Abstract
Rapid diagnosis is essential for the management of Staphylococcus aureus infections. A host recognition protein from S. aureus bacteriophage phiSLT was recombinantly produced and used to coat streptavidin latex beads to develop a latex agglutination test (LAT). The diagnostic accuracy of this bacteriophage-based test was compared with that of a conventional LAT, Pastorex Staph-Plus, by investigating a clinical collection of 86 S. aureus isolates and 128 coagulase-negative staphylococci (CoNS) from deep tissue infections. All of the clinical S. aureus isolates were correctly identified by the bacteriophage-based test. While most of the CoNS were correctly identified as non-S. aureus isolates, 7.9% of the CoNS caused agglutination. Thus, the sensitivity of the bacteriophage-based LAT for identification of S. aureus among clinical isolates was 100%, its specificity was 92.1%, its positive predictive value (PPV) was 89.6%, and its negative predictive value (NPV) was 100%. The sensitivity, specificity, PPV, and NPV of the Pastorex LAT for the identification of S. aureus were 100%, 99.2%, 98.9%, and 100%, respectively. Among the additionally tested 35 S. aureus and 91 non-S. aureus staphylococcal reference and type strains, 1 isolate was false negative by each system; 13 and 8 isolates were false positive by the bacteriophage-based and Pastorex LATs, respectively. The ability of the phiSLT protein to detect S. aureus was dependent on the presence of wall teichoic acid (WTA) and corresponded to the production of ribitol phosphate WTA, which is found in most S. aureus clones but only a small minority of CoNS. Bacteriophage-based LAT identification is a promising strategy for rapid pathogen identification. Finding more specific bacteriophage proteins would increase the specificity of this novel diagnostic approach.
INTRODUCTION
Rapid identification of microbial pathogens improves patient management by providing an earlier basis for the choice of an optimal antimicrobial agent (1–3). This is of particular importance in cases of acute and life-threatening infections, such as diseases caused by Staphylococcus aureus (4). Pathogen identification is complicated in situations where causative and rather saprophytic microorganisms of related species may co-occur in diagnostic specimens because of colonization of the same habitats or contamination during specimen collection, transport, or processing. One example with major diagnostic relevance is cocolonization of the skin and mucous membranes by methicillin-susceptible S. aureus (MSSA) and methicillin-resistant coagulase-negative staphylococci (CoNS), which may lead to false-positive results in nucleic acid amplification assays based on the multiple-locus approach designed for the screening of methicillin-resistant S. aureus (MRSA) (5). While S. aureus is a major cause of skin, soft tissue, respiratory, bone, joint, and endovascular infections, CoNS are considered less pathogenic bacteria affecting mainly immunocompromised patients or those with indwelling devices (6). While matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) considerably accelerated the identification of microbes (7), latex agglutination tests (LATs) remain valuable, e.g., for preliminary, very rapid differentiation between S. aureus and CoNS directly while reading cultures on solid media (8, 9). Recent LATs for S. aureus identification are based on the detection of coagulase activity due to the clumping factor, protein A, and capsular polysaccharides 5 and 8. These so-called third-generation LATs are characterized by increased sensitivity; however, problems due to false-positive reactions remain (10–12).
Wall teichoic acid (WTA) is a surface-exposed glycopolymer with a species-specific structure that has been proposed as a target molecule for rapid species detection (13, 14). Because several bacteriophages use WTA to recognize specific host bacteria, corresponding phage-encoded WTA-binding proteins may be suitable tools for rapid diagnostic tests. In this study, we investigated a novel LAT based on an engineered bacteriophage host recognition protein.
(This work was presented in part at the Joint Annual Meeting of the German Society for Hygiene and Microbiology and the German Society for Infectious Diseases, Rostock, Germany, 22 to 25 September 2013 [DVP08].)
MATERIALS AND METHODS
Bacterial strains.
