Abstract
Three unrelated sequence type 131 (ST131), ST58, and ST83 Escherichia coli isolates with low-level resistance to imipenem and resistance to ertapenem were recovered in a Spanish hospital from July to October 2012. They were positive for blaOXA-48 carried by an IncL/M conjugative plasmid, which may have been acquired from Klebsiella pneumoniae.
TEXT
Resistance to carbapenem antibiotics in Gram-negative bacteria has become an increasingly important problem worldwide over the last 2 decades (1). The most relevant mechanism for this resistance is the production of carbapenemases, with the responsible genes often carried by plasmids which contribute to their spread by horizontal gene transfer (2). OXA-48 is a class D β-lactamase which confers resistance to penicillins and weak, but nonetheless significant, resistance to carbapenems but not to extended-spectrum cephalosporins (3). This enzyme was first identified in 2004 in Turkey and since then has been reported in many other countries, mainly those of the Middle East, North Africa, and Europe, but also in the United States (4–6). OXA-48 is most often detected in Klebsiella pneumoniae, although other members of the Enterobacteriaceae family, including Escherichia coli, also produce it (3). In K. pneumoniae, the blaOXA-48 gene has been located in a conjugative plasmid of about 60 kb assigned to the IncL/M group (7). Here, we report the characterization of three E. coli OXA-48-producing isolates, each obtained from a different patient at a Spanish hospital (Hospital Universitario Central de Asturias [HUCA]) over a 3-month period (Table 1). These are the first OXA-48-producing E. coli isolates detected in our region, and only one was previously reported in Spain (8). The three patients underwent surgery, were hospitalized for long periods, were coinfected with other bacteria, and received prolonged treatment with numerous antimicrobials.
TABLE 1.
Characteristics of patients and clinical samples positive for E. coli or for E. coli and K. pneumoniae isolates carrying the blaOXA-48 gene and recovered in a Spanish hospital
| Patient no. (sexa/age [yr]) | Hospital unit | Disease/outcome | Sample origin | Isolate(s)b | Date of isolation (day/mo/yr) | Final therapy |
|---|---|---|---|---|---|---|
| 1 (F/46) | General surgery | Surgical wound infection/discharged | Colostomy | Ec-HUCA 1, Kp-HUCA 4 (Morganella morganii) | 16/7/2012 | Ciprofloxacin |
| 2 (M/57) | ICUc | Surgical wound infection/discharged | Wound exudate | Ec-HUCA 2 (Enterococcus faecalis) | 15/10/2012 | Ciprofloxacin, cefoxitin, vancomycin |
| 3 (M/68) | Reanimation | Septic shockd/discharged | Urine | Ec-HUCA 3 | 26/10/2012 | Colistin, meropenem, amikacin |
F, female; M, male.
Ec, Escherichia coli; Kp, Klebsiella pneumoniae; HUCA, Hospital Universitario Central de Asturias.
ICU, intensive care unit.
Septic shock was caused by Pseudomonas aeruginosa, which was not recovered from urine.
The first E. coli isolate (Ec-HUCA 1) was recovered in July 2012 from a colostomy specimen from a 46-year-old female (patient 1) with postsurgical septic shock. The colostomy specimen was also positive for K. pneumoniae (isolate Kp-HUCA 4, which was additionally detected in blood cultures for the same patient and partially characterized in this study) and Morganella morganii. The second isolate (Ec-HUCA 2) was recovered in October 2012 from a surgical wound in a 57-year-old male (patient 2) with intestinal obstruction. After empirical treatment with meropenem, the isolate was detected in the wound drainage, as was Enterococcus faecalis. The third isolate (Ec-HUCA 3) was found in a urine sample from a 68-year-old male (patient 3) with several complications after septic shock caused by multidrug-resistant Pseudomonas aeruginosa secondary to a surgical wound infection (Table 1).
