Abstract
Rapid and accurate diagnosis of influenza is important for infection control, as well as for patient management. Alere i Influenza A&B is an isothermal nucleic acid amplification-based integrated system for detection and differentiation of influenza virus A and influenza virus B. The performance of the Alere i Influenza A&B was screened using frozen nasopharyngeal-swab specimens collected in viral transport medium (VTM) that were originally tested fresh with the FilmArray Respiratory Panel (RP) assay during the 2012–2013 influenza outbreak. In total, 360 VTM specimens were selected for Alere i Influenza A&B testing: 40 influenza virus A H1N1-2009 (influenza virus A-1), 40 influenza virus A H3N2 (influenza virus A-3), 37 influenza virus A “equivocal” or “no subtype detected” (influenza virus A-u), 41 influenza virus B, and 202 influenza virus-negative specimens, as initially determined by the FilmArray RP assay. The Alere assay showed sensitivities of 87.2%, 92.5%, 25.0%, and 97.4% for influenza virus A-1, influenza virus A-3, influenza virus A-u, and influenza virus B, respectively, after discordant resolution by Prodesse ProFLU+ PCR. The specificities were 100% for both influenza virus A and influenza virus B. In general, the Alere i Influenza A&B provided good sensitivity, although the assay did show poorer sensitivity with samples determined to have low influenza virus A titers by Prodesse ProFlu+ PCR (a mean real-time PCR threshold cycle [CT] value of 31.9 ± 2.0), which included the majority of the samples called influenza virus A “equivocal” or “no subtype detected” by a single BioFire FilmArray RP test. The integrated, rapid, and simple characteristics of the Alere i Influenza A&B assay make it a potential candidate for point-of-care testing, with a test turnaround time of less than 15 min.
INTRODUCTION
Influenza is a contagious respiratory illness caused by influenza viruses A and B in humans. The clinical presentation of the disease ranges from asymptomatic infection to severe complications, including viral pneumonia and death, especially in immunocompromised patients, patients with underlying comorbidities, and those at the extremes of age (1–3). Since the 2009 pandemic, influenza virus A 2009 H1N1 (influenza virus A-1) has quickly become a dominant influenza virus strain circulating throughout the world along with seasonal influenza virus A H3N2 (influenza virus A-3) and influenza virus B (4, 5). Typical symptoms of influenza include fever, cough, sore throat, rhinorrhea, and nasal congestion, symptoms that overlap those of infections with other viruses circulating at the same time (6, 7). Unlike other viruses, rapid and accurate diagnosis of influenza is necessary for prompt administration of antiviral therapy, mainly oseltamivir, which should be administered within 48 h of symptom onset (8). Additional benefits of rapid identification are infection control, public health notification and tracking, and prevention of unnecessary use of antibiotics, hospital procedures, and laboratory tests (9–11).
Current diagnostic techniques for the detection and identification of influenza virus include rapid influenza antigen detection tests (RIDTs), direct fluorescent-antibody assays (DFAs), viral culture, and nucleic acid amplification tests (NAAT). Commercially available RIDTs are widely used in clinical practice as point-of-care tests because they are simple to use and provide results within 15 to 30 min (12, 13). However, their sensitivities vary widely depending on the manufacturer and can be as low as 10%, with specificities ranging from 90 to 100% (14). DFAs are more sensitive than RIDTs and can be accomplished within 3 h but require skilled technologists for correct interpretation of results (15, 16). Similarly, culture has increased sensitivity over both RIDTs and DFAs but requires skilled technologists and specialized laboratory settings and has a long turnaround time (2 to 14 days) (17). NAAT are highly sensitive and are gradually replacing culture as the gold standard, but these tests are generally expensive; depending on the manufacturer, require highly skilled molecular technologists; and have turnaround times of up to 24 h from receipt to results (1, 7, 18–22). As a result, specimens with negative RIDTs are usually tested subsequently by more sensitive culture or molecular assays. PCR-based molecular assays have shown excellent clinical utility for the detection and identification of influenza viruses; numerous FDA-cleared commercial devices are now available (18, 23, 24).