A clinical collection of 86 S. aureus and 128 CoNS sequential isolates (1 isolate per patient) recovered from deep tissue infections (e.g., bone, joint, cardiovascular, and soft tissues) during 2012 was used (Table 1). MALDI-TOF MS (15) and species-specific PCR and/or universal PCR and sequencing approaches (16) were used as reference methods for identification to the species level. Additionally, a collection of 126 staphylococcal reference and type strains including 35 S. aureus and 91 non-S. aureus strains and comprising 55 species and subspecies was tested (see Table S1 in the supplemental material). All bacterial isolates were subcultivated overnight on Columbia blood agar prior to testing.
TABLE 1.
Species distribution among 214 clinical staphylococcal isolates
| Species | No. of isolates |
|---|---|
| S. aureus | 86 |
| Non-S. aureus | 128 |
| S. epidermidis | 89 |
| S. capitis | 15 |
| S. haemolyticus | 9 |
| S. lugdunensis | 6 |
| S. caprae | 4 |
| S. pettenkoferi | 1 |
| S. saprophyticus | 1 |
| S. sciuri | 1 |
| S. simulans | 1 |
| S. warneri | 1 |
Bacteriophage-based LAT.
A host recognition protein, H-SA-BP-1, from S. aureus bacteriophage phiSLT, modified for better solubility and binding affinity, was recombinantly produced in Escherichia coli. The biotinylated protein was used to coat streptavidin latex (Microcoat Biotechnologie GmbH) beads to develop an agglutination test (Hyglos GmbH, Bernried, Germany). We evaluated the diagnostic accuracy of this phage-based test and compared it with that of a conventional LAT, Pastorex Staph-Plus (referred to here as Pastorex; Bio-Rad, Marnes-la-Coquette, France), which is based on the simultaneous detection of the fibrinogen affinity antigen (clumping factor), protein A, and capsular polysaccharides 5 and 8 of S. aureus (17). For each test isolate, one drop of test latex from a dropper bottle was placed into a circle of a paper test card and one drop of control latex as a negative control was placed into another circle of the test card. One to several colonies were removed from the agar with an inoculating loop and emulsified in the latex drop by spreading it over the surface of the circle. The same procedure was performed for Pastorex. The test was considered positive if clearly visible agglutination was observed with the test latex. The bacteriophage-based test was performed in triplicate independently by three investigators, who were blinded to the identities of the isolates tested, to the result of other investigators, and to the Pastorex results.
Flow cytometric analysis of S. aureus cells.
Wild-type S. aureus USA300, its isogenic tagO mutant, and the complemented mutant (18) were used for flow cytometric analysis. To detect bound H-SA-BP-1 protein, 100 μl S. aureus cells (suspension with an optical density at 578 nm of 0.5) were incubated with biotin-labeled H-SA-BP-1 protein (5 μg) in 100 μl Tris-buffered saline–Tween 20-CaCl2–bovine serum albumin buffer, washed, and then incubated with Alexa Fluor 488-streptavidin (1:200; Invitrogen) at 4°C for 1 h. Washed S. aureus cells were sonicated for 15 s to disperse clumped cells before examination by flow cytometry (Accuri C6; Beckman Coulter).
RESULTS
All of the S. aureus isolates from the clinical collection were correctly identified by both the bacteriophage-based test and Pastorex. Among CoNS, nonspecific agglutination of test and control samples in both the phage-based test and Pastorex was observed with one S. epidermidis isolate, which was excluded from the analysis for this reason. While most of the clinical CoNS isolates were correctly identified as non-S. aureus isolates by the phage-based test, 7.9% (10/127) of the CoNS (7 S. epidermidis, 1 S. pettenkoferi, 1 S. saprophyticus, and 1 S. warneri) caused agglutination. Pastorex produced a false-positive result in 0.8% (1/127). The only falsely identified isolate was S. lugdunensis. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the bacteriophage-based test and Pastorex for identification of S. aureus among clinical staphylococcal isolates are shown in Table 2. For 96.9% (124/128) of the clinical isolates, the results of the bacteriophage-based test read by three independent investigators was the same, demonstrating that reading of results was technically easy and unambiguous in most cases.
TABLE 2.
