Abstract
We characterized penicillin-susceptible group B streptococcal (PSGBS) clinical isolates exhibiting no growth inhibition zone around a ceftibuten disk (CTBr PSGBS). The CTBr PSGBS isolates, for which augmented MICs of cefaclor and ceftizoxime were found, shared a T394A substitution in penicillin-binding protein 2X (PBP 2X) and a T567I substitution in PBP 2B, together with an additional G429S substitution in PBP 2X or a T145A substitution in PBP 1A, although the T145A substitution in the transglycosidase domain of PBP 1A would have no effect on the level of resistance to ceftibuten.
TEXT
Group B streptococcus (GBS) is one of the principal causes of neonatal sepsis and meningitis. GBS also causes cutaneous and invasive infections in adults, including pregnant women, elderly people, and immunocompromised individuals. Serotypes III and Ia are predominant in isolates from neonates with invasive infections (1, 2). In adults with invasive infections, the most common serotype reported in Japan is Ib, but this serotype is not frequent in other countries (1, 3–5). Penicillin is the first-line antibiotic for treating GBS disease and for intrapartum chemoprophylaxis. We recently reported GBS isolates with reduced penicillin susceptibility (PRGBS), where at least two key amino acid substitutions, V405A and/or Q557E, in penicillin-binding protein 2X (PBP 2X) contribute to a considerable reduction in β-lactam susceptibility (6). Besides these two key substitutions, multiple amino acid substitutions were also found in PBP 2X, PBP 2B, and PBP 1A among PRGBS isolates, depending on the penicillin MIC levels (6–8). After our aforementioned study, PRGBS isolates harboring amino acid substitutions in PBPs were also reported in the United States and Canada (9–11). The prevalence of penicillin nonsusceptibility among GBS isolates from various clinical sources has increased from around 4.5% between 2007 and 2012 to 6.6% in 2013, according to the Japan Nosocomial Infections Surveillance (JANIS) of the Ministry of Health, Labour and Welfare (see http://www.nih-janis.jp). Since a low to moderate increase in penicillin MICs for PRGBS isolates has been observed, it is still very difficult to distinguish them from penicillin-susceptible GBS (PSGBS) isolates in routine susceptibility tests using disk diffusion or microdilution methods (12). However, a reduction in growth inhibition zone diameters or an elevation of the MICs of ceftizoxime and ceftibuten was found to be a good marker for screening of PRGBS (6, 13). Most GBS isolates displaying no growth inhibition zone around a ceftibuten disk were identified to be PRGBS isolates. However, we occasionally noticed a very small number of PSGBS isolates that exhibited no growth inhibition zone around the ceftibuten disk (CTBr PSGBS). The objective of the present study was to characterize these CTBr PSGBS isolates for a better understanding of the mechanisms underlying the reduced susceptibility to cephalosporins and penicillins in GBS.
In May 2011 and January 2012, six clinical isolates of CTBr PSGBS, B1 to B6, were isolated clinically in a general hospital located in Chiba Prefecture, Japan. All of those isolates were serotype Ib, so an additional two PRGBS isolates (A1 and A2) and a PSGBS isolate (B7) with serotype Ib were selected for comparative analyses (Table 1). The diameters of the growth inhibition zones produced around disks containing 30 μg of ceftibuten per disk (6.35-mm diameter; Eiken Chemical Co., Ltd., Tokyo, Japan) were measured by Kirby-Bauer's disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (13, 14). MICs were determined by a broth microdilution method and were interpreted by following CLSI guidelines (14, 15). Ceftizoxime MICs were determined by Etest according to the manufacturer's instructions (Sysmex bioMérieux Co., Ltd., Tokyo, Japan). Sequence analyses of the pbp2x, -2b, and -1a genes were performed as previously described (8, 16). Pulsed-field gel electrophoresis (PFGE) of chromosomal digests with SmaI was performed as described previously by Nagano et al. (8). The sequence type (ST) of each isolate was determined by the protocol for multilocus sequence typing (MLST) as described previously (16, 17). Phylogenetic analysis was performed on the newly identified CTBr PSGBS isolates together with previously reported PRGBS and PSGBS clinical isolates (8, 16) and three reference strains (2603V/R, NEM316, and COH1) to explore the molecular phylogeny of pbp genes among the PSGBS, CTBr PSGBS, and PRGBS isolates. The concatenated pbp2x, pbp2b, and pbp1a sequences were subjected to phylogenetic analyses as described previously (8, 18).
