Abstract
Campylobacteriosis is the most commonly reported form of human bacterial gastroenteritis in the world. Sound identification of infectious sources requires subtyping, but the most widely used methods have turnaround times measured in days and require specialist equipment and skills. A multiplex ligation-dependent probe amplification-binary typing (MBiT) assay was developed for subtyping Campylobacter jejuni and Campylobacter coli. It was tested on 245 isolates, including recent isolates from Belgium and New Zealand, and compared to multilocus sequence typing (MLST). When used in an outbreak setting, MBiT identified the predominant genotype and possible additional cases days before pulsed-field gel electrophoresis (PFGE) results were available. MBiT was more discriminatory than MLST and, being a single assay with results produced within 6 h, was more rapid and cost-effective than both MLST and PFGE. In addition, MBiT requires only basic molecular biology equipment and skills.
INTRODUCTION
Campylobacter species, notably Campylobacter jejuni and Campylobacter coli, are the most commonly reported bacterial causes of human gastroenteritis in the world (1). They are widely distributed in nature (2), and ingestion of contaminated food or water is a major route of human infection (3). Identification of sources and infectious trends is often vital for informing control measures, and this process requires subtyping of isolates. A variety of subtyping methods are available for Campylobacter, with the most widely used being multilocus sequence typing (MLST) (4) and pulsed-field gel electrophoresis (PFGE) (5). Neither of these techniques is particularly rapid, with analysis times of several days (6, 7). Data processing can also be complex (7, 8).
With the aim of making subtyping of all C. jejuni isolates achievable for laboratories globally, our group previously developed an 18-target PCR binary typing (P-BIT) system for C. jejuni based on pathogenicity- or survival-associated genes that required no specialized laboratory resources (9). P-BIT does, however, require 18 individual PCRs per isolate. Multiplex ligation-dependent probe amplification (MLPA) is a modification of PCR that allows up to 40 targets to be detected in a single reaction (10). It requires a standard thermal cycler, and PCR products can be visualized using a range of detection platforms. This paper reports on the conversion of the P-BIT system to the MLPA-binary typing (MBiT) format, which allows all 18 products to be examined in a single reaction. We also demonstrate its utility in an outbreak setting.
MATERIALS AND METHODS
MLPA probes were designed for the 18 discriminatory gene targets used in the Campylobacter P-BIT system (9) according to the instructions provided by the pioneers of MLPA, MRC-Holland (Amsterdam, The Netherlands). The probes were manufactured as described by Schouten and colleagues (11) at MRC-Holland. The target genes, specific probe sequences, primer sequences, and product sizes are listed in Table 1.
TABLE 1.
Target genes, specific probe sequences, and primer sequences used in Campylobacter MBiT
| No. | Target gene | Left probe oligonucleotide | Right probe oligonucleotide | Product size (nt)a |
|---|---|---|---|---|
| 1 | tetO | CAGATACAATGAATTTGGAGCGTCAAAGG | GGAATCACTATCCAGACAGCAGTGACATC | 124 |
| 2 | virB8 | GTAAAGGTAGAACAAGCAGGAATGGATATGAG | AGCCGATGAGAATTTATTAAAATCAATTCTCGCAGGCT | 142 |
| 3 | cgtA | CTTTGGATAGCAGGCAATATACTTTCAAGAGAA | GCTTTTAAGGTAATAACTTCATTTTTTACTCGTATAAAAGCCC | 160 |
| 4 | Cj1136 | GTTTTTTGCCTCTTTAGCATTTTCTCCAAAAA | ATTCATACAAGTCTTTAAAATACTCTGACACATTTGCC | 178 |
| 5 | panB | GATCGTGACGCTATCTTAAATGCCTCAAG | ATTTATCAAAGAAAGCCACGCAAATGGGGTAAAAGTGGAAG | 196 |