The ideal diagnostic test for influenza would have the fast turnaround time and simplicity of an RIDT with the sensitivity and specificity of a NAAT. The Alere i Influenza A&B (Alere Scarborough, Scarborough, ME) assay incorporates a nicking endonuclease amplification reaction (NEAR) technique for the detection and differentiation of influenza virus A and influenza virus B in nasopharyngeal swab (NPS) specimens. The system requires only 2 min of total hands-on time to process and set up one sample, and results are available within 15 min. The objective of this study was to evaluate the clinical performance of the Alere i Influenza A&B assay on NPS specimens soaked in viral transport medium (VTM) that were previously tested by the FilmArray Respiratory Panel (RP) (BioFire Diagnostics, Salt Lake City, UT).
(This study was presented in part at the Association for Molecular Pathology 2013 Annual Meeting, Phoenix, AZ, 14 to 16 November 2013.)
MATERIALS AND METHODS
Clinical specimens.
A retrospective study was conducted on banked NPS specimens collected from inpatients presenting with influenza-like symptoms at Memorial Sloan-Kettering Cancer Center (MSKCC) between 15 December 2012 and 1 March 2013 during an influenza outbreak. These NPS specimens were soaked in 3 ml of VTM, and the leftovers were stored at −80°C after the single FilmArray RP assay was performed. Based on the FilmArray results, up to 40 specimens positive for each virus and genotype, i.e., influenza virus A-1, influenza virus A-3, influenza virus A “equivocal” or “untypeable” (influenza virus A-u), and influenza virus B, were selected. For each influenza virus-positive specimen, 1 or 2 upstream and/or downstream influenza virus-negative specimens, based on their accession numbers, were also selected. Duplicate specimens from the same patient were later excluded. The study was approved by the MSKCC Institutional Review Board with waiver of Health Insurance Portability and Accountability Act of 1996 (HIPAA) authorization and informed consent.
Alere i Influenza System.
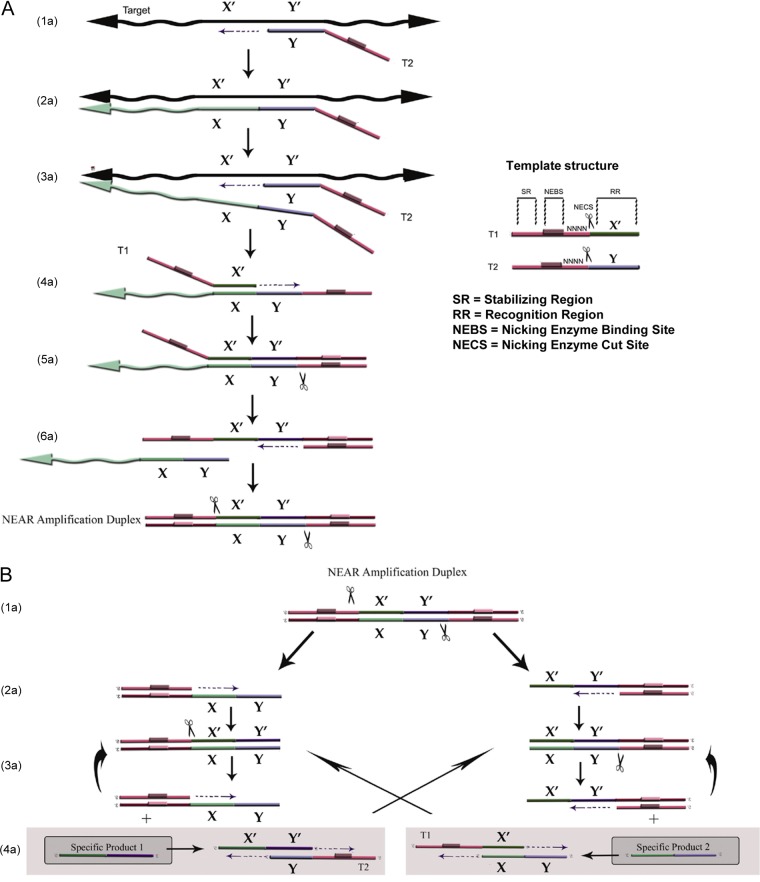
The Alere i Influenza System deploys the NEAR technique to detect and distinguish influenza virus A and influenza virus B. NEAR incorporates an isothermal nucleic acid amplification technology to provide ultrarapid DNA or RNA amplification, with results in 10 min or less when coupled with fluorescence-based detection (25, 26). Two templates (primers) and three enzymes (a thermostable DNA polymerase, a reverse transcriptase, and a thermostable nicking endonuclease) provide the driving force behind NEAR when the target is RNA. In conjunction with the two templates, these enzymes provide a method for exponential amplification of short amplicons that combines satisfactory specificity with a very short time frame. Detection is accomplished in real time, using fluorescently labeled molecular beacons (Fig. 1).