TABLE 1.
Origins and antimicrobial susceptibilities of GBS isolates
| Strain or categorya and isolate no. | Date of isolation (mo/day/yr) | Patient statusb | Wardc | Age (yr) | Sexd | Specimen typee | Serotype | CTB inhibition zone diameterf (mm) | MIC (μg/ml) ofg: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEC | PEN | AMP | CTM | CTX | CRO | CDN | FEP | CFM | ZOXh | MEM | ERY | CLR | CLI | LVX | TET | VAN | |||||||||
| 2603V/R ATCC BAA-611 | V | 19.0 | 1 | 0.06 | 0.12 | 0.5 | ≤0.06 | 0.12 | ≤0.06 | ≤0.5 | 0.5 | 0.064 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | 0.5 | >4 | 0.5 | ||||||
| NEM316 ATCC 12403 | III | 18.7 | 1 | 0.06 | 0.12 | ≤0.5 | ≤0.06 | ≤0.12 | ≤0.06 | ≤0.5 | 0.5 | 0.094 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | 0.5 | >4 | 0.5 | ||||||
| PRGBS | |||||||||||||||||||||||||
| A1 | 12/5/2011 | Out | EM | 99 | M | TTA | Ib | No zone | 16 | 0.5 | 0.25 | >4 | 1 | 0.5 | 0.5 | 1 | 1 | >32 | 0.5 | >1i | >1i | >1i | >8 | ≤0.5 | 0.5 |
| A2 | 1/12/2012 | In | IM 2C | 77 | M | TTA | Ib | No zone | 16 | 0.5 | 0.25 | 4 | 1 | 1 | 0.5 | 1 | >1 | >32 | 0.25 | 1i | 1i | >1i | >8 | ≤0.5 | 0.5 |
| CTBr PSGBS | |||||||||||||||||||||||||
| B1 | 1/8/2012 | In | IM 2C | 77 | M | TTA | Ib | No zone | 8 | 0.12 | 0.12 | 2 | 0.25 | 0.25 | 0.12 | ≤0.5 | 1 | 1.5 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | >8 | ≤0.5 | 0.5 |
| B2 | 5/16/2011 | In | S 4C | 80 | M | Pus (oral cavity) | Ib | No zone | 16 | 0.06 | 0.12 | 1 | 0.12 | ≤0.12 | ≤0.06 | ≤0.5 | 1 | 0.75 | ≤0.12 | >1j | >1j | >1j | >8 | >4 | 0.5 |
| B3 | 9/12/2011 | In | IM 2C | 68 | M | TTA | Ib | No zone | 16 | 0.06 | 0.12 | 1 | 0.12 | ≤0.12 | ≤0.06 | ≤0.5 | 1 | 0.75 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | >8 | ≤0.5 | 0.5 |
| B4 | 9/14/2011 | In | NS 3B | 92 | F | Urine | Ib | No zone | 8 | 0.06 | 0.12 | 0.5 | 0.12 | ≤0.12 | ≤0.06 | ≤0.5 | 0.5 | 0.75 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | >8 | ≤0.5 | 0.5 |
| B5 | 10/18/2011 | In | IM 2C | 64 | M | TTA | Ib | No zone | 8 | 0.06 | 0.12 | 2 | 0.25 | ≤0.12 | 0.12 | ≤0.5 | 1 | 1.5 | ≤0.12 | ≤0.12 | ≤0.12 | 0.12 | >8 | ≤0.5 | 0.5 |
| B6 | 1/18/2012 | In | IM 4W | 94 | M | TTA | Ib | No zone | 8 | 0.06 | 0.12 | 1 | 0.12 | ≤0.12 | ≤0.06 | ≤0.5 | 0.5 | 1 | ≤0.12 | >1k | >1k | >1k | >8 | >4 | 0.5 |
| PSGBS | |||||||||||||||||||||||||
| B7 | 10/17/2011 | Out | EM | 73 | M | Urine | Ib | 17.6 | 2 | 0.06 | 0.12 | 1 | 0.12 | ≤0.12 | ≤0.06 | ≤0.5 | 1 | 0.19 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | >8 | ≤0.5 | 0.5 |
PRGBS, GBS isolates with reduced penicillin susceptibility; CTBr PSGBS, PSGBS with no growth inhibition zones around a ceftibuten disk; PSGBS, penicillin-susceptible GBS.