Antimicrobial susceptibility tests were performed by disk diffusion assays (Becton Dickinson, Sparks, MD, USA). The Microscan system (Neg Combo Panel Type 53; Siemens Healthcare Diagnostics, Deerfield, IL, USA) was applied for the identification of bacterial isolates and the determination of MICs. The Etest (bioMérieux, Marcy-l'Étoile, France) was used to determine the MIC for tigecycline and to confirm the MIC for ertapenem. The results of the disk diffusion assays (not shown) and the MICs (Table 2) were interpreted according to CLSI breakpoints that were updated in January 2013 (9). As shown in Table 2, the resistance pattern was different for each of the three E. coli isolates. Note that each isolate showed intermediate susceptibility to imipenem (MICs of 2 mg/liter) and resistance to ertapenem (MICs of 2 mg/liter), and they were positive for the modified Hodge test recommended for the detection of carbapenemases (9). Ec-HUCA 2 was also resistant to third-generation cephalosporins. The K. pneumoniae isolate recovered from patient 1 was resistant to third-generation cephalosporins and ertapenem but susceptible to imipenem. Apart from resistance to β-lactam antibiotics, resistance to nalidixic acid, ciprofloxacin, tobramycin, and/or trimethoprim-sulfamethoxazole was detected in some of the isolates (Table 2).
TABLE 2.
MICs for blaOXA-48-positive isolates and their transconjugantsa
| Antimicrobial agent | MIC (μg/ml) for: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ec-HUCA 1 (ST131) | Tc-1/1 | Tc-1/2 | Ec-HUCA 2 (ST58) | Tc-2/1 | Tc-2/2 | Ec-HUCA 3 (ST83) | Tc-3/1 | Tc-3/2 | Kp-HUCA 4 | Tc-4/1 | Tc-4/2 | |
| Amoxicillin | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Amoxicillin-clavulanic acid | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 | >16/8 |
| Piperacillin-tazobactam | >64 | 64 | 64 | >64 | >64 | >64 | >64 | 64 | >64 | >64 | 16 | 32 |
| Cefazolin | >16 | ≤8 | ≤8 | >16 | >16 | ≤8 | >16 | ≤8 | >16 | >16 | ≤8 | ≤8 |
| Cefuroxime | >16 | ≤8 | ≤8 | >16 | >16 | ≤8 | ≤8 | 8 | ≤8 | >16 | ≤8 | ≤8 |
| Cefoxitin | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | < = 8 | ≤8 | ≤8 |
| Cefotaxime | ≤1 | ≤1 | ≤1 | >32 | >32 | ≤1 | ≤1 | ≤1 | ≤1 | >32 | ≤1 | ≤1 |
| Ceftazidime | ≤1 | ≤1 | ≤1 | >16 | >16 | ≤1 | ≤1 | ≤1 | ≤1 | >16 | ≤1 | ≤1 |
| Cefepime | ≤1 | ≤1 | ≤1 | >16 | >16 | ≤1 | ≤1 | ≤1 | ≤1 | >16 | ≤1 | ≤1 |
| Aztreonam | ≤1 | ≤1 | ≤1 | >16 | >16 | ≤1 | ≤1 | ≤1 | ≤1 | >16 | ≤1 | ≤1 |
| Imipenem | 2 | ≤1 | ≤1 | 2 | 2 | 2 | 2 | ≤1 | 2 | ≤1 | ≤1 | 1 |
| Ertapenem | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 1 | 1 |
| Gentamicin | ≤2 | ≤2 | ≤2 | 8 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| Tobramycin | ≤2 | ≤2 | ≤2 | >8 | >8 | ≤2 | ≤2 | ≤2 | ≤2 | >8 | ≤2 | ≤2 |
| Amikacin | ≤8 | ≤8 | ≤8 | 32 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 |
| Acid nalidixic | >16 | ≤16 | ≤16 | ≤16 | ≤16 | ≤16 | ≤16 | ≤16 | ≤16 | >16 | ≤16 | ≤16 |