Performance of the bacteriophage-based LAT in comparison with that of the conventional LAT Pastorex for distinguishing S. aureusa from CoNSb
| Method | % Sensitivity | % Specificity | % PPV | % NPV |
|---|---|---|---|---|
| Phage-based test | 100 | 92.1 | 89.6 | 100 |
| Pastorex | 100 | 99.2 | 98.9 | 100 |
n = 86.
n = 128. One S. epidermidis isolate, which caused nonspecific agglutination of test and control latex in both the phage-based test and Pastorex, was excluded from the analysis.
The performance of both tests was more variable when a collection of 126 staphylococcal reference and type strains was tested (see Table S1 in the supplemental material). The bacteriophage-based test and Pastorex each produced one false-negative result. The phage-based test did not identify reference S. aureus strain ATCC 27664, and Pastorex failed to detect type strain S. aureus subsp. anaerobius DSM 20714. The result was uninterpretable because of the nonspecific agglutination in control latex with four and eight of the non-S. aureus reference strains in the bacteriophage-based test and Pastorex, respectively. Nonspecific agglutination was not observed with S. aureus strains. Overall, 14.9% (13/87) and 9.6% (8/83) of the interpretable non-S. aureus reference and type strains were false positive in the bacteriophage-based test and Pastorex, respectively (see Table S1 in the supplemental material).
Because most staphylococcal bacteriophages are known to bind teichoic acids (19, 20), the phiSLT H-SA-BP-1 protein from the bacteriophage-based test was studied for its capacity to bind to S. aureus USA300 and an isogenic WTA-deficient tagO mutant. Of note, the mutant failed to bind H-SA-BP-1 but complementation with a wild-type copy of tagO restored binding (Fig. 1), thereby demonstrating that H-SA-BP-1 requires WTA for binding. Typical S. aureus produces ribitol phosphate WTA, while most CoNS have different WTA types, such as glycerol phosphate WTA (21). In order to determine if the ability of the bacteriophage-based test to detect S. aureus depends on the presence of ribitol phosphate WTA, the binding of H-SA-BP-1 to the rare S. aureus sequence type 395 (ST395) lineage, which produces CoNS-related glycerol phosphate WTA (21), was analyzed. ST395 prototype strain PS187 failed to bind H-SA-BP-1, thereby providing evidence that ribitol phosphate WTA is the specific diagnostic target of H-SA-BP-1.
FIG 1.
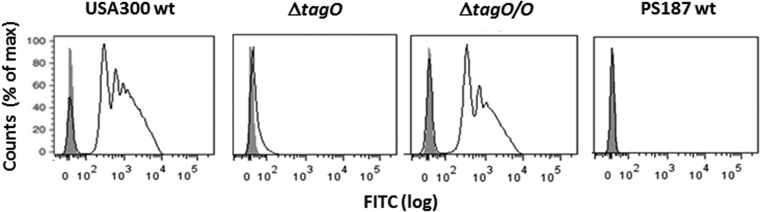
Ribitol phosphate WTA is required for H-SA-BP-1 binding to S. aureus. Results of flow cytometric analyses of H-SA-BP-1 binding to wild-type S. aureus (USA300wt), isogenic WTA-deficient mutant (ΔtagO), plasmid-complemented mutant (ΔtagO/O), and wild-type S. aureus (PS187wt) cells are shown. Shaded and open peaks correspond to samples without and with H-SA-BP-1, respectively. FITC, fluorescein isothiocyanate.
DISCUSSION
Bacteriophages, or phages, are viruses that bind to specific receptors on the bacterial surface to eventually infect the host (22). Phages are utilized for patient treatment in some parts of the world (23), and recombinant bacteriophage endolysins have recently been suggested for use as therapeutic agents against bacterial infections (24–26). Also in diagnostics, the use of phages as a tool for S. aureus typing because of their high specificity has a long history (27, 28). Although it is still in use in some countries (29), this method has been widely replaced by new molecular methods. Recently, novel bacteriophage-based diagnostic applications have appeared, as reviewed elsewhere (22, 30). They include susceptibility testing with phage amplification assays to detect multidrug-resistant pathogens (31–33). Tailspike proteins, which are used by many phages to bind specifically to the cell surface, have recently been suggested for use as molecular probes for bacterial detection (34). Our study investigated a novel application of engineered bacteriophage host-binding protein as a rapid diagnostic tool in the form of a LAT.