FIG 1.
NEAR mechanism. (A) Mechanism of NEAR amplification duplex formation. (1a and 2a) The recognition region of T2 binds to the complementary target region and is extended by polymerase along the target. (3a) A second T2 binds to the same target and is extended, displacing the first T2. (4a) The recognition region of T1 binds to its complement in the released strand and is extended to the 5′ end, creating a double-stranded nicking enzyme recognition site. (5a) Nicking enzyme binds and nicks (indicated by scissors). (6a) polymerase synthesizes off the cleaved 3′ OH along T1, displacing the remaining target complement, and the final extended double-stranded complex is termed the NEAR amplification duplex. (B) Mechanism of product formation. (1b and 2b) Nicking enzymes bind to both nicking enzyme recognition sites on the NEAR duplex; cleavage and strand displacement amplification at both sites creates two complexes, each consisting of a duplex stability region, a nicking enzyme recognition region, and a single-stranded target. (3b and 4b) Repeated nicking, polymerization, and strand displacement result in the amplification of products 1 and 2. Cleaved complexes are regenerated (3b), while products 1 and 2 can anneal to T1 and T2, respectively (4b), resulting in bidirectional extension and creating duplexes that generate the opposite product upon cleavage. The products continue to recycle until the templates, deoxynucleoside triphosphates (dNTPs), or enzymes are depleted.
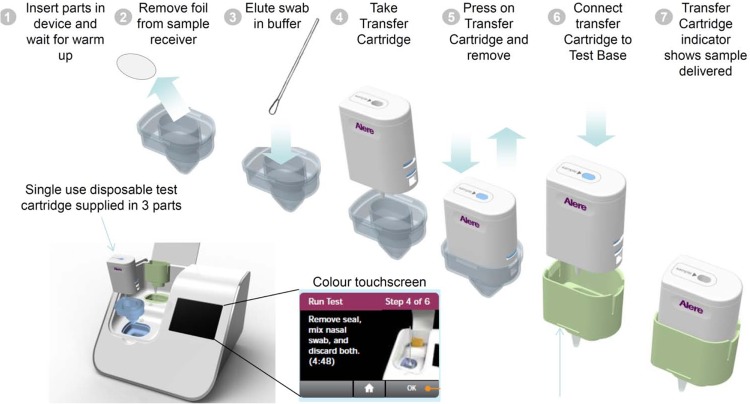
The Alere system was performed as instructed by the manufacturer. Briefly, the sample receiver and test base were inserted into the instrument, and the lysis buffer in the sample receiver was automatically heated by the instrument (Fig. 2). After approximately 3 min, an aliquot of sample from the VTM (0.2 ml) was transferred into the sample receiver and mixed by pipetting. Two 0.1-ml aliquots of the eluate from the sample receiver were then manually transferred via the transfer cartridge to the test base to rehydrate the lyophilized NEAR InfA and InfB reaction mixes and initiate target amplification and detection. Heating, agitation, and detection by fluorescence are performed automatically by the instrument. The internal control results are automatically checked by the reader to ensure that the test result is valid. Results are reported for influenza A and B viruses as negative or positive within 10 to 12 min. As required by the study protocol, a positive-control and a negative-control swab provided by the manufacturer were run each day before patient specimens were tested.