Out, outpatient; In, inpatient.
S, surgery; IM, internal medicine; NS, neurosurgery; EM, emergency.
M, male; F, female.
TTA, transtracheal aspirate.
Determined by standard disk diffusion test. CTB, ceftibuten.
CEC, cefaclor; PEN, penicillin; AMP, ampicillin; CTM, cefotiam; CTX, cefotaxime; CRO, ceftriaxone; CDN, cefditoren; FEP, cefepime; CFM, cefixime; ZOX, ceftizoxime; MEM, meropenem; ERY, erythromycin; CLR, clarithromycin; CLI, clindamycin; LVX, levofloxacin; TET, tetracycline; VAN, vancomycin.
MIC results of the Etest.
Positive for ermT/R.
Positive for ermB, ermT/R, and mefA/E.
Positive for ermB.
Eight clinical isolates, including six CTBr PSGBS isolates and two PRGBS isolates, exhibited no growth inhibition zones around the ceftibuten disks. Those isolates showed increased cefaclor MICs (8 to 16 μg/ml) compared to those for the reference PSGBS isolates (Table 1). However, considerable differences were observed in ceftizoxime MIC levels among the CTBr PSGBS, PRGBS, and reference isolates. CTBr PSGBS and the reference isolates were all susceptible to penicillin. Nonsusceptibility to penicillin, cefotaxime, and cefepime was found in two of the PRGBS isolates tested. As shown in Table 2, two amino acid substitutions, T394A in PBP 2X and T567I in PBP 2B, were shared among the CTBr PSGBS isolates. A unique amino acid substitution, G429S in PBP 2X, was also observed among CTBr PSGBS isolates B1, B3, and B5. Moreover, a T145A substitution in PBP 1A was also found among CTBr PSGBS isolates B1, B3, and B5. The PRGBS isolates A1 and A2 shared A400V, V405A, and Q557E substitutions in PBP 2X and a T567I substitution in PBP 2B (Table 2).
TABLE 2.
MLST and amino acid substitutions in PBPs among serotype Ib GBS clinical isolates
| Categorya and isolate no. | MLST sequence type | Amino acid substitution inb: |
||||||
|---|---|---|---|---|---|---|---|---|
| PBP 2X | PBP 2B | PBP 1A | ||||||
| PRGBS | ||||||||
| A1 | 1 | —c | A400V | V405A | — | Q557E | T567I | — |
| A2 | 1 | — | A400V | V405A | — | Q557E | T567I | — |
| CTBr PSGBS | ||||||||
| B1 | 1 | T394A | — | — | G429S | — | T567I | — |
| B2 | 1 | T394A | — | — | — | — | T567I | T145A |
| B3 | 1 | T394A | — | — | G429S | — | T567I | — |
| B4 | 1 | T394A | — | — | — | — | T567I | T145A |
| B5 | 1 | T394A | — | — | G429S | — | T567I | — |
| B6 | 1 | T394A | — | — | — | — | T567I | T145A |
| PSGBS | ||||||||
| B7 | 10 | — | — | — | — | — | — | — |
PRGBS, GBS isolates with reduced penicillin susceptibility; CTBr PSGBS, PSGBS with no growth inhibition zones around a ceftibuten disk; PSGBS, penicillin-susceptible GBS.
Amino acid substitutions compared to the sequences of Streptococcus agalactiae strains 2603V/R and NEM316.
—, no substitution was observed on the basis of the amino acid sequences of the reference strains described above.
PFGE patterns of the CTBr PSGBS isolates were mutually different, although a probable close genetic relatedness was suggested between clinical isolates B1 and B5. PRGBS isolates A1 and A2, which were recovered separately from an outpatient and an inpatient, respectively, shared the same PFGE profile. Note that CTBr PSGBS isolate B1 and PRGBS isolate A2 showed different PFGE patterns, despite being isolated from the same inpatient. CTBr PSGBS and PRGBS isolates were assigned to ST1, a founder of clonal complex 1 (CC1) (Table 2).