Providing results within seconds, manual rapid on-plate tests are useful for preliminary identification of colonies cultivated both from primary sterile body sites, as well as from samples that normally contain colonizing microflora. If staphylococci are found in normally sterile samples, the S. aureus or CoNS differentiation result can swiftly be provided to a clinician by entering it into the clinical computer-based information system directly while reading the culture. Thus, a physician can make decisions on antimicrobial treatment based on local susceptibility data even before confirmative identification (e.g., by MALDI-TOF) and antimicrobial susceptibility testing (AST) have taken place. For critically relevant samples, rapid identification of S. aureus will allow a microbiologist to inform a clinician immediately by phone. In the case of samples from nonsterile body sites, rapid identification of presumable S. aureus colonies within mixed indigenous flora may directly guide the microbiologist's decision to proceed with further diagnostic procedures, i.e., confirmative MALDI-TOF MS identification and AST, rather than to report a sample as containing only normal flora. Even in a well-organized microbiological laboratory in the era of MALDI-TOF MS, it can take a longer time until the cultures in question are forwarded to the MALDI-TOF MS station, the analysis is performed, and the results are validated. In contrast, S. aureus LATs are performed directly during routine plate reading and differentiation between S. aureus and CoNS is available within seconds.
Similar to various other currently available conventional third-generation S. aureus LATs, Pastorex, used as a comparator in this study, represents a rapid LAT based on the detection of clumping factor, protein A, and the type 5 and 8 capsular polysaccharides of S. aureus (8, 9, 17, 35).
While all clinical S. aureus isolates were unambiguously identified by the phage-based test, the rate at which false-positive results were obtained with CoNS was 7.9%. The phage-based test contains latex particles sensitized with a binding molecule highly specific for its WTA target, which is conserved in the cell wall of S. aureus. Although bacteriophages are, in general, highly specific, it was previously shown with recombinant bacteriophage proteins that this specificity is not absolute (24, 25) and that S. aureus bacteriophages can occasionally infect certain CoNS (21). In line with the notion that H-SA-BP-1 binds ribitol phosphate WTA, some of the false-positive CoNS, S. saprophyticus and S. xylosus, are known to produce S. aureus-related ribitol phosphate WTA (36). Among the clinical strains, Pastorex produced only one false-positive result, with an S. lugdunensis isolate. Positive third-generation S. aureus LAT results for CoNS have been previously reported for S. lugdunensis (17) and were attributed to the ability of this species to produce clumping factor (37).
It is noteworthy that both recent studies that evaluated a commercially available phage amplification assay for S. aureus detection and determination of methicillin susceptibility directly from positive blood cultures demonstrated the excellent specificity but moderate sensitivity of this approach (31, 32). This is quite contrary to the results obtained with the test used here, since we observed perfect sensitivity but moderate specificity of the phage-based test. This might be explained by the different characteristics of the phages on which the tests are based. Finding more suitable phages by screening natural habitats, followed by determination of the binding characteristics, or alternative use of molecularly engineered cell wall-binding domains of bacteriophage endolysins (24) would increase the diagnostic utility of phage-based detection methods. An important factor that could contribute to the reduced sensitivity of the phage amplification assay (31, 32) is that it was performed directly with positive blood cultures, which is obviously more challenging than identification from isolated colonies. Application of the bacteriophage-based LAT might theoretically also be possible for direct detection of S. aureus in positive blood cultures. However, the relatively small amount of bacterial cells in positive blood culture broth (compared to the colony biomass), as well as blood and broth components, may hinder detection. Nevertheless, preprocessing of positive blood cultures, e.g., bacterial cell pellet preparation by lysis and centrifugation, could improve detection. Direct determination of methicillin susceptibility would not be possible with such a phage-based LAT, but it might be combined, e.g., with a penicillin binding protein 2a LAT for detection of MRSA. These approaches warrant further studies.