| 6 | maf5 | CAGCCAGTTCATAAGCCATATGAGATACACTCATG | CCTGCGTTAATATATCCATAATCGTCTAGCCC | 214 |
| 7 | Cj1135 | CGCTTTCATTTTTATTTAAAATTACTCTTGAACCTTGTAAAATTACTTTG | CGTTTTGCAAATTCTAAATGATTTTTTACAAAGTCTTTTTCTAAAATCATATC | 232 |
| 8 | Cj0265 | GGATTTACATAACCATCAATTTTTAGCAAAACCCTGTTA | TTTTCGCTTTTTAATACTTCAAAAGGATTTGTAGGTAAAAGTC | 250 |
| 9 | CJE1733 | CTTTATGTCTTATTTTGATGATGTATCCGCCTTTG | GCTAAGGTTGATTATGCAAAGCTTTCAAAGGT | 268 |
| 10 | Cj0122 | CTAAGCTCTTTTAATAGATATTTTGGAAACAATCCTTTGCAA | ACACTTACCAAAATAAGAGATGAAAGTATTGAAAATGGAAATC | 286 |
| 11 | gmhA2 | GTTAATATGCTAGTATCTGTAGTAAGTGCAATACTTGCGATA | CCGGGTCTATCAAAATAAAACCTACTTACAAATTCCC | 311 |
| 12 | flgE2 | GTTCTTCTTTGTGGACAGCTACAAATATTACATTTACAC | CACAACCTCCTCAAGCAGCTACGAATGT | 338 |
| 13 | CJE1500 | GTTTTGATGATGGTCTAAAAGATCATTACGATTTCG | TTTTCCCTAAACTCTTAGAGCATAAAATTTTTGGATTATTTTTTATACC | 365 |
| 14 | Cj0423 | CTGGCTGATAATTTCTTTGCTAAATTTGTTTTTTGG | TTGGACTATTATTGTTTGGCTTGTTTGTTTAATATGGTCT | 391 |
| 15 | wlaN | CCACAATCATCTACTACAATGATTTCTATATCTTTAAAAGTTTGGTTA | ATGCAACTTTCTAATGCTCTAGCAATATATTTTTCCACAT | 418 |
| 16 | cfrA | CCAAAAAAGTAAGTGATAAATGGGAAACTTCTGTAAG | TTTGGATGCTCTTTTAAATGAAAATAAAGATTGGGGTAATACTT | 445 |
| 17 | Cj1321 | CATGTAATTTTATCACTTTTATACATCCTCAATCTTTTGTTTCA | AAAGAAGCTAAAATAGGACAAGGGGTTATAGTGTGTGT | 473 |
| 18 | Cj0008 | GAAAATAATGTCATAGAATGCCGTAAAAATAGTCCTGAAG | CGTTTGCTGTGTTATCTTTACTTTATCCAAATTTAGATTATAAAAATAATAATTTT | 503 |
| SALSA PCR primers | FAM-GGGTTCCCTAAGGGTTGGA | GTGCCAGCAAGATCCAATCTAGA | NAb |
nt, nucleotides.
NA, not applicable. SALSA PCR primers are used to amplify all MLPA probes once the left and right probe oligonucleotides are ligated.
Each of the 222 C. jejuni, 22 C. coli, and 1 Campylobacter lari isolates listed in Data Set S1 (in the supplemental material) were grown on Columbia sheep blood agar plates at 42°C under microaerobic conditions for 48 to 72 h. Suspensions were prepared in phosphate-buffered saline (BR0014G; Oxoid, Basingstoke, England) to a density of 0.8 using a turbidity meter (Dade International, West Sacramento, CA), and DNA was extracted using the method described previously (9). Each DNA extract was diluted to 20 ng/µl in Tris-EDTA (TE) buffer (Life Technologies) and analyzed using the standard MLPA protocol, SALSA MLPA reagent kits (MRC-Holland), and Campylobacter MBiT probe mixes. The hybridization time was shortened from 16 h to 1 to 2 h. The MLPA products were diluted 1:100 in water and separated using the ABI genetic analyzer 3130XL with POP-7 polymer and GeneScan 600 LIZ size standard (Life Technologies). The data were analyzed using GeneMapper (Life Technologies). The presence or absence of each product was scored as 0 or 1 to make a binary code and converted to a 6-digit type, as described for the earlier P-BIT assay (9).
The ability of a variety of detection methods to differentiate MBiT products was evaluated using three fully sequenced C. jejuni isolates (NCTC11168, RM1221, and RM1864 [81-176]), one fully sequenced C. coli isolate (RM2228), and the control plasmids supplied with the Campylobacter MBiT probe mixes. Each sample was tested using probe mixes containing (i) all 18 probe pairs, (ii) the odd-numbered probe pairs, and (iii) the even-numbered probe pairs (Table 1). The remainder of the MLPA assay was performed as described above. The products were detected (i) diluted 1 in 100 in water on the ABI genetic analyzer as above, (ii) undiluted on the MCE-202 MultiNA microchip electrophoresis system (Shimadzu Corporation, Kyoto, Japan) using standard operating procedures for on-chip mixing with the DNA 1000 reagent kit (Shimadzu), and (iii) undiluted in a 2% agarose gel in 1× Tris-borate-EDTA (TBE) at 110 V for 70 min and visualized using ethidium bromide.