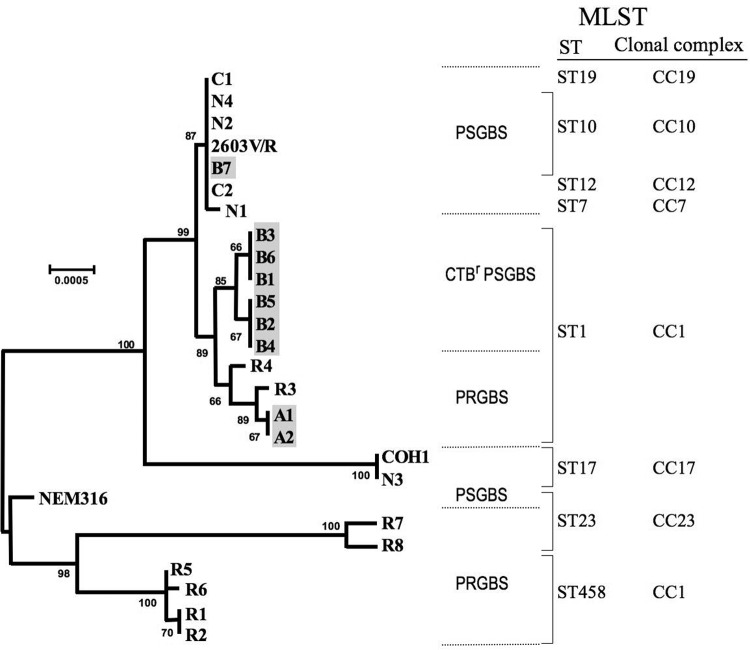
In the phylogenetic analyses of concatenated pbp genes shown in Fig. 1, CTBr PSGBS isolates B1 through B6 formed a clade and then formed a sister clade with four PRGBS isolates, A1 and A2 from this study and R3 and R4 from a previous report (8). All 10 of these isolates belonged to ST1. The PSGBS isolate B7 formed a clade together with other PSGBS isolates, including those from reference strain 2603V/R, which were assigned to ST10.
FIG 1.
Correlation between phylogenetic analysis of GBS isolates based on concatenated alignment of sequenced pbp2x, pbp2b, and pbp1a genes and MLST sequence types. The CTBr PSGBS isolates B1 to B6, PRGBS isolates A1 and A2, and PSGBS isolate B7, characterized in the present study, are shaded. Other isolates, such as PRGBS isolates R1 to R8 and PSGBS isolates N1 to N4, were reported previously (8). Ten PRGBS clinical isolates (strain numbers 1, 2-1, 2-2, 3, 4-1, 4-2, 5, 6, 7, and 8) caused a probable nosocomial transmission in a general hospital (16) and were identical to isolate R5 in the viewpoint of pbp phylogeny. Three reference strains, 2603V/R (ATCC BAA-611, GenBank accession number NC004116), NEM316 (ATCC 12403, GenBank accession number NC004368), and COH1 (GenBank accession number AAJR01000000) are included. Bootstrap support values (500 replicates) are shown as percentages. Scale bars indicate the expected number of changes per sequence position.