Among the large collection of staphylococcal reference and type strains, one S. aureus reference strain was not identified by each test (see Table S1 in the supplemental material), in contrast to clinical strains, where no false-negative results were obtained with either system. It is noteworthy that S. aureus ATCC 27664, which produced a false-negative result, belongs to the rare ST395 lineage (38) with CoNS-related WTA (21). The proportion of false-positive findings was higher in the reference strain collection than in the clinical strains, too (see Table S1). Some of those species, e.g., S. lugdunensis and S. schleiferi, have been previously reported as falsely identified by Pastorex because of the production of clumping factor (8). The following reasons for false-positive LAT results have also been described: reactivity of S. haemolyticus and S. hominis strains with monoclonal antibodies to S. aureus capsular polysaccharides (11, 17), a noncapsular heat-stable antigen in S. epidermidis (12), unspecific aggregation of latex particles caused by S. saprophyticus (39), cross-reaction of some streptococci with latex because of a protein with an affinity for the Fc fragments of immunoglobulins (40), nonspecific latex agglutination by Candida spp. (10), etc. The worse performance of both tests with the reference strain collection has only limited clinical importance because most of the non-S. aureus species tested are not relevant to humans. However, the potential occurrence of both false-positive and false-negative results demonstrates that microbiologists should be watchful while evaluating LAT results. Additional diagnostics are needed in situations when an unexpected LAT result is observed with isolates exhibiting typical morphology.
Nonspecific agglutination in test and control latex samples of both the phage-based test and Pastorex occurred with one S. epidermidis isolate in the clinical collection (Table 2) and was also observed among reference strains (see Table S1 in the supplemental material). Such nonspecific reactions render the results uninterpretable and have also been reported for S. aureus LATs by others (8, 9, 35, 39). Attention should be paid to such results of diagnostic tests since nonspecific agglutination can also occur with nonrelated microorganisms erroneously picked up from the agar with mixed cultures, e.g., streptococci or Candida spp. (10, 40).
Our study had several limitations. First, this proof-of-principle study was retrospective, using previously characterized staphylococci. A prospective clinical trial is required to understand the true performance characteristics of this assay. This is especially important since medical laboratory personnel using the test would first need to identify suitable colonies on a plate for testing. Second, organisms other than S. aureus and CoNS were not included in the analysis. Characterization of the performance characteristics of this assay using other organisms with Gram stain characteristics or colony morphologies similar to those of staphylococci would be useful and should be performed in the framework of a prospective clinical evaluation. Third, further applications of this assay to the direct detection of S. aureus in clinical specimens would clearly have more impact on early clinical decision making than identification from cultivated colonies. One such application may be S. aureus detection from positive blood cultures, as described above.
In conclusion, bacteriophage-based identification by a LAT is a promising strategy for rapid pathogen identification directly from agar plates. Finding more specific phage proteins would increase the specificity of this novel diagnostic approach.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Barbara Grünastel and Katrin Blaschke for excellent technical assistance.
This study was supported in part by the DZIF (German Center for Infection Research grant TTU 08.807 to K.B., G.P., A.P., and G.X.) and by the German Research Council (grants TRR34 and SFB766 to G.X. and A.P.).
T.W. is an employee of Hyglos GmbH, and S.M. was an employee of Hyglos GmbH. Hyglos GmbH is developing bacteriophage-based bacterial detection tests. The rest of us have no conflict of interest to declare.
Footnotes
Published ahead of print 16 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01432-14.