| Ciprofloxacin | 1 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | >2 | ≤0.5 | ≤0.5 |
| Trimethoprim-sulfamethoxazole | ≤2/38 | ≤2/38 | ≤2/38 | >2/38 | >2/38 | ≤2/38 | ≤2/38 | ≤2/38 | ≤2/38 | >2/38 | ≤2/38 | ≤2/38 |
| Tigecycline | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| β-Lactamase gene(s) | blaOXA-48, blaTEM-1 | blaOXA-48 | blaOXA-48 | blaOXA-48, blaTEM-1, blaCTX-M-15 | blaOXA-48, blaCTX-M-15 | blaOXA-48 | blaOXA-48 | blaOXA-48 | blaOXA-48 | blaOXA-48, blaCTX-M-15 | blaOXA-48 | blaOXA-48 |
| Relevant plasmid(s) (kb) | ca. 60 | ca. 60 | ca. 60 | ca. 60, <150 | ca. 60, <150 | ca. 60 | ca. 60 | ca. 60 | ca. 60 | ca. 60, >150 | ca. 60 | ca. 60 |
Four and 20 transconjugants (Tc) were analyzed in matings using as donors clinical isolates of Escherichia coli (Ec) and Klebsiella pneumoniae (Kp) recovered in the Hospital Universitario Central de Asturias (HUCA). A rifampin-resistant derivative of E. coli J53 was used as the recipient, and selection was performed with rifampin and ertapenem. For each mating, results from two independent transconjugants are shown (see the text for details).
The genes responsible for resistance to carbapenems and extended-spectrum cephalosporins were identified by PCR amplification (10, 11) followed by sequencing. The presence of blaOXA-48 was demonstrated in the three E. coli isolates and Kp-HUCA 4, and blaCTX-M-15 was detected in Ec-HUCA 2 and Kp-HUCA 4. To establish the location of these genes, plasmid DNA extracted from each isolate (12) was hybridized with probes specific for the two genes amplified from Kp-HUCA 4 and labeled as previously reported (13). The same gel was also hybridized with phage λ DNA (Fermentas GmbH, Madrid, Spain), included as a control. The λ probe was generated by random-primed DNA labeling with digoxigenin-dUTP using the DIG DNA labeling kit (Roche Applied Sciences). As shown in Fig. 1a, each isolate displayed a distinct profile, including a common plasmid of ca. 60 kb accompanied by one or more plasmids of various sizes. The ca. 60-kb plasmid hybridized with the blaOXA-48 probe in the four isolates (Fig. 1b), while the blaCTX-M-15 mapped on plasmids slightly larger or slightly smaller than 150 kb in Kp-HUCA 4 and Ec-HUCA 2, respectively (Fig. 1c).
FIG 1.
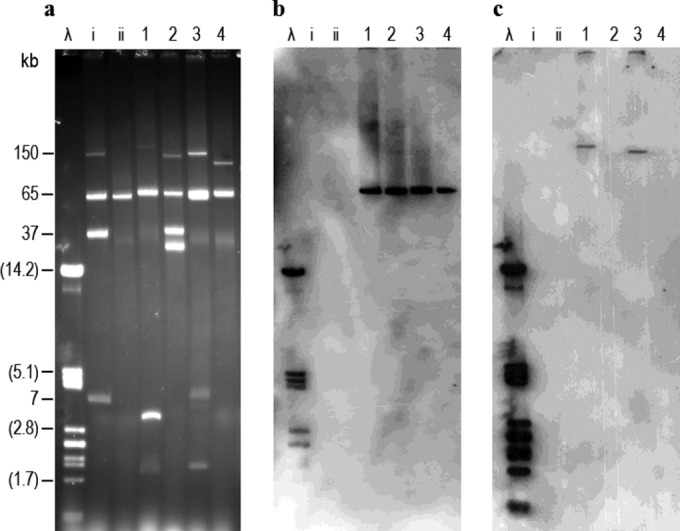
Plasmid profiles of Klebsiella pneumoniae (Kp) and Escherichia coli (Ec) isolates producing OXA-48 carbapenemase (a) and hybridization with blaOXA-48 (b) and blaCTX-M-15 (c) probes. Lanes λ, phage λ DNA digested with PstI, used as reference for the hybridization experiments (the sizes of some of the linear fragments are shown in parentheses); lanes i and ii, plasmids obtained from E. coli V517 (NCTC 50192) and plasmid RP4 used as molecular size standards for undigested DNA; lanes 1, Kp-HUCA 4; lanes 2, Ec-HUCA 1; lanes 3, Ec-HUCA 2; lanes 4, Ec-HUCA 3.
