20), a larger number of antimicrobial compounds can be simultaneously accommodated if automated dispensing techniques are applied. Another significant advantage of the plurality of wells is the possibility to include numerous replicates of each experimental condition. Combined with an accuracy of 98.3%, the numerous replicates dramatically limit the error rate in susceptibility pattern identification.
The ability to individually study replicates of bacterial cultures exposed to different antibiotic concentrations revealed significant bacterial heterogeneity within the population. The mixed growth pattern (some wells showing growth and others inhibition) observed for strain ATCC 25922 among the 12 wells at the intermediate concentration of 4 μg/ml of ampicillin led us to set the MIC breakpoint at 8 μg/ml, defining the strain as resistant. If it had not been for the large number of wells on the nanowell AST device, this heterogeneity might have been overlooked, posing a risk of defining an inaccurately low MIC. It is highly unlikely that growth heterogeneity appeared by virtue of unsuccessful diffusion of the precoated antibiotic into the medium. The large surface-to-volume ratio of each well allows fast diffusion, thereby preventing delayed growth due to adapted resistance in heterogenic environments (25).
In the present study, absorbance recordings were performed every 5 or 10 min in 336 wells per device, a number that can be greatly increased if desired. Generation of data sets this size calls for novel methods of handling and presentation. For the first time known to us, a heatmap was applied to present bacterial growth. This was appealing from several perspectives. The color-coded growth curves indicate intuitively the MICs of several antibiotics simultaneously for a given strain. Additionally, the wealth of information available in the heatmap, such as the emergence of new susceptibility patterns in heterogeneous pathogen populations, can be exploited to identify new resistance patterns.
Before proceeding to clinical implementations, follow-up studies are needed to extend the use of the nanowell AST device to clinical isolates of both Gram-negative and Gram-positive origins. This is important since the diagnostic times may vary among different bacterial species (10). Moreover, once the manufacturing procedure is fully automated, the multiplexing capabilities of the nanowell AST device will be fully exploited by enabling its coating with a larger number of antibiotics. This may be a collection of general antibiotics, or devices can be designed to include specific combinations of antibiotics relevant for given infections. Whereas this study focused on infection of the urinary tract, which predominantly is of monomicrobial origin, future designs can be customized to include groups of antibiotics relevant for bacterial species found in other locales. We expect that the nanowell AST device will be broadly applied to determine antimicrobial susceptibility in a variety of bacterial infections, thereby addressing the clinician's need for diagnostic speed and accuracy. Given the current efforts of both European (26, 27) and American (28) organizations to combat antimicrobial resistance, we anticipate that the nanowell AST device will also serve as a useful tool in basic research laboratories devoted to putting an end to the threat of resistant pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. K. P. Rajeswari for help with RStudio and for the code used to create the heatmaps, C. Meuli for experimental help, and B. Libberton and K. Melican for critically reading the manuscript.
This project was funded by Swedish Research Council-NT and -3R (A.R.-D.), Carl Bennet AB (A.R.-D.), the Swedish Governmental Agency for Innovation Systems (VINNOVA) (A.R.-D. and H.A.-S.), and the Swedish Foundation for Strategic Research (SSF) (H.A.-S.).
The intellectual property rights of the described AST are protected by patent application SE1450045-8. E.W., H.A., P.K., H.A.-S., and A.R.-D. are coinventors. A.B. declares no competing interests.
Footnotes
Published ahead of print 2 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01161-14.