REFERENCES
- 1.Barenfanger J, Drake C, Kacich G. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doern GV, Vautour R, Gaudet M, Levy B. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trenholme GM, Kaplan RL, Karakusis PH, Stine T, Fuhrer J, Landau W, Levin S. 1989. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J. Clin. Microbiol. 27:1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11–16. 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 5.Becker K, Pagnier I, Schuhen B, Wenzelburger F, Friedrich AW, Kipp F, Peters G, von Eiff C. 2006. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 44:229–231. 10.1128/JCM.44.1.229-231.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr W. 2001. Staphylococcus aureus and Staphylococcus epidermidis: emerging pathogens in nosocomial infections. Contrib. Microbiol. 8:102–107. 10.1159/000060402. [DOI] [PubMed] [Google Scholar]
- 7.Patel R. 2013. MALDI-TOF mass spectrometry: transformative proteomics for clinical microbiology. Clin. Chem. 59:340–342. 10.1373/clinchem.2012.183558. [DOI] [PubMed] [Google Scholar]
- 8.van Griethuysen A, Bes M, Etienne J, Zbinden R, Kluytmans J. 2001. International multicenter evaluation of latex agglutination tests for identification of Staphylococcus aureus. J. Clin. Microbiol. 39:86–89. 10.1128/JCM.39.1.86-89.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weist K, Cimbal AK, Lecke C, Kampf G, Ruden H, Vonberg RP. 2006. Evaluation of six agglutination tests for Staphylococcus aureus identification depending upon local prevalence of meticillin-resistant S. aureus (MRSA). J. Med. Microbiol. 55:283–290. 10.1099/jmm.0.46225-0. [DOI] [PubMed] [Google Scholar]
- 10.Becker K, Almasri AS, von Eiff C, Peters G, Heilmann C, Fegeler W. 2007. Systematic survey of nonspecific agglutination by Candida spp. in latex assays. J. Clin. Microbiol. 45:1315–1318. 10.1128/JCM.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poutrel B, Mendolia C, Sutra L, Fournier JM. 1990. Reactivity of coagulase-negative staphylococci isolated from cow and goat milk with monoclonal antibodies to Staphylococcus aureus capsular polysaccharide types 5 and 8. J. Clin. Microbiol. 28:358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blake JE, Metcalfe MA. 2001. A shared noncapsular antigen is responsible for false-positive reactions by Staphylococcus epidermidis in commercial agglutination tests for Staphylococcus aureus. J. Clin. Microbiol. 39:544–550. 10.1128/JCM.39.2.544-550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown S, Santa Maria JP, Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67:313–336. 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276–287. 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 15.Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics—identification of microorganisms and beyond (minireview). Appl. Microbiol. Biotechnol. 93:965–974. 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 16.Becker K, Harmsen D, Mellmann A, Meier C, Schumann P, Peters G, von Eiff C. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988–4995. 10.1128/JCM.42.11.4988-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier JM, Bouvet A, Mathieu D, Nato F, Boutonnier A, Gerbal R, Brunengo P, Saulnier C, Sagot N, Slizewicz B, Mazie J-C 1993. New latex reagent using monoclonal antibodies to capsular polysaccharide for reliable identification of both oxacillin-susceptible and oxacillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 31:1342–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. 2012. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U. S. A. 109:18909–18914. 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193:4006–4009. 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia G, Wolz C. 2014. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 21:593–601. 10.1016/j.meegid.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 21.Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Broker BM, Penades JR, Nubel U, Holst O, Dandekar T, Peschel A, Xia G. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun. 4:2345. 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smartt AE, Xu T, Jegier P, Carswell JJ, Blount SA, Sayler GS, Ripp S. 2012. Pathogen detection using engineered bacteriophages. Anal. Bioanal. Chem. 402:3127–3146. 10.1007/s00216-011-5555-5. [DOI] [PubMed] [Google Scholar]