FIG 2.
Flowchart of the Alere i Influenza A&B assay. The total elapsed time was within 15 min, and the total hands-on time was within 2 min.
FilmArray RP assay.
The specimens were transported within 2 h to the microbiology laboratory, where they were processed immediately for routine respiratory-pathogen diagnosis. The FilmArray RP (version 1.6) was performed according to the manufacturer's instructions and as previously described (19). Briefly, 1 ml of hydration solution was injected into the FilmArray pouch to rehydrate the reagents. Using a transfer pipette, approximately 300 μl of respiratory sample was added to the sample buffer vial, and the resulting mixture was transferred to the pouch using the sample-loading syringe. The pouch was then placed in the FilmArray instrument, and a preprogrammed PCR run was initiated.
Prodesse ProFLU+.
The ProFLU+ assay (GenProbe/Hologic H44VK00) was performed on a subset of enrolled NPS/VTM specimens according to the manufacturer's instructions, with the following modifications (27, 28). Sample processing was performed on the Qiacube work station (Qiagen, Valencia, CA) using the QiaAmp Viral RNA minikit. PCR was performed on the Roche LightCycler 480 (Roche Diagnostics, Indianapolis, IN). The data analysis and report were done blinded to FilmArray and Alere results.
Evaluation standards and data analysis.
Specimens were considered positive or negative when results from the Alere assay matched those of the FilmArray RP. Discrepant results were resolved based on the Prodesse ProFLU+ results. McNemar's test was used to compare the Alere and the reference results, and a Student t test was used to compare cycle threshold (CT) value means. Statistical analysis was performed using QuickCalcs (GraphPad Software). A P value of ≤0.05 was considered statistically significant.
RESULTS
During the study period, 3,675 NPS specimens were submitted and tested by a single FilmArray RP assay. Among them, 45, 425, 37, and 77 tested positive for influenza virus A-1, influenza virus A-3, influenza virus A-u, and influenza virus B, respectively. Based on the selection criteria, a total of 360 specimens were enrolled in the study. The specimens were collected from 193 female and 167 male patients, with 69 (19.2%) children (<18 years old) and 291 (80.8%) adults. They included 40 influenza virus A-1, 40 influenza virus A-3, 37 influenza virus A-u (21 equivocal and 16 untypeable), 41 influenza virus B, and 202 influenza virus-negative specimens, as determined by the FilmArray RP assay. Among the 202 influenza virus-negative samples tested by FilmArray, 60 were positive for one or more non-influenza viruses, including 21 rhinoviruses/enteroviruses, 13 respiratory syncytial viruses, and 14 coronaviruses (229E, 5; OC43, 4; NL63, 4; HKU1, 1).
Among the 360 enrolled specimens, the single Alere assay reported 79 influenza virus A, 37 influenza virus B, and 240 negative and 4 invalid results (failure of internal controls) (Table 1). A single ProFLU+ assay was performed on all 42 discordant specimens and 37 concordant specimens (based on Alere and FilmArray). The results from all 37 concordant samples were confirmed by ProFLU+. Among the 42 discordant specimens, ProFLU+ detected 31/41 (8 influenza virus A, 21 influenza virus A-u, and 2 influenza virus B) that were positive by the FilmArray RP and 1/1 positive (influenza virus A) by the Alere assay.
TABLE 1.