For comparison, C. jejuni and C. coli strains were also analyzed using C. jejuni multilocus sequence typing (MLST) (12). Amplification was performed in a 25-μl volume using AmpliTaq Gold master mixture (Life Technologies) and 5 pmol of each primer. Products were sequenced with the ABI genetic analyzer and alleles were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/) (13). Novel alleles and sequence types (ST) were submitted for clonal complex (CC) designations.
To demonstrate the usefulness of MBiT in an outbreak setting, 51 human C. jejuni and C. coli isolates recovered by Christchurch clinical laboratories over the period in which an outbreak of waterborne gastroenteritis was occurring in the nearby town of Darfield were subtyped using PFGE (14) and MBiT. Either a single colony from the primary isolation plate or a small section of a PFGE plug (1/8 of the size used for PFGE) was added to 5 μl of TE buffer and used as the sample for MBiT analysis.
RESULTS
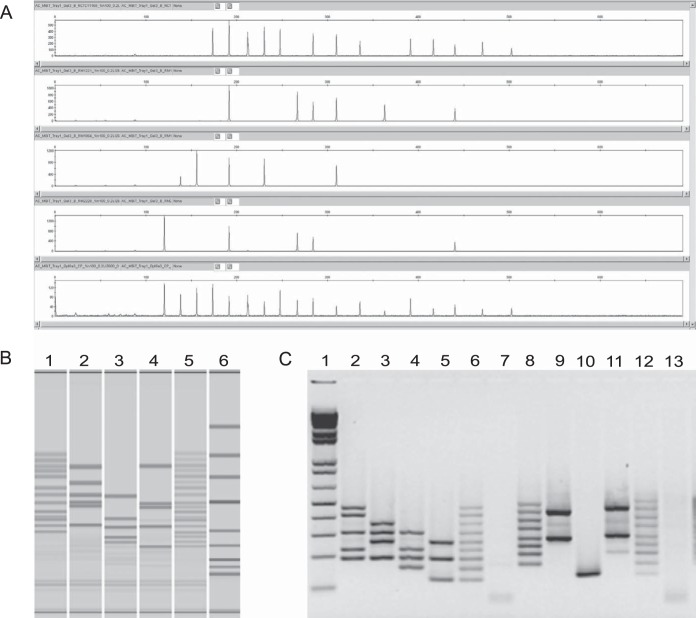
Strain MBiT profiles derived from the genetic analyzer or MultiNA systems were consistent for all samples and probe mixes. MBiT products, with length differences as small as 18 nucleotides, were detectable in as little as 5 min with the MultiNA. Furthermore, the individual MLPA were distinguished using agarose gel electrophoresis when the odd- and even-numbered probes were tested in separate tubes (see Fig. 1 for examples).
FIG 1.
Illustration of alternative Campylobacter MBiT detection methods using fully sequenced isolates and control plasmids. In each image the order is NCTC11168 (MBiT 073767), RM1221 (MBiT 024311), RM1864 (MBiT 621200), RM2228 (MBiT 164101), and control plasmids (MBiT 777777). (A) ABI genetic analyzer; (B) MultiNA with lane 1, Fermentas GeneRuler Low Range DNA Ladder; (C) agarose gel electrophoresis with lane 1, Life Technologies 1 kb Plus DNA Ladder; lanes 7 and 13, TE; lanes 2 to 7, odd-numbered probes only; lanes 8 to 13, even-numbered probes only.
From the 245 strains examined, MBiT produced 120 types with a Simpson's index of diversity (ID) (http://darwin.phyloviz.net/ComparingPartitions) of 0.987. Using MLST, 105 sequence types (ID, 0.979) and 26 clonal complexes (ID, 0.897) were identified.