- 23.Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659. 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idelevich EA, von Eiff C, Friedrich AW, Iannelli D, Xia G, Peters G, Peschel A, Wanninger I, Becker K. 2011. In vitro activity against Staphylococcus aureus of a novel antimicrobial agent, PRF-119, a recombinant chimeric bacteriophage endolysin. Antimicrob. Agents Chemother. 55:4416–4419. 10.1128/AAC.00217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, Yagyu K, Muraoka A, Sugai M, Hiramatsu K, Honke K, Matsuzaki S. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phiMR11. J. Infect. Dis. 196:1237–1247. 10.1086/521305. [DOI] [PubMed] [Google Scholar]
- 26.Pastagia M, Schuch R, Fischetti VA, Huang DB. 2013. Lysins: the arrival of pathogen-directed anti-infectives. J. Med. Microbiol. 62:1506–1516. 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- 27.Blair JE, Williams RE. 1961. Phage typing of staphylococci. Bull. World Health Organ. 24:771–784. [PMC free article] [PubMed] [Google Scholar]
- 28.Marples RR, Rosdahl VT, Members of the Subcommittee IUMS 1997. International quality control of phage typing of Staphylococcus aureus. J. Med. Microbiol. 46:511–516. 10.1099/00222615-46-6-511. [DOI] [PubMed] [Google Scholar]
- 29.Kali A, Stephen S, Sivaraman U, Kumar S, Joseph NM, Srirangaraj S, Easow JM. 2013. Bacteriophage types of methicillin-resistant Staphylococcus aureus in a tertiary care hospital. Australas. Med. J. 6:496–503. 10.4066/AMJ.2013.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tawil N, Sacher E, Mandeville R, Meunier M. 2014. Bacteriophages: biosensing tools for multi-drug resistant pathogens. Analyst 139:1224–1236. 10.1039/c3an01989f. [DOI] [PubMed] [Google Scholar]
- 31.Bhowmick T, Mirrett S, Reller LB, Price C, Qi C, Weinstein MP, Kirn TJ. 2013. Controlled multicenter evaluation of a bacteriophage-based method for rapid detection of Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 51:1226–1230. 10.1128/JCM.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan KV, Turner NN, Roundtree SS, McGowan KL. 2013. Rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible Staphylococcus aureus (MSSA) using the KeyPath MRSA/MSSA blood culture test and the BacT/ALERT system in a pediatric population. Arch. Pathol. Lab Med. 137:1103–1105. 10.5858/arpa.2012-0422-OA. [DOI] [PubMed] [Google Scholar]
- 33.Albert H, Trollip AP, Mole RJ, Hatch SJ, Blumberg L. 2002. Rapid indication of multidrug-resistant tuberculosis from liquid cultures using FASTPlaqueTB-RIF, a manual phage-based test. Int. J. Tuberc. Lung Dis. 6:523–528. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Arya SK, Glass N, Hanifi-Moghaddam P, Naidoo R, Szymanski CM, Tanha J, Evoy S. 2010. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 26:131–138. 10.1016/j.bios.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Personne P, Bes M, Lina G, Vandenesch F, Brun Y, Etienne J. 1997. Comparative performances of six agglutination kits assessed by using typical and atypical strains of Staphylococcus aureus. J. Clin. Microbiol. 35:1138–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endl J, Seidl HP, Fiedler F, Schleifer KH. 1983. Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch. Microbiol. 135:215–223. 10.1007/BF00414483. [DOI] [PubMed] [Google Scholar]
- 37.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, Nervi C, Fleurette J. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. Syst. Bacteriol. 38:168–172. 10.1099/00207713-38-2-168. [DOI] [Google Scholar]
- 38.Yamada K, Wanchun J, Ohkura T, Murai A, Hayakawa R, Kinoshita K, Mizutani M, Okamoto A, Namikawa T, Ohta M. 2013. Detection of methicillin-resistant Staphylococcus aureus using a specific anti-PBP2a chicken IgY antibody. Jpn. J. Infect. Dis. 66:103–108. 10.7883/yoken.66.103. [DOI] [PubMed] [Google Scholar]
- 39.Gregson DB, Low DE, Skulnick M, Simor AE. 1988. Problems with rapid agglutination methods for identification of Staphylococcus aureus when Staphylococcus saprophyticus is being tested. J. Clin. Microbiol. 26:1398–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kronvall G. 1973. A surface component in group A, C, and G streptococci with non-immune reactivity for immunoglobulin G. J. Immunol. 111:1401–1406. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