Performance of the Alere i Influenza A&B on VTM specimens compared to reference results
| Virus | Genotype | No. detected (Alere/reference)a |
Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance P value | |||
|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/+ | −/− | |||||
| Influenza virus A | Total | 79 b | 0 | 29 | 248 | 73.2 (64.1, 80.61) | 100 (98.47, 100) | <0.001 |
| H1N1-2009 | 34 | 5 | 87.2 (73.29, 94.4) | 100 (98.8,100) | 0.0736 | |||
| H3N2 | 37 | 3 | 92.5 (80.14,97.42) | 100 (98.8,100) | 0.2482 | |||
| Untypeable | 7 | 21 | 25 (12.68, 43.36) | 100 (98.84,100) | <0.001 | |||
| Influenza virus B | 37 | 0 | 1 | 318 | 97.4 (86.5, 99. 53) | 100 (98.88, 100) | 1.0 | |
Four invalid Alere results were excluded from the statistical analysis, leaving a total of 356. +, positive; −, negative.
One FilmArray-negative sample was identified as influenza virus A by the Alere and confirmed by ProFLU+.
Using a combination standard defined as concordant results from two or more of the Alere, FilmArray, and ProFLU+ assays, the clinical performance of the Alere assay was determined (Table 1). The performances of the Alere and reference assays for influenza virus A-1, influenza virus A-3, and influenza virus B were not statistically different; those for influenza virus A-u were statistically different. The specificity for influenza virus A was 100% (95% confidence interval [CI], 98.5 to 100). The sensitivities for the three influenza virus A genotypes were 87.2% for influenza virus A-1, 92.5% for influenza virus A-3, and 25% for influenza virus A-u. For influenza virus B testing, the Alere assay showed excellent agreement with reference results. Only 1 of 38 influenza virus B-positive specimens was missed by the Alere assay. The sensitivity and specificity for influenza virus B were 97.4% (95% CI, 86.5 to 99.5) and 100% (95% CI, 98.9 to 100), respectively.
The greatest comparative difference in sensitivity was observed for influenza virus A-u; only 7 of 28 (25%) of the specimens were positive by the Alere assay. Discordant results were revealed more frequently for specimens with higher CT values in the FilmArray and ProFLU+ assays (Table 2). Among the 29 influenza virus A reference-positive, Alere-negative specimens, 22 (75.9%) had CT values of 25 to 30 by FilmArray RP and 26 (89.7%) had CT values of 30 to 40 by the ProFLU+ assay. The mean CT values in FilmArray RP (27.0 ± 2.6) and ProFLU+ (31.9 ± 2.0) in specimens with discordant results were significantly higher than those obtained for concordant results (17.1 ± 5.4 for FilmArray RP, P < 0.01; 30.1 ± 1.9 for ProFLU+, P = 0.018).
TABLE 2.
Positivity of Alere influenza virus A results and CT values in FilmArray RP and Pro FLU+
| Alere result | No. with a CT value ofa: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FilmArray RP |
Pro FLU+ |
||||||||
| <15.0 | 15.0–19.9 | 20.0–24.9 | 25.0–30.0 | Mean ± SD | <25.0 | 25.0–29.9 | 30.0–40.0 | Mean ± SD | |
| Positiveb | 30 | 23 | 15 | 10 | 17.1 ± 5.4 | 1 | 6 | 4 | 29.5 ± 2.8 |
| Negative | 0 | 0 | 7 | 22 | 27.0 ± 2.6 | 0 | 6 | 23 | 32.0 ± 2.0 |
CT values of 30 and 40 were used to represent FilmArray RP and ProFLU+ negative results, respectively.
One influenza virus A-positive sample determined by Alere and ProFLU+ was excluded.