PRGBS isolates were reported in the United States and Canada after our first investigation of their emergence and molecular mechanisms, but they are still rare (6–11). Domestically, our subsequent investigations revealed the increase of PRGBS among clinical GBS isolates, their tendency toward resistance to multiple drugs, and a probable association of such a multidrug-resistant PRGBS clone with nosocomial spread (16). Most of those PRGBS isolates belonged to the MLST CC1, which includes the ancestral genotype ST1 (16, 19). In the present study, we report the molecular characteristics of CTBr PSGBS. To date, disappearance of a growth inhibition zone around a ceftibuten disk or an elevation in the cefaclor MIC has not been described in penicillin-susceptible β-hemolytic streptococci. The common substitutions found in the CTBr PSGBS isolates (T394A in PBP 2X and T567I in PBP 2B) have already been found in several PRGBS clinical isolates (6, 8, 16), but the contribution of these amino acid substitutions to β-lactam resistance remains unclear. The G429S substitution in PBP 2X was found to be a different amino acid substitution at the same position with the G429D substitution that was previously identified in PRGBS isolates (7). The T145A substitution in PBP 1A, which was the substitution on the N-terminal side of the transglycosylase domain, was not detected in previously identified PRGBS isolates, although the T145A substitution would have no effect on the ceftibuten resistance phenotype. Because the PFGE patterns of six CTBr PSGBS isolates differed somewhat from each other, the hospital transmission of a single CTBr PSGBS clone during a short period would be rather unlikely. The PFGE profiles of the CTBr PSGBS isolates also differed from those of the serotype Ib PRGBS isolates A1 and A2, which were isolated during the same study period and had the apparent key substitutions V405A and Q557E in PBP 2X. Interestingly, PRGBS isolate A2 was isolated 4 days after the isolation of CTBr PSGBS isolate B1 from transtracheal aspirate cultures from the same patient, and both isolates shared a T567I substitution in PBP 2B, but they did not have any common substitutions in PBP 2X, as shown in Table 2. These findings contradicted our initial assumption that the CTBr PSGBS isolate might change to a PRGBS isolate after acquisition of reduced susceptibility to penicillin through accumulating mutations in its pbp genes. The CTBr PSGBS and PRGBS isolates, which have emerged independently from an ancestral PSGBS strain, may have cocolonized in the respiratory tract of the host. This speculation would be supported by the findings that six CTBr PSGBS isolates and two PRGBS isolates were all assigned to ST1, and these two groups formed sister clades with each other in the phylogenetic tree of the pbp genes. The EUCAST recently established clinical breakpoints for penicillin and streptococcus groups A, B, C, and G (http://www.eucast.org/clinical_breakpoints/), including a resistance criterion for penicillin at an MIC of >0.25 μg/ml. The EUCAST also notes that β-lactam susceptibility, including cefaclor and ceftibuten susceptibility in β-hemolytic streptococcus groups A, B, C, and G, is inferred from penicillin susceptibility. Cefaclor is one of the most widely used oral cephalosporins for the empirical treatment of acute upper respiratory infections in Japan, thus allowing for various bacterial species to be exposed to this drug. Cefaclor and ceftibuten are not included in the therapeutic options for GBS infections. However, the emergence of CTBr PSGBS clones would pose a possible problem because GBS isolates susceptible to penicillin would not necessarily be susceptible to other β-lactams, including cephalosporins, depending on the amino acid substitution(s) acquired in their PBPs.
To our knowledge, this is the first report describing the presence of a serotype Ib PSGBS lineage (i.e., CTBr PSGBS) demonstrating unique susceptibility profiles to several cephalosporins. Lineages of CTBr PSGBS and PRGBS, which displayed a sister clade relationship, shared an amino acid substitution (T567I) in the transpeptidase domain of PBP 2B and were assigned to ST1, suggesting a common origination. Since PBPs play an important role in bacterial viability through maintaining cellular integrity and shape, amino acid substitutions randomly acquired in PBPs would usually provide disadvantages for bacterial growth. Once a pbp gene mutation resulting in an amino acid substitution relevant to the resistance to β-lactams occurs in GBS, it might change to PRGBS by accumulating mutations in its PBPs, or it might evolve into CTBr PSGBS at a low enough frequency to survive in the presence of oral cephalosporins, such as cefaclor and ceftibuten. Our findings will contribute to a better understanding of the future development of resistance to β-lactams in GBS.
Nucleotide sequence accession numbers.
The pbp gene sequences from representative CTBr PSGBS isolates B1 and B2 determined in this study have been deposited in GenBank under accession numbers AB819280 to AB819285.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Health, Labour and Welfare of Japan (grants H21-Shinkou-Ippan-008 and H24-Shinkou-Ippan-010) and in part by research grants for medical science from the Takeda Science Foundation (2012).
We are deeply grateful to Satowa Suzuki of the Department of Bacteriology II, National Institute of Infectious Diseases, for scientific advice in relation to epidemiology. We also thank Kyoko Osaki of Funabashi Futawa Hospital for her excellent technical assistance.