To compare genomic differences between recently isolated human C. jejuni and C. coli that are geographically distant, we compared MBiT results for the New Zealand strains isolated in 2009 with the Belgian strains isolated in 2010. Of interest, tetO, which encodes tetracycline resistance, and CJE1733, which encodes an arsenical-resistance protein (provisional), were observed more frequently in Belgian isolates than New Zealand isolates (Belgium, 40 isolates; New Zealand, 4 isolates [Fisher Exact, 2-tailed P = 0] and Belgium, 42 isolates; New Zealand, 25 isolates [P = 0.014], respectively). Conversely, Cj1135, a putative two-domain glycosyltransferase, was observed more frequently in isolates from New Zealand (Belgium, 27 isolates; New Zealand, 47 isolates [P = 0.041]).
Of the 51 human C. jejuni and C. coli isolates received by the Institute of Environmental Science and Research (ESR) from Christchurch clinical laboratories during the Darfield waterborne gastroenteritis outbreak, 27 (representing 25 cases) were isolated from cases with home addresses in Darfield. The majority (22/27, 81%) of the Darfield isolates were C. coli with the Sm0131:Kp0132 PFGE profile and 064103 MBiT type, 1 was a C. coli with the Sm0431:Kp0564 PFGE profile and 024101 MBiT type, and the remaining 4 were C. jejuni with different PFGE profiles and MBiT types (Fig. 2).
FIG 2.
Cluster analysis of Campylobacter MBiT data for the isolates submitted during the Darfield waterborne gastroenteritis outbreak, prepared using simple matching and unweighted-pair group method with arithmetic mean (UPGMA).
Isolates from two cases that did not live in Darfield were C. coli with the outbreak PFGE profile and MBiT type. One of these cases, with a Christchurch address, was contacted by a local health protection officer, and it was confirmed that this individual had been working in Darfield and drinking from the town water supply during the exposure period of the outbreak. The other case with the outbreak PFGE profile and MBiT type was not contacted. Three clusters of 2 to 4 strains, each with indistinguishable PFGE and MBiT profiles, from Ashburton or Christchurch cases were also identified. Two other clusters of 2 to 3 strains with indistinguishable profiles comprised cases from different geographical locations.
DISCUSSION
This pilot study demonstrates that MBiT achieved several of the performance criteria first proposed by Struelens (15) and updated by van Belkum and colleagues (16). It has high typeability, with all 245 isolates producing positive results for at least one target gene. It has high discriminatory power, with an ID greater than that of MLST. It has clear applicability to the investigation and identification of outbreaks. Campylobacter MBiT produces results within 6 h and uses readily accessible reagents, equipment, and skills. For laboratories with access to multichannel pipettes and automated electrophoresis equipment, such as the genetic analyzer and MultiNA, the Campylobacter MBiT assay offers an easy, single-tube assay with the potential for high-throughput subtyping of large numbers of isolates. The two-tube testing option required for detection by gel electrophoresis is still easy to perform but uses more reagents and consumables. It also requires controls to run more frequently in order to compensate for fragment migration fluctuations on the gel. Thus, the two-tube system is more expensive than the single-tube option and has limited throughput capability. Nonetheless, by designing the Campylobacter MBiT assay with single- and two-tube options, we have designed a system that is accessible to a wide range of laboratories from (i) reference laboratories with expensive equipment, software, and highly skilled practitioners testing hundreds of isolates a week to (ii) front-line public health laboratories, food industry laboratories, and laboratories in developing countries that are likely to have limited equipment, practitioners with less-developed skills, and fewer isolates to test. The six-digit MBiT code is highly portable and also amenable to computerized analysis and incorporation into electronic databases. All of these features make MBiT a very convenient subtyping system (16). In addition to these performance and convenience criteria, the choice of virulence- and survival-associated genes means the Campylobacter MBiT assay may generate data useful for molecular risk assessment (8) and evaluation of the genetic effects of intervention strategies or changes in industry practices over time.
The differences in carriage of the antibiotic resistance genes tetO and CJE1733 in human isolates from Belgium and New Zealand reflects the prevalence of antimicrobial resistance phenotypes in the two countries. Of the 318 human Campylobacter isolates tested in New Zealand during 2009, none were resistant to erythromycin and 5 (1.6%) were resistant to fluoroquinolone (https://surv.esr.cri.nz/PDF_surveillance/Antimicrobial/AR/National_AR_2009.pdf). Conversely, of a set of 354 human Campylobacter isolates tested in Belgium during 2010, 11 (3.1%), 186 (52.5%), and 204 (57.6%) were resistant to erythromycin, ciprofloxacin (a fluoroquinolone), and ampicillin, respectively (17). Similar differences in antimicrobial resistance rates have been observed in isolates from meat and poultry in the two countries (17, 18).