DISCUSSION
The Alere i Influenza A&B assay described in this study is a novel isothermal-amplification-based integrated system for detection and differentiation of influenza virus A and influenza virus B. It is a fast and user-friendly procedure with a time from specimen collection to results of less than 15 min, including 2 min hands-on time. In this study, we evaluated the performance of the Alere i assay on a panel of archived, frozen NPS VTM samples. Perfect specificity was observed for both influenza virus A and influenza virus B detection, which provides clinicians with the confidence to act appropriately when a positive result is obtained. The influenza virus A sensitivity observed here would translate to 87% after adjusting for the incidence of each subtype in the overall collection (n = 3,675). The lowest sensitivity was observed for the small subset of specimens that were “equivocal” or “untypeable” by the FilmArray RP test; without them, the overall influenza virus A sensitivity would have been 92%. Reference-positive, Alere-negative discordant results were observed more frequently for specimens with higher CT values (i.e., lower viral loads) in FilmArray assays, suggesting that the lower detection rate by the Alere assay for FilmArray influenza virus A-u specimens was associated with the lower viral titer in the diluted NPS VTM. Furthermore, in our study, the 291 (80.8%) NPS specimens tested were from adults, who probably showed less viral shedding than children for many detection methods (7, 29). The Alere assay had excellent performance in influenza virus B detection; only 1 out of 38 influenza virus B-positive specimens was not detected by Alere i Influenza A&B, giving a high sensitivity of 97.4%.
This study has two important limitations. First, the Alere assay accommodates crude swab specimens eluted directly, rather than swabs first diluted in VTM, which may reduce detection of very low-titer VTM samples. Second, the enrolled specimens had been stored frozen for 6 to 9 months before the Alere and Prodesse testing (a circumstance not permitted in the FilmArray package insert), perhaps further reducing the already low titers of the specimens and expanding the impact of sampling statistics on the result distribution. It should be noted, however, that the samples called influenza virus A-u in this study included Biofire “equivocal” and “no subtype detected,” both of which require at least one Biofire retest before declaring the Biofire influenza virus A status, according to the FilmArray RP Instruction booklet (i.e., the influenza virus status was ambiguous), and since this retest was not performed, the Biofire reference assay status was uncertain. Taken together, the results reported here suggest that the performance of the Alere i assay against the Film Array RP assay may depend upon sample handling and the frequency of FilmArray influenza virus A equivocal/untypeable specimens.
The Alere i Influenza A&B assay evaluated in this study has the potential to serve as an alternative to RIDTs, with significantly improved sensitivity. The Alere i system has the advantage of a significantly shorter test time than any currently available molecular assay. The simple, pipette-free procedure runs on a fully integrated, closed, small-footprint system, making the Alere assay potentially suitable for point-of-care testing.
ACKNOWLEDGMENTS
We thank the Clinical Microbiology Service staff of the Memorial Sloan-Kettering Cancer Center for help in collecting clinical specimens. Prodesse PCR was performed by Brion Mermer and Nancy Turcotte at Alere Scarborough.
This study was supported in part by a research agreement between MSKCC and Alere Scarborough (SK2013-0262).
Richard B. Roth is an employee of Ionian Technologies, a subsidiary of Alere Scarborough, the commercial manufacturer of the Alere i Influenza A&B system.