To investigate the self-transfer ability of the blaOXA-48 plasmids, conjugation experiments were performed using each of the four isolates as donors and a rifampin-resistant derivative of E. coli J53 as the recipient. Transconjugants were selected on eosin methylene blue (EMB) agar (Oxoid, Madrid, Spain) containing rifampin (100 mg/liter) plus ertapenem (0.5 mg/liter). At least four independent transconjugants were tested per conjugation with regard to plasmid content and antimicrobial susceptibility (see Table 2 for representative examples). As expected, each transconjugant carried the ca. 60-kb plasmid, either alone or together with other plasmid(s), and each was PCR positive for the blaOXA-48 gene. These self-transferable blaOXA-48 plasmids were assigned to incompatibility group IncL/M by PCR amplification using transconjugants carrying only this plasmid as the source of the template DNA and primers targeting the repA, traU, and parA genes specific for this group (7). Three out of four transconjugants tested from crosses involving Ec-HUCA 2 harbored the blaOXA-48 plasmid together with the larger blaCTX-M-15 plasmid, and they were resistant to cefotaxime and other extended-spectrum cephalosporins (Table 2). In contrast, all transconjugants analyzed from matings involving Kp-HUCA 4 (up to 20) were positive for the blaOXA-48 gene and plasmid only and showed a resistance pattern consistent with this (Table 2).
Multilocus sequence typing (MLST) (see mlst.warwick.ac.uk/mlst/dbs/Ecoli/) assigned Ec-HUCA 1, Ec-HUCA 2, and Ec-HUCA 3 to ST131, ST58, and ST83, respectively (Table 2). Pulsed-field gel electrophoresis (PFGE) performed with XbaI (14) revealed a different profile for each of the three E. coli isolates, and the XbaI profile of Kp-HUCA 4 was also different (Fig. 2). Note that the cluster of E. coli isolates carrying the blaOXA-48 gene was detected in our hospital during the national outbreak of K. pneumoniae producing the OXA-48 carbapenemase (4, 8). As the responsible gene appeared to be carried by the same conjugative IncL/M plasmid in the two species, it may have been transferred from K. pneumoniae into E. coli isolates which had been circulating at the time in the same hospital. Such a transfer may have independently occurred more than once, as the E. coli isolates were not clonally related according to their STs. The presence of blaOXA-48 in Ec-HUCA 1 (ST131) is particularly worrisome, considering the ability of the pandemic ST131 clone to acquire genes encoding extended-spectrum β-lactamases, especially blaCTX-M (15). In Spain, the only reported OXA-48-producing E. coli isolate was also ST131 (8). Note that the detection of carbapenem resistance was difficult, because the three E. coli isolates showed intermediate resistance to imipenem, and that treatment of the patients was complicated by additional resistances accompanying OXA-48 production in two out of the three isolates and by coinfection of the patients with other bacteria. Altogether, these facts highlight the risk of high antimicrobial pressure, in conjunction with long-term hospitalization, for the emergence of new resistant bacteria. Nevertheless, after the therapy indicated in Table 1, further cultures tested negative for OXA-48-producing Enterobacteriaceae, and the three patients were eventually discharged from the hospital.
FIG 2.
