LETTER
We read with particular attention the article “Isolation and Characterization of Porcine Epidemic Diarrhea Viruses Associated with the 2013 Disease Outbreak among Swine in the United States” by Chen and colleagues (1), published in January 2014 in the Journal of Clinical Microbiology. Chen et al. proposed that the full-length spike (S) gene or the S1 portion of it is a suitable phylogenetic marker to reflect evolutionary relationships between emergent U.S. and Chinese strains of porcine epidemic diarrhea viruses (PEDVs). This viewpoint proposed by the authors deserves more discussion (1).
To our knowledge, earlier work demonstrates that phylogenetic analysis based on the nucleotide sequences of the complete nucleocapsid (N) gene is very useful for differentiating closely related PEDV strains (2, 3). In addition to the N gene, open reading frame 3 (ORF3) is an important virulence gene that can be used as a valuable tool for studying the molecular epidemiology of PEDV (4, 5). In recent published studies, phylogenetic analyses of the N and ORF3 genes indicated that Chinese PEDVs circulating between 2010 and 2013 during PED outbreaks have originated mostly from earlier domestic PEDV strains (6, 7, 8). Based on these results, it is reasonable to reconsider the epidemiological status of U.S. PEDVs in outbreaks of sever acute PED.
To characterize more precisely the genetic relationships among PEDV strains isolated in areas of epidemicity of the United States and China, the complete nucleotide sequences of 72 N and 54 ORF3 genes from representative PEDV strains available in GenBank were independently used to construct respective phylogenetic trees.
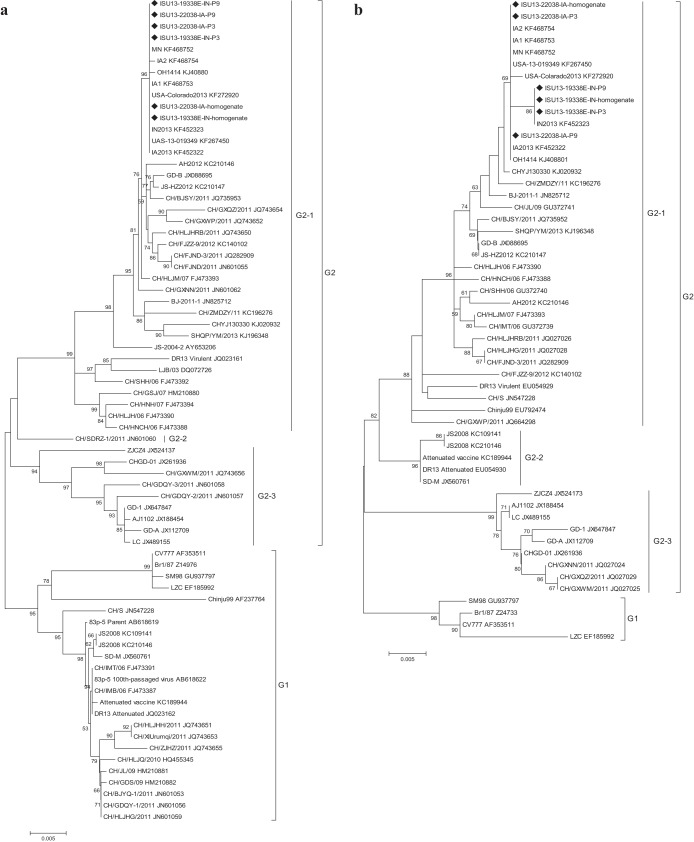
Figure 1 shows the phylogenetic relationship of PEDV strains isolated in the United States and China. All U.S. PEDV strains are highly related to each other and grouped tightly among themselves in a well-supported cluster across the two genes compared. The phylogenetic analysis of the N gene nucleotide sequences revealed 2 major groups (G1 and G2) (Fig. 1a). G2 was composed of three subgroups (G2-1, G2-2, and G2-3). Interestingly, all U.S. strains and some Chinese strains in 2011 to 2012 were placed in the same subcluster with one early Chinese strain (China [CH]/HLJM/07_FJ473393) within subgroup G2-1. We speculate that all U.S. PEDV strains probably have a Chinese origin and that the N genes originated from CH/HLJM/07, previously identified in northeast China before large-scale outbreaks of PEDV-induced diarrhea. The phylogenetic tree based on the ORF3 nucleotide sequence showed that the strains could also be divided into two groups (G1 and G2) (Fig. 1b). G2 was further divided into three subgroups (G2-1, G2-2, and G2-3). It is noteworthy that all U.S. PEDV strains and CHYJ130330_KJ020932 clustered closely, forming a separate subcluster that falls into G2-1. Of special note, the highly virulent CHYJ130330 strain of PEDV was collected previously from Guangdong (March 2013) Province in south China (9).
FIG 1.
Phylogenetic analyses of U.S. and Chinese PEDV strains based on N gene nucleotide sequences (a) and ORF3 gene nucleotide sequences (b). The trees were generated by the distance-based neighbor-joining method of the software MEGA5.0. Numbers at nodes represent the percentage of 1,000 bootstrap replicates (values <50 are not shown). The U.S. PEDV strains identified by Chen et al. (1) are indicated by diamonds. Scale bar, 0.005 inferred substitution per site.
In summary, the phylogenetic reconstructions may indicate that the N and ORF3 genes should be given significant focus in the evaluation of the genetic diversity of PEDV strains emerging in the United States. Additionally, the fact that all 10 U.S. PEDV strains arose from unpredictable recombinant events seems to have involved early and current Chinese PEDV variants.
Footnotes
For the author reply, see doi:10.1128/JCM.01747-14.
REFERENCES
- 1.Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. 2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52:234–243. 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Chen F, Yuan Y, Zeng X, Wei Z, Zhu L, Sun B, Xie Q, Cao Y, Xue C, Ma J, Bee Y. 2013. Sequence and phylogenetic analysis of nucleocapsid genes of porcine epidemic diarrhea virus (PEDV) strains in China. Arch. Virol. 158:1267–1273. 10.1007/s00705-012-1592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Liu X, Lang H, Wang Z, Shi D, Shi H, Zhang X, Feng L. 2013. Genetic variation of nucleocapsid genes of porcine epidemic diarrhea virus field strains in China. Arch. Virol. 158:1397–1401. 10.1007/s00705-013-1608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Wang C, Shi H, Qiu H, Liu S, Chen X, Zhang Z, Feng L. 2010. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 155:1471–1476. 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Zeng L, Yang J, Yu F, Ge J, Guo Q, Gao X, Song T. 2013. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010-2012. Viruses 5:2375–2383. 10.3390/v5102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Y, Tian X, Li W, Zhou Q, Wang D, Bi Y, Chen F, Song Y. 2012. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 9:195. 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, Qiao S, Yang Y, Su Y, Zhao P, Zhou E, Zhang G. 2014. Phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in central China based on the ORF3 gene and the main neutralization epitopes. Arch. Virol. 159:1057–1065. 10.1007/s00705-013-1929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang DQ, Ge FF, Ju HB, Wang J, Liu J, Ning K, Liu PH, Zhou JP, Sun QY. May 2014. Whole-genome analysis of porcine epidemic diarrhea virus (PEDV) from eastern China. Arch. Virol. 10.1007/s00705-014-2102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia A, Feng X, Liu Q, Zhou R, Wang G. 2014. Complete genome sequence of CHYJ130330, a highly virulent strain of porcine epidemic diarrhea virus in south China. Genome Announc. 2:e00165–14. 10.1128/genomeA.00165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]