Footnotes
Published ahead of print 2 July 2014
REFERENCES
- 1.Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL. 2012. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J. Clin. Microbiol. 50:364–371. 10.1128/JCM.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond SP, Gagne LS, Stock SR, Marty FM, Gelman RS, Marasco WA, Poritz MA, Baden LR. 2012. Respiratory virus detection in immunocompromised patients with FilmArray respiratory panel compared to conventional methods. J. Clin. Microbiol. 50:3216–3221. 10.1128/JCM.00538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Principi N, Esposito S. 2009. Antigen-based assays for the identification of influenza virus and respiratory syncytial virus: why and how to use them in pediatric practice. Clin. Lab. Med. 29:649–660. 10.1016/j.cll.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 4.De Witte E, Goossens H, Ieven M. 2012. Evaluation of the ESPLINE(R) Influenza A & B-N assay for the detection of influenza A and B in nasopharyngeal aspirates. Eur. J. Clin. Microbiol. Infect. Dis. 31:761–766. 10.1007/s10096-011-1372-1. [DOI] [PubMed] [Google Scholar]
- 5.Deng YM, Caldwell N, Barr IG. 2011. Rapid detection and subtyping of human influenza A viruses and reassortants by pyrosequencing. PLoS One 6:e23400. 10.1371/journal.pone.0023400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal AS, Sarkar M, Chakrabarti S, Rajendran K, Kaur H, Mishra AC, Chatterjee MK, Naik TN, Chadha MS, Chawla-Sarkar M. 2009. Comparative evaluation of real-time PCR and conventional RT-PCR during a 2 year surveillance for influenza and respiratory syncytial virus among children with acute respiratory infections in Kolkata, India, reveals a distinct seasonality of infection. J. Med. Microbiol. 58:1616–1622. 10.1099/jmm.0.011304-0. [DOI] [PubMed] [Google Scholar]
- 7.Ginocchio CC. 2011. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens. Clin. Infect. Dis. 52(Suppl 4):S312–S325. 10.1093/cid/cir046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki FY, Macleod MD, Paggiaro P, Carewicz O, El Sawy A, Wat C, Griffiths M, Waalberg E, Ward P. 2003. Early administration of oral oseltamivir increases the benefits of influenza treatment. J. Antimicrob. Chemother. 51:123–129. 10.1093/jac/dkg007. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2012. Evaluation of 11 commercially available rapid influenza diagnostic tests—United States, 2011–2012. MMWR Morb. Mortal. Wkly. Rep. 61:873–876. [PubMed] [Google Scholar]
- 10.Singh BK, Savill NJ, Ferguson NM, Robertson C, Woolhouse ME. 2010. Rapid detection of pandemic influenza in the presence of seasonal influenza. BMC Public Health 10:726. 10.1186/1471-2458-10-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak-Weekley SM, Marlowe EM, Poulter M, Dwyer D, Speers D, Rawlinson W, Baleriola C, Robinson CC. 2012. Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J. Clin. Microbiol. 50:1704–1710. 10.1128/JCM.06520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Doorn HR, Kinh N, Tuan HM, Tuan TA, Minh NN, Bryant JE, Hang V, Uyen le TT, Thinh le Q, Anh T, Lan NP, Trung NV, Taylor W, Merson L, Wertheim HF, Farrar J, Wolbers M, Chau N, de Jong MD. 2012. Clinical validation of a point-of-care multiplexed in vitro immunoassay using monoclonal antibodies (the MSD influenza test) in four hospitals in Vietnam. J. Clin. Microbiol. 50:1621–1625. 10.1128/JCM.00085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale SE, Mayer C, Mayer MC, Menegus MA. 2008. Analytical and clinical sensitivity of the 3M rapid detection influenza A+B assay. J. Clin. Microbiol. 46:3804–3807. 10.1128/JCM.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG. 2007. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J. Clin. Virol. 39:132–135. 10.1016/j.jcv.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Hannoun C, Tumova B. 2000. Survey on influenza laboratory diagnostic and surveillance methods in Europe. European Scientific Working Group on Influenza. Eur. J. Epidemiol. 16:217–222. [DOI] [PubMed] [Google Scholar]