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core Surveillance/Emerging Infections Program Network 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299:2056–2065. 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 2.Poyart C, Réglier-Poupet H, Tazi A, Billoët A, Dmytruk N, Bidet P, Bingen E, Raymond J, Trieu-Cuot P. 2008. Invasive group B streptococcal infections in infants, France. Emerg. Infect. Dis. 14:1647–1649. 10.3201/eid1410.080185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins ER, Melo-Cristino J, Ramirez M. 2012. Dominance of serotype Ia among group B streptococci causing invasive infections in nonpregnant adults in Portugal. J. Clin. Microbiol. 50:1219–1227. 10.1128/JCM.05488-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murayama SY, Seki T, Sakata H, Sunaoshi K, Nakayama E, Iwata S, Sunakawa K, Ubukata K, Invasive Streptococcal Disease Working Group 2009. Capsular type and antibiotic resistance in Streptococcus agalactiae isolates from patients, ranging from newborns to the elderly, with invasive infections. Antimicrob. Agents Chemother. 53:2650–2653. 10.1128/AAC.01716-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tazi A, Morand PC, Réglier-Poupet H, Dmytruk N, Billoët A, Antona D, Trieu-Cuot P, Poyart C. 2011. Invasive group B streptococcal infections in adults, France (2007–2010). Clin. Microbiol. Infect. 17:1587–1589. 10.1111/j.1469-0691.2011.03628.x. [DOI] [PubMed] [Google Scholar]
- 6.Kimura K, Suzuki S, Wachino J, Kurokawa H, Yamane K, Shibata N, Nagano N, Kato H, Shibayama K, Arakawa Y. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:2890–2897. 10.1128/AAC.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagano N, Kimura K, Nagano Y, Yakumaru H, Arakawa Y. 2009. Molecular characterization of group B streptococci with reduced penicillin susceptibility recurrently isolated from a sacral decubitus ulcer. J. Antimicrob. Chemother. 64:1326–1328. 10.1093/jac/dkp374. [DOI] [PubMed] [Google Scholar]
- 8.Nagano N, Nagano Y, Kimura K, Tamai K, Yanagisawa H, Arakawa Y. 2008. Genetic heterogeneity in pbp genes among clinically isolated group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:4258–4267. 10.1128/AAC.00596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahesh S, Hensler ME, Van Sorge NM, Gertz RE, Jr, Schrag S, Nizet V, Beall BW. 2008. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 52:2915–2918. 10.1128/AAC.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudreau C, Lecours R, Ismaïl J, Gagnon S, Jetté L, Roger M. 2010. Prosthetic hip joint infection with a Streptococcus agalactiae isolate not susceptible to penicillin G and ceftriaxone. J. Antimicrob. Chemother. 65:594–595. 10.1093/jac/dkp458. [DOI] [PubMed] [Google Scholar]
- 11.Longtin J, Vermeiren C, Shahinas D, Tamber GS, McGeer A, Low DE, Katz K, Pillai DR. 2011. Novel mutations in a patient isolate of Streptococcus agalactiae with reduced penicillin susceptibility emerging after long term oral suppressive therapy. Antimicrob. Agents Chemother. 55:2983–2985. 10.1128/AAC.01243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura K, Nagano N, Nagano Y, Wachino J, Shibayama K, Arakawa Y. 2013. Ability of the VITEK 2 system to detect group B streptococci with reduced penicillin susceptibility (PRGBS). J. Antimicrob. Chemother. 68:1442–1444. 10.1093/jac/dkt008. [DOI] [PubMed] [Google Scholar]
- 13.Kimura K, Wachino J, Kurokawa H, Suzuki S, Yamane K, Shibata N, Arakawa Y. 2009. Practical disk diffusion test for detecting group B streptococcus with reduced penicillin susceptibility. J. Clin. Microbiol. 47:4154–4157. 10.1128/JCM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard—8th ed. CLSI M7-A8 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Nagano N, Nagano Y, Toyama M, Kimura K, Tamura T, Shibayama K, Arakawa Y. 2012. Nosocomial spread of multidrug-resistant group B streptococci with reduced penicillin susceptibility belonging to clonal complex 1. J. Antimicrob. Chemother. 67:849–856. 10.1093/jac/dkr546. [DOI] [PubMed] [Google Scholar]
- 17.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530–2536. 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150–163. 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 19.Kimura K, Nagano N, Nagano Y, Wachino J, Suzuki S, Shibayama K, Arakawa Y. 2011. Predominance of sequence type 1 group with serotype VI among group B streptococci with reduced penicillin susceptibility identified in Japan. J. Antimicrob. Chemother. 66:2460–2464. 10.1093/jac/dkr352. [DOI] [PubMed] [Google Scholar]