REFERENCES
- 1.US Department of Health and Human Services, Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 2.European Centre for Disease Prevention and Control (ECDC) and European Medicines Agency (EMEA). 2009. The bacterial challenge: time to react. ECDC/EMEA joint technical report; ECDC/EMEA, Stockholm, Sweden. [Google Scholar]
- 3.Willing BP, Russell SL, Finlay BB. 2011. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9:233–243. 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Ericsson H, Tunevall G, Wickman K. 1960. The paper disc method for determination of bacterial sensitivity to antibiotics. Relationship between the diameter of the zone of inhibition and the minimum inhibitory concentration. Scand. J. Clin. Lab. Invest. 12:414–422. [DOI] [PubMed] [Google Scholar]
- 6.Bolmstrom A, Arvidson S, Ericsson M, Karlsson A. 1988. A novel technique for direct quantitation of antimicrobial susceptibility of microorganisms, p 325, abstr 1209 Program Abstr. Twenty-Eighth Intersci. Conf. Antimicrob. Agents Chemother., Los Angeles, CA. American Society for Microbiology, Washington, DC. [Google Scholar]
- 7.Kelly MT, Leicester C. 1992. Evaluation of the Autoscan Walkaway system for rapid identification and susceptibility testing of gram-negative bacilli. J. Clin. Microbiol. 30:1568–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doern GV, Brueggemann AB, Perla R, Daly J, Halkias D, Jones RN, Saubolle MA. 1997. Multicenter laboratory evaluation of the bioMerieux Vitek antimicrobial susceptibility testing system with 11 antimicrobial agents versus members of the family Enterobacteriaceae and Pseudomonas aeruginosa. J. Clin. Microbiol. 35:2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leverstein-van Hall MA, Fluit AC, Paaum A, Box ATA, Brisse S, Verhoef J. 2002. Evaluation of the Etest ESBL and the BD Phoenix, VITEK 1, and VITEK 2 automated instruments for detection of extended-spectrum beta-lactamases in multiresistant Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 40:3703–3711. 10.1128/JCM.40.10.3703-3711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligozzi M, Bernini C, Bonora MG, de Fatima M, Zuliani J, Fontana R. 2002. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 40:1681–1686. 10.1128/JCM.40.5.1681-1686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eigner U, Schmid A, Wild U, Bertsch D, Fahr A-M. 2005. Analysis of the comparative workflow and performance characteristics of the VITEK 2 and Phoenix systems. J. Clin. Microbiol. 43:3829–3834. 10.1128/JCM.43.8.3829-3834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossney AS, Herra CM, Fitzgibbon MM, Morgan PM, Lawrence MJ, O'Connell B. 2007. Evaluation of the IDI-MRSA assay on the SmartCycler real-time PCR platform for rapid detection of MRSA from screening specimens. Eur. J. Clin. Microbiol. Infect. Dis. 26:459–466. 10.1007/s10096-007-0303-7. [DOI] [PubMed] [Google Scholar]
- 13.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair A, Arnold C, Woodford N. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in enterococci. Antimicrob. Agents Chemother. 47:3620–3622. 10.1128/AAC.47.11.3620-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43:2291–2302. 10.1128/JCM.43.5.2291-2302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodford N, Sundsfjord A. 2005. Molecular detection of antibiotic resistance: when and where? J. Antimicrob. Chemother. 56:259–261. 10.1093/jac/dki195. [DOI] [PubMed] [Google Scholar]
- 17.Eun YJ, Utada AS, Copeland MF, Takeuchi S, Weibel DB. 2011. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem. Biol. 6:260–266. 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinn I, Kinnunen P, Albertson T, McNaughton BH, Newton DW, Burns MA, Kopelman R. 2011. Asynchronous magnetic bead rotation (AMBR) biosensor in microfluidic droplets for rapid bacterial growth and susceptibility measurements. Lab Chip 11:2604–2611. 10.1039/c0lc00734j. [DOI] [PubMed] [Google Scholar]
- 19.Lindström S, Eriksson M, Vazin T, Sandberg J, Lundeberg J, Frisén J, Andersson-Svahn H. 2009. High-density microwell chip for culture and analysis of stem cells. PLoS One 4:e6997. 10.1371/journal.pone.0006997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindström S, Larsson R, Andersson-Svahn H. 2008. Towards high-throughput single cell/clone cultivation and analysis. Electrophoresis 29:1219–1227. 10.1002/elps.200700536. [DOI] [PubMed] [Google Scholar]
- 21.Lindström S, Hammond M, Brismar H, Andersson-Svahn H, Ahmadian A. 2009. PCR amplification and genetic analysis in a microwell cell culturing chip. Lab Chip 9:3465–3471. 10.1039/b912596e. [DOI] [PubMed] [Google Scholar]
- 22.Madigan MT, Martinko JM, Parker J, ed. 2000. Brock biology of microorganisms. p 135–162 Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- 23.Meletiadis J, te Dorsthorst DTA, Verweij PE. 2003. Use of turbidimetric growth curves for early determination of antifungal drug resistance of filamentous fungi. J. Clin. Microbiol. 41:4718–4725. 10.1128/JCM.41.10.4718-4725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs MR. 2001. Optimisation of antimicrobial therapy using pharmacokinetic and pharmacodynamic parameters. Clin. Microbiol. Infect. 7:589–596. 10.1046/j.1198-743x.2001.00295.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK, Pourmand N, Austin RH. 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333:1764–1767. 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 26.European Commission, Directorate-General for Enterprise and Industry. 2010. EU-funded FP6 research projects on antimicrobial drug resistance. European Commission, Brussels, Belgium. 10.2777/22731. [DOI] [Google Scholar]
- 27.European Commission. 2011. Action plan against the rising threats from antimicrobial resistance. European Commission, Brussels, Belgium: http://ec.europa.eu/health/antimicrobial_resistance/policy/index_en.htm [Google Scholar]
- 28.National Institutes of Health. NIH categorical spending. Estimates of funding for various research, condition, and disease categories (RCDC). National Institutes of Health, Bethesda, MD: http://report.nih.gov/categorical_spending.aspx. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.