- 16.Leonardi GP. 2010. Rapid identification of 2009 H1N1 influenza A virus using fluorescent antibody methods. Am. J. Clin. Pathol. 134:910–914. 10.1309/AJCPR7LTR5UUUYDT. [DOI] [PubMed] [Google Scholar]
- 17.Lee GC, Jeon ES, Kim WS, Le DT, Yoo JH, Chong CK. 2010. Evaluation of a rapid diagnostic test, NanoSign(R) Influenza A/B Antigen, for detection of the 2009 pandemic influenza A/H1N1 viruses. Virol. J. 7:244. 10.1186/1743-422X-7-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teo J, Di Pietro P, San Biagio F, Capozzoli M, Deng YM, Barr I, Caldwell N, Ong KL, Sato M, Tan R, Lin R. 2011. VereFlu: an integrated multiplex RT-PCR and microarray assay for rapid detection and identification of human influenza A and B viruses using lab-on-chip technology. Arch. Virol. 156:1371–1378. 10.1007/s00705-011-0999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babady NE, Mead P, Stiles J, Brennan C, Li H, Shuptar S, Stratton CW, Tang YW, Kamboj M. 2012. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J. Clin. Microbiol. 50:2282–2288. 10.1128/JCM.06186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chidlow G, Harnett G, Williams S, Levy A, Speers D, Smith DW. 2010. Duplex real-time reverse transcriptase PCR assays for rapid detection and identification of pandemic (H1N1) 2009 and seasonal influenza A/H1, A/H3, and B viruses. J. Clin. Microbiol. 48:862–866. 10.1128/JCM.01435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandt D, Monecke S, Scott C, Gall A, Hoffmann B, Ehricht R. 2012. Economic high-throughput-identification of influenza A subtypes from clinical specimens with a DNA-oligonucleotide microarray in an outbreak situation. Mol. Cell. Probes 26:6–10. 10.1016/j.mcp.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Tang YW, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, White-Abell J, Quinn CD, Li H, Washington CA, Cromwell J, Giamanco CM, Forman M, Holden J, Rothman RE, Parker ML, Ortenberg EV, Zhang L, Lin YL, Gaydos CA. 2013. Clinical accuracy of a PLEX-ID flu device for simultaneous detection and identification of influenza viruses A and B. J. Clin. Microbiol. 51:40–45. 10.1128/JCM.01978-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jannetto PJ, Buchan BW, Vaughan KA, Ledford JS, Anderson DK, Henley DC, Quigley NB, Ledeboer NA. 2010. Real-time detection of influenza A, influenza B, and respiratory syncytial virus A and B in respiratory specimens by use of nanoparticle probes. J. Clin. Microbiol. 48:3997–4002. 10.1128/JCM.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Bose ME, Beck ET, Fan J, Tiwari S, Metallo J, Jurgens LA, Kehl SC, Ledeboer N, Kumar S, Weisburg W, Henrickson KJ. 2009. Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin. J. Clin. Microbiol. 47:2772–2778. 10.1128/JCM.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maples BK, Holmberg RC, Miller AP, Provins JW, Roth RB, Mandell JG. March 2009. Nicking and extension amplification reaction for the exponential amplification of nucleic acids. US patent US2009/0017453.
- 26.Maples BK, Holmberg RC, Miller AP, Provins TW, Roth RB, Mandell JG. January 2009. Nicking and extension amplification reaction for the exponential amplification of nucleic acids. US patent US2009/0081670.
- 27.Legoff J, Kara R, Moulin F, Si-Mohamed A, Krivine A, Belec L, Lebon P. 2008. Evaluation of the one-step multiplex real-time reverse transcription-PCR ProFlu-1 assay for detection of influenza A and influenza B viruses and respiratory syncytial viruses in children. J. Clin. Microbiol. 46:789–791. 10.1128/JCM.00959-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeffelholz MJ, Pong DL, Pyles RB, Xiong Y, Miller AL, Bufton KK, Chonmaitree T. 2011. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J. Clin. Microbiol. 49:4083–4088. 10.1128/JCM.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewandrowski K, Tamerius J, Menegus M, Olivo PD, Lollar R, Lee-Lewandrowski E. 2013. Detection of influenza A and B viruses with the Sofia analyzer: a novel, rapid immunofluorescence-based in vitro diagnostic device. Am. J. Clin. Pathol. 139:684–689. 10.1309/AJCP7ZTLJCP3LLMA. [DOI] [PubMed] [Google Scholar]