The carriage of Cj1135, which encodes an enzyme that may be involved in the production of surface-associated sugars, may reflect bacterial cell surface features that increase its ability to infect human intestinal cells and thus may be partially responsible for the high rate of campylobacteriosis in New Zealand (1). Interestingly, Cj1135 has been found to have a higher carriage rate among enteritis-associated strains from The Netherlands, Curaçao, and Japan (19), suggesting a possible human intestinal cell-specific interaction.
The utility of the MBiT assay in outbreak settings was observed in real time during the waterborne gastroenteritis outbreak in Darfield. MBiT results for the first set of samples received by ESR were available early the following morning. These results demonstrated that the majority of isolates from Darfield residents had a single MBiT type and that a Christchurch resident also had the outbreak MBiT type. Rapid follow-up by a health protection officer was possible and confirmed the patient had consumed Darfield town water during the exposure period, and so this case was formally included in the outbreak. PFGE results, which were available 2 days later, confirmed the MBiT results.
Although the P-BIT assay, from which the Campylobacter MBiT was adapted, was designed for C. jejuni, it appears also to provide useful subtyping information for C. coli, as demonstrated by the 12 different MBiT types observed for the 22 C. coli isolates in the validation study and the ability to identify the predominant type in an outbreak involving C. coli. Different MBiT types have so far been observed for C. jejuni and C. coli isolates. The C. lari isolate from the P-BIT paper was also typeable using the Campylobacter MBiT assay. The utility of the assay on Campylobacter sp. other than C. jejuni and C. coli is yet to be established. In future developments of the Campylobacter MBiT assay, we aim to identify targets that provide additional subtyping information for C. coli and potentially other Campylobacter sp.
The identification of several other small clusters of cases with indistinguishable PFGE profiles and MBiT types suggests that small outbreaks may be relatively common. No follow-up investigation was undertaken to identify common infection sources for these small clusters of cases. A small 2002 study of human cases in the same catchment area as the current study also observed numerous small clusters of cases with indistinguishable PFGE profiles (20). A larger 2009 to 2010 study in the same geographical location (21), which also observed clustering of cases based on PFGE, demonstrated how, even with typing data from human, environmental, and food-related isolates, source identification can be difficult. The authors of that paper suggested that a more rapid subtyping method, with turnaround time measured in hours rather than days, would allow more efficient use of epidemiological resources (21), thus facilitating a more rapid response with consequent benefits to public health and economics. Monitoring of small clusters may also facilitate the identification of point sources linked to recurrent events that, taken together, represent significant numbers of human cases and thus are worthy of additional public health resources and interventions. An example would be a producer, supplier, or restaurant linked to several small clusters of divergent types over a period of several months. MBiT is rapid and cheap enough that, for the first time, subtyping of all C. jejuni/C. coli isolated from human cases is possible. This will make it possible to use subtyping data alongside, or potentially to focus, epidemiological data collection to identify, investigate, and monitor clusters of all sizes.
In conclusion, the speed and cost of the Campylobacter MBiT assay make it an ideal candidate for use as a front-line subtyping system for both industrialized and developing countries and make possible the real-time subtyping of all C. jejuni and C. coli isolates. This assay is commercially available from MRC-Holland.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ESR Core Funding from the New Zealand Ministry for Science and Innovation.
Jan Schouten, the founder and owner of MRC-Holland, is the European patent holder for multiplex ligation-dependent probe amplification (MLPA), which is the basis for the assay described in the paper and is now commercially available. Paul van Vught works at MRC-Holland. ESR entered into a collaborative agreement with MRC-Holland on March 5, 2014, facilitating payment of royalties to ESR for the sale of this assay by MRC-Holland.
Footnotes
Published ahead of print 2 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00815-14.