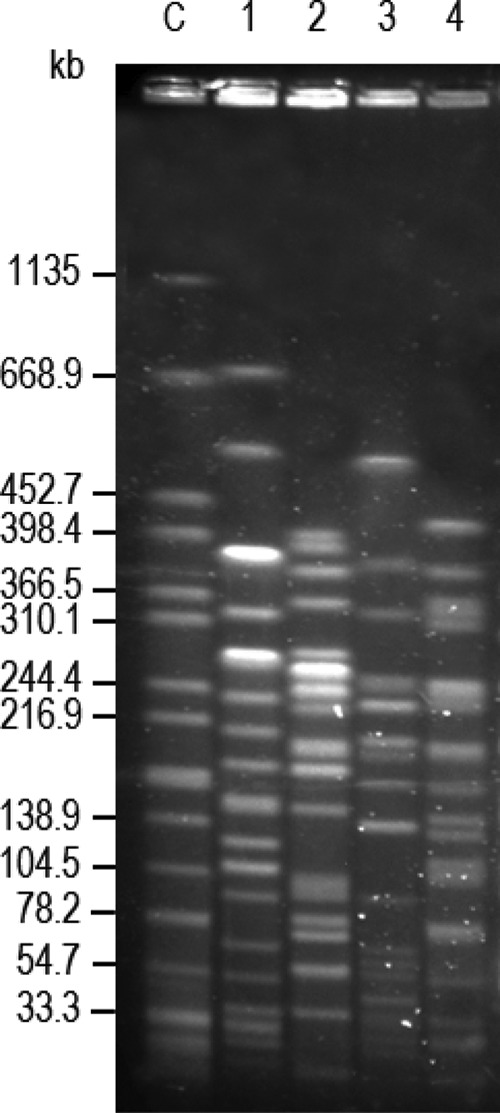
XbaI pulsed-field electrophoresis profiles of Klebsiella pneumoniae (Kp) and Escherichia coli (Ec) isolates producing OXA-48 carbapenemase. Lane C, XbaI-digested DNA of Salmonella enterica serovar Braenderup H9812 used as size standard; lane 1, Kp-HUCA 4; lane 2, Ec-HUCA 1; lane 3, Ec-HUCA 2; lane 4, Ec-HUCA 3.
ACKNOWLEDGMENTS
We are grateful to Irene Rodriguez, Hospital Universitario Ramón y Cajal, Madrid, for helpful advice.
This work has been supported by project FIS PI11-00808 (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Spain), cofunded by the European Regional Development Fund of the European Union: a Way to Making Europe. I.M. was the recipient of a predoctoral grant from the Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología (FICYT BP09-069).
Footnotes
Published ahead of print 20 June 2014
REFERENCES
- 1.Patel G, Bonomo RA. 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272. 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606. 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 4.Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P, European Network on Carbapenemases 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431. 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 5.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57:130–136. 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J. Clin. Microbiol. 51:680–683. 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562. 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oteo J, Saez D, Bautista V, Fernandez-Romero S, Hernandez-Molina JM, Perez-Vazquez M, Aracil B, Campos J, Spanish Collaborating Group for the Antibiotic Resistance Surveillance P 2013. Carbapenemase-producing Enterobacteriaceae in Spain in 2012. Antimicrob. Agents Chemother. 57:6344–6347. 10.1128/AAC.01513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing. NCCLS approved standard 23rd informational supplement, M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, Liebana E. 2005. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49:1319–1322. 10.1128/AAC.49.4.1319-1322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 12.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero A, Rodicio MR, Echeita MA, Mendoza MC. 2008. Salmonella enterica serotype Typhimurium carrying hybrid virulence-resistance plasmids (pUO-StVR): a new multidrug-resistant group endemic in Spain. Int. J. Med. Microbiol. 298:253–261. [DOI] [PubMed] [Google Scholar]
- 14.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 15.Qureshi ZA, Doi Y. 2014. Escherichia coli sequence type 131: epidemiology and challenges in treatment. Expert Rev. Anti Infect. Ther. 12:597-609. 10.1586/14787210.2014.899901. [DOI] [PubMed] [Google Scholar]