REFERENCES
- 1.World Health Organization. 2012. The global view of campylobacteriosis: report of expert consultation, Utrecht, Netherlands, 9 to 11 July 2012. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Brown PE, Christensen OF, Clough HE, Diggle PJ, Hart CA, Hazel S, Kemp R, Leatherbarrow AJ, Moore A, Sutherst J, Turner J, Williams NJ, Wright EJ, French NP. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501–6511. 10.1128/AEM.70.11.6501-6511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller WG, Mandrell RE. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p 101–163 In Ketley JM, Konkel ME. (ed), Campylobacter. Molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom. [Google Scholar]
- 4.On SLW. 2013. Isolation, identification and subtyping of Campylobacter: where to from here? J. Microbiol. Methods 95:3–7. 10.1016/j.mimet.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 5.On SLW, McCarthy N, Miller WG, Gilpin BJ. 2008. Molecular epidemiology of Campylobacter species, p 191–211 In Blaser MJ, Szymanski CM, Nachamkin I. (ed), Campylobacter, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 6.Ahmed MU, Dunn L, Ivanova EP. 2012. Evaluation of current molecular approaches for genotyping of Campylobacter jejuni strains. Foodborne Pathog. Dis. 9:375–385. 10.1089/fpd.2011.0988. [DOI] [PubMed] [Google Scholar]
- 7.Allerberger F. 2012. Molecular typing in public health laboratories: from an academic indulgence to an infection control imperative. J. Prev. Med. Public Health 45:1–7. 10.3961/jpmph.2012.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taboada EN, Clark CG, Sproston EL, Carrillo CD. 2013. Current methods for molecular typing of Campylobacter species. J. Microbiol. Methods 95:24–31. 10.1016/j.mimet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Cornelius AJ, Gilpin B, Carter P, Nicol C, On SLW. 2010. Comparison of PCR binary typing (P-BIT), a new approach to epidemiological subtyping of Campylobacter jejuni, with serotyping, pulsed-field gel electrophoresis and multilocus sequence typing methods. Appl. Environ. Microbiol. 76:1533–1544. 10.1128/AEM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sellner LN, Taylor GR. 2004. MLPA and MAPH: new techniques for detection of gene deletions. Hum. Mutat. 23:413–419. 10.1002/humu.20035. [DOI] [PubMed] [Google Scholar]
- 11.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acid Res. 30:e57. 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23. 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolley KA, Chan M-S, Maiden MCJ. 2004. mlstbNet—distributed multilocus sequence typing (MLST) databases. BMC Bioinformatics 5:86. 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartholomew N, Brunton C, Gilpin B, Mitchell P, Williamson J. 3 February 2014. A waterborne outbreak of campylobacteriosis in the South Island of New Zealand due to a failure to implement a multibarrier approach. J. Water Health. [DOI] [PubMed] [Google Scholar]
- 15.Struelens MJ. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2–11. 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 16.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13:1–46. 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 17.Federal Agency for the Safety of the Food Chain Scientific Institute of Public Health Veterinary and Agrochemical Research Centre. 2012. Trends and sources 2010 to 2011. Report on zoonotic agents in Belgium. Federal Agency for the Safety of the Food Chain Scientific Institute of Public Health Veterinary and Agrochemical Research Centre, Brussels, Belgium. [Google Scholar]
- 18.Heffernan H, Wong T, Lindsay J, Bowen B, Woodhouse R. 2011. A baseline survey of antimicrobial resistance in bacteria from selected New Zealand foods, 2009 to 2010. MAF Technical Paper 2011/53. Ministry of Agriculture and Forestry, Wellington, New Zealand. [Google Scholar]
- 19.Taboada EN, van Belkum A, Yuki N, Acedillo RR, Godschalk PC, Koga M, Endtz HP, Gilbert M, Nash JH. 2007. Comparative genomic analysis of Campylobacter jejuni associated with Guillain-Barre and Miller Fisher syndromes: neuropathogenic and enteritis-associated isolates can share high levels of genomic similarity. BMC Genomics 8:359. 10.1186/1471-2164-8-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilpin B, Cornelius AJ, Robson B, Boxall N, Ferguson A, Nicol C, Henderson T. 2006. Application of pulsed-field gel electrophoesis to identify potential outbreaks of campylobacteriosis in New Zealand. J. Clin. Microbiol. 44:406–412. 10.1128/JCM.44.2.406-412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilpin BJ, Walshe G, On SL, Smith D, Marshall JC, French NP. 2013. Application of molecular epidemiology to understanding campylobacteriosis in the Canterbury region of New Zealand. Epidemiol. Infect. 141:1253–1266. 10.1017/S0950268